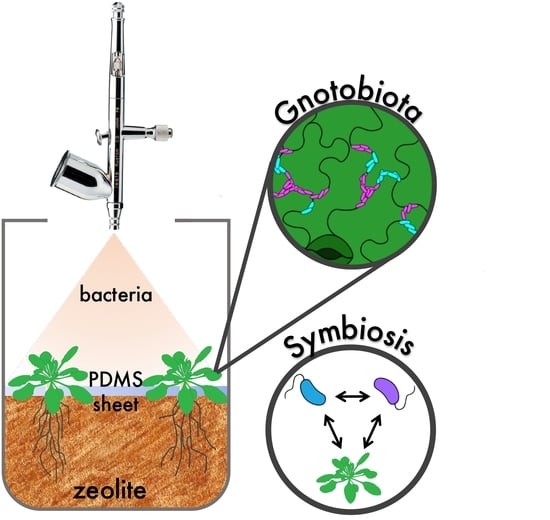
Litterbox—A gnotobiotic Zeolite-Clay System to Investigate Arabidopsis–Microbe Interactions
Abstract
1. Introduction
2. Materials and Methods
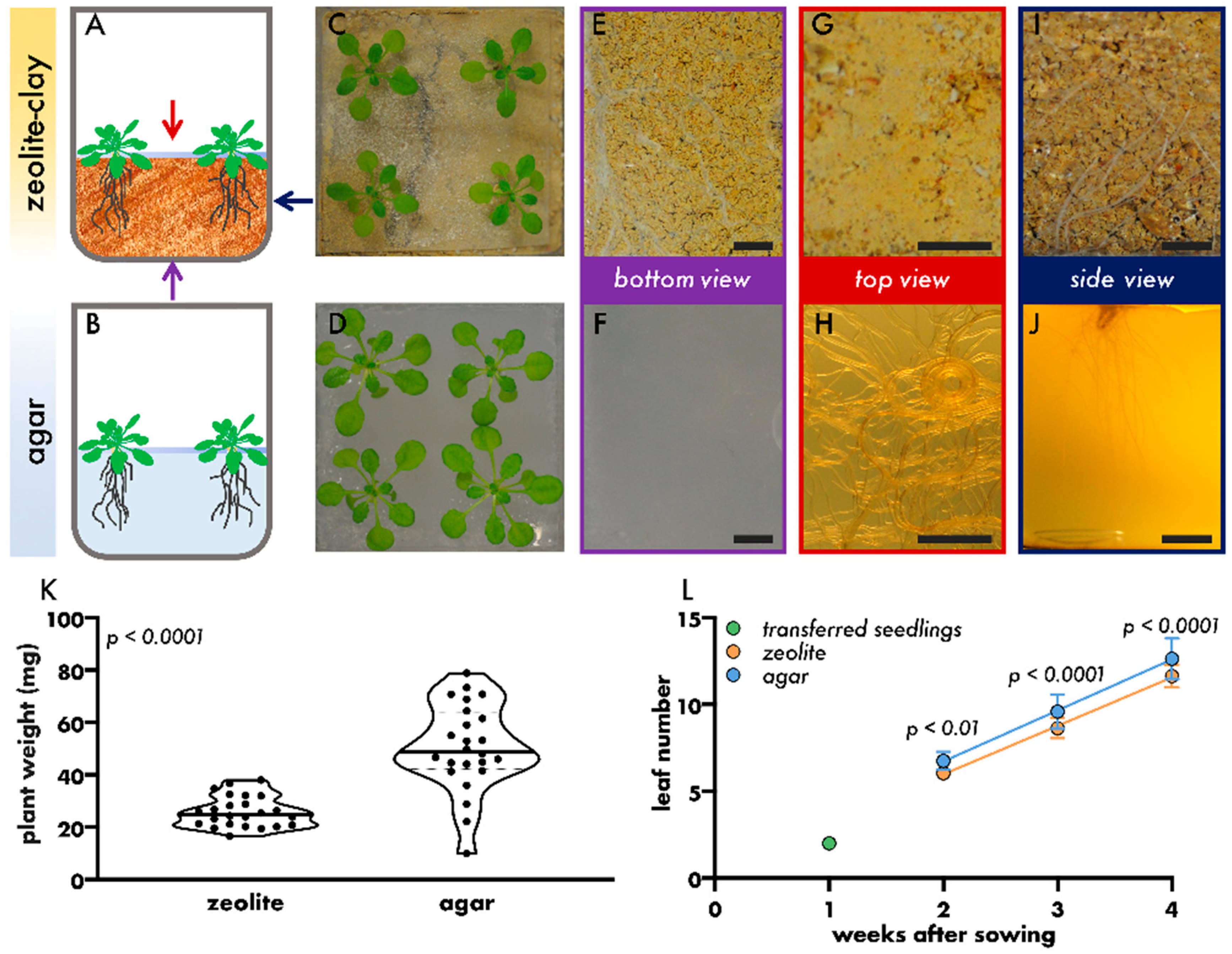
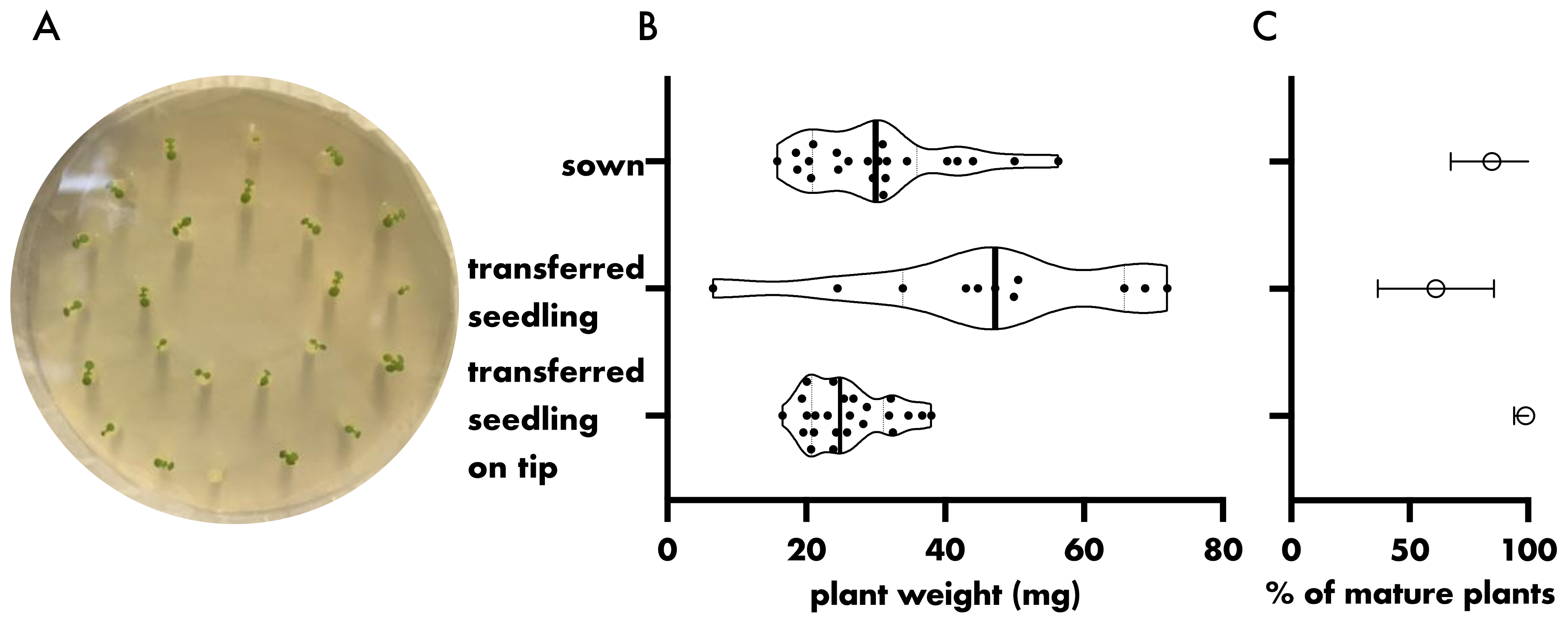
2.1. Plant Growth
2.2. Fabrication of PDMS Sheets
2.3. Plant Inoculation
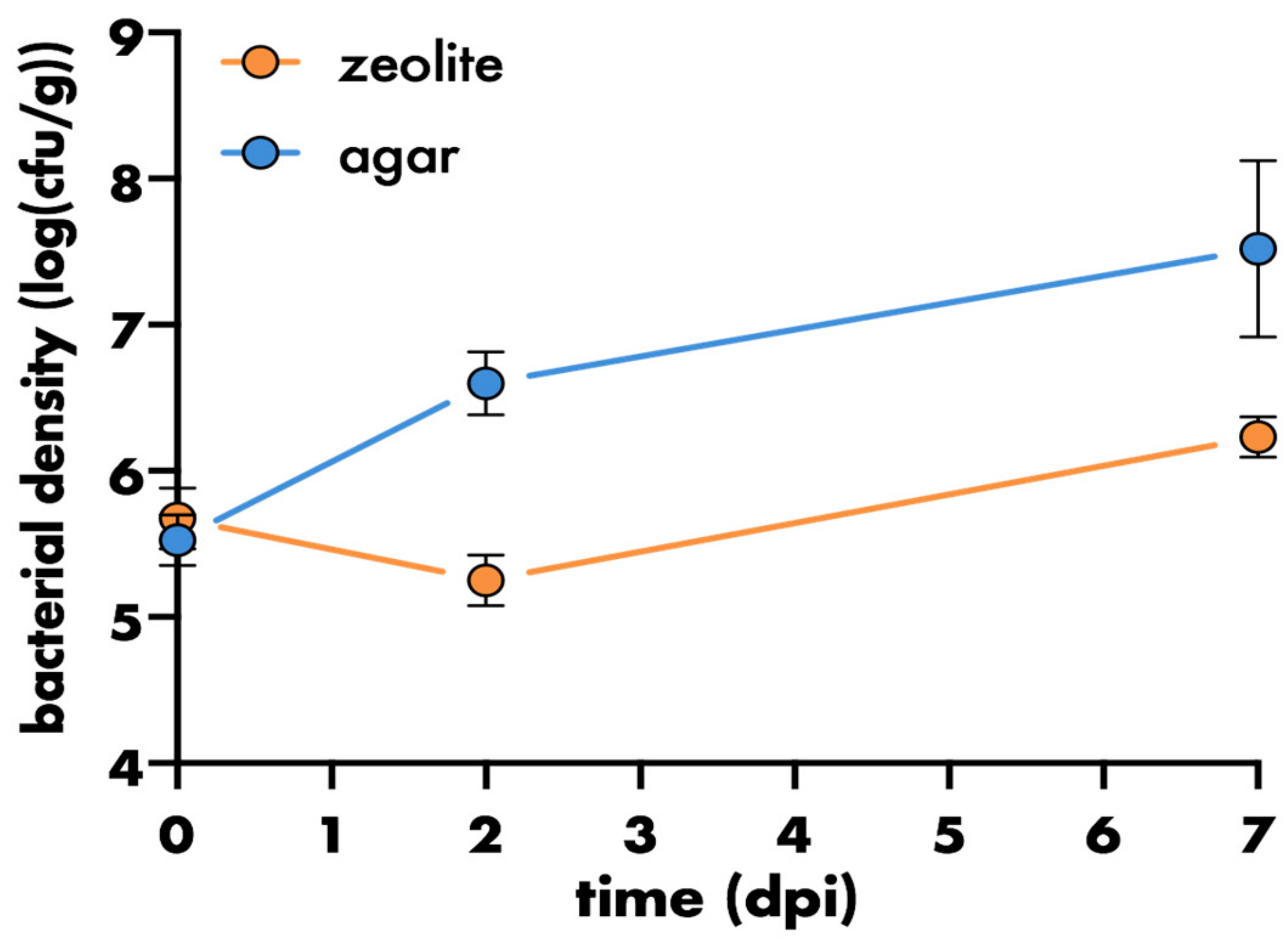
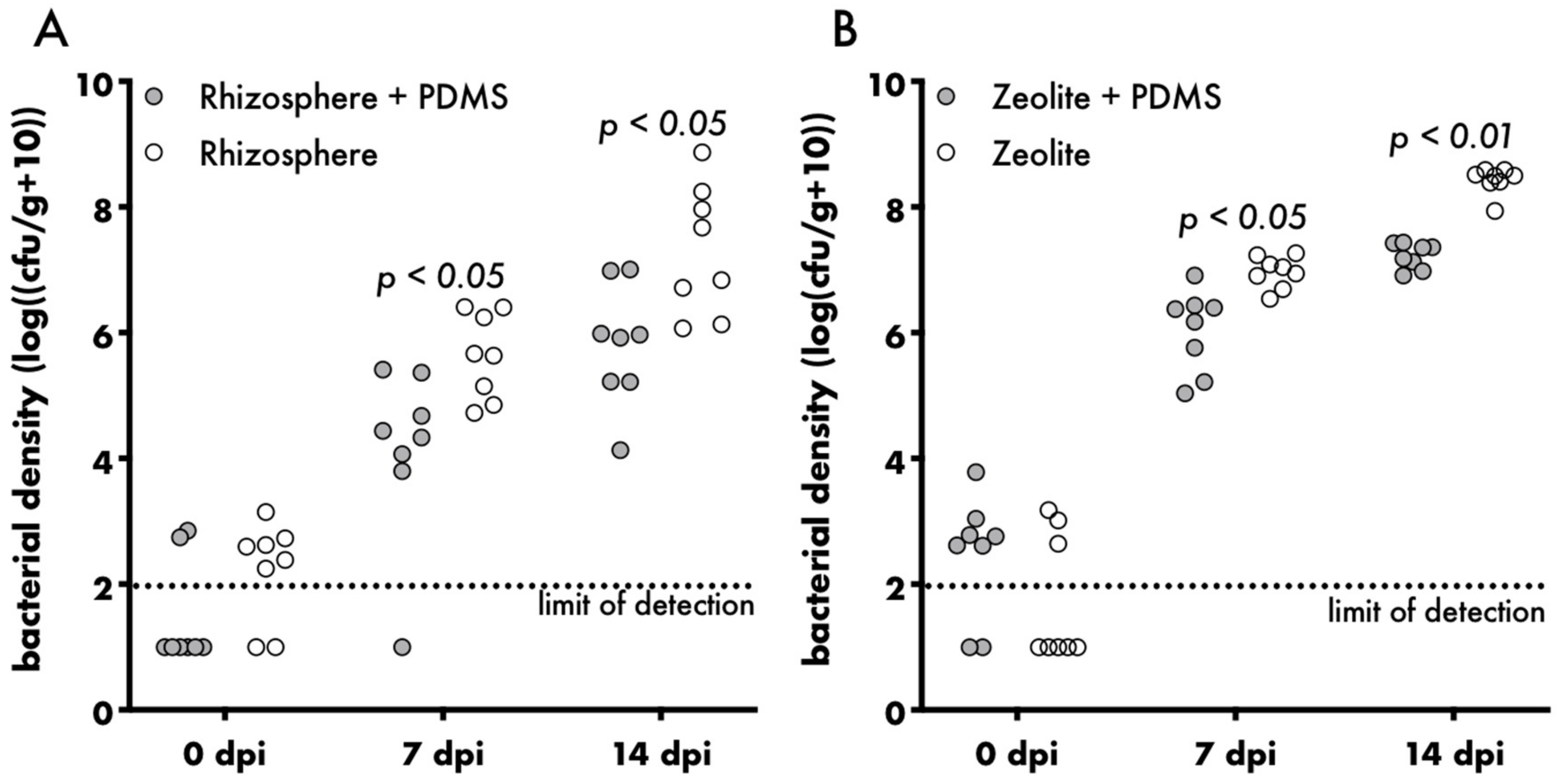
2.4. Enumeration of Bacteria Recovered from Varying Environments
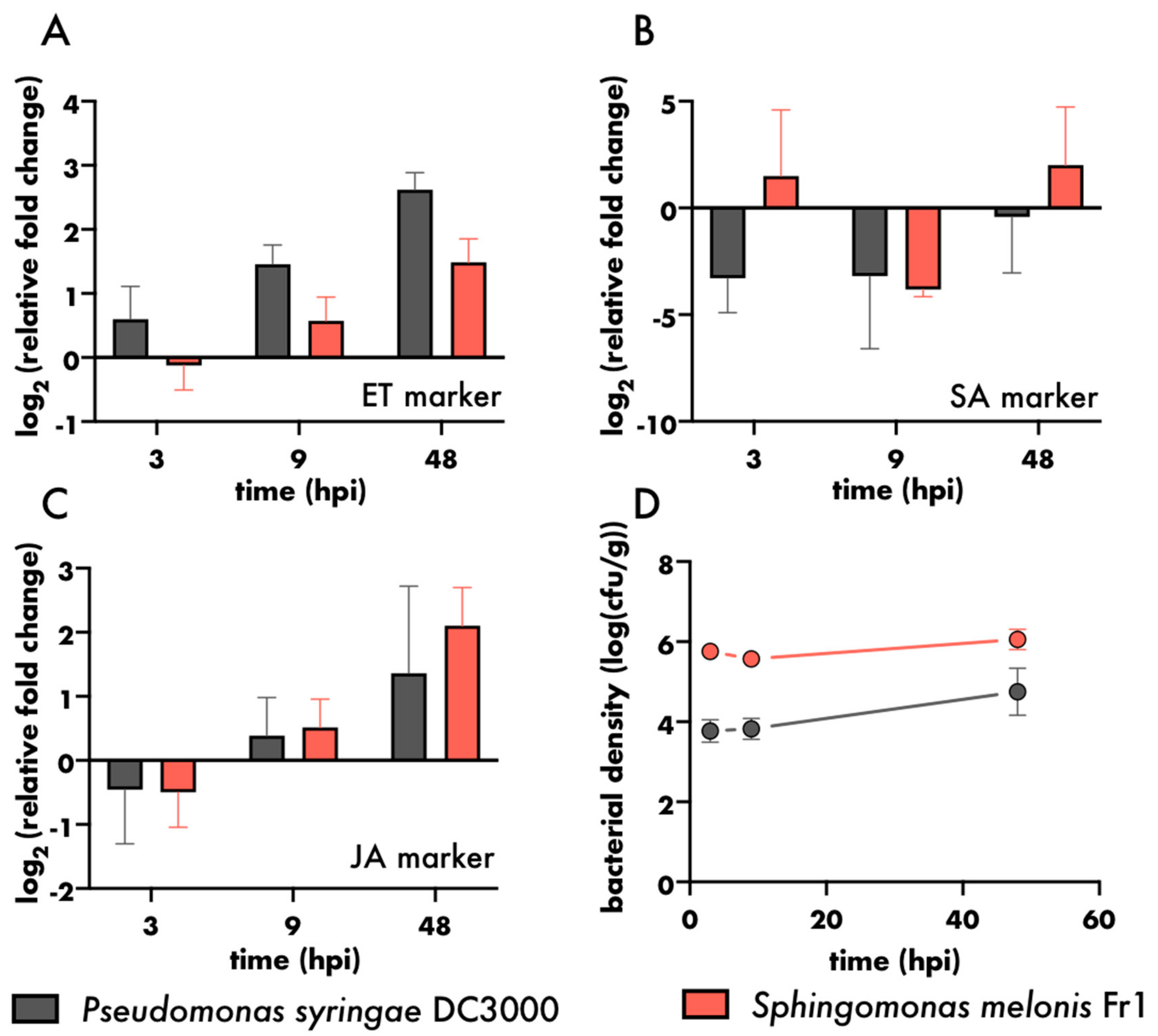
2.5. Gene Expression Analysis
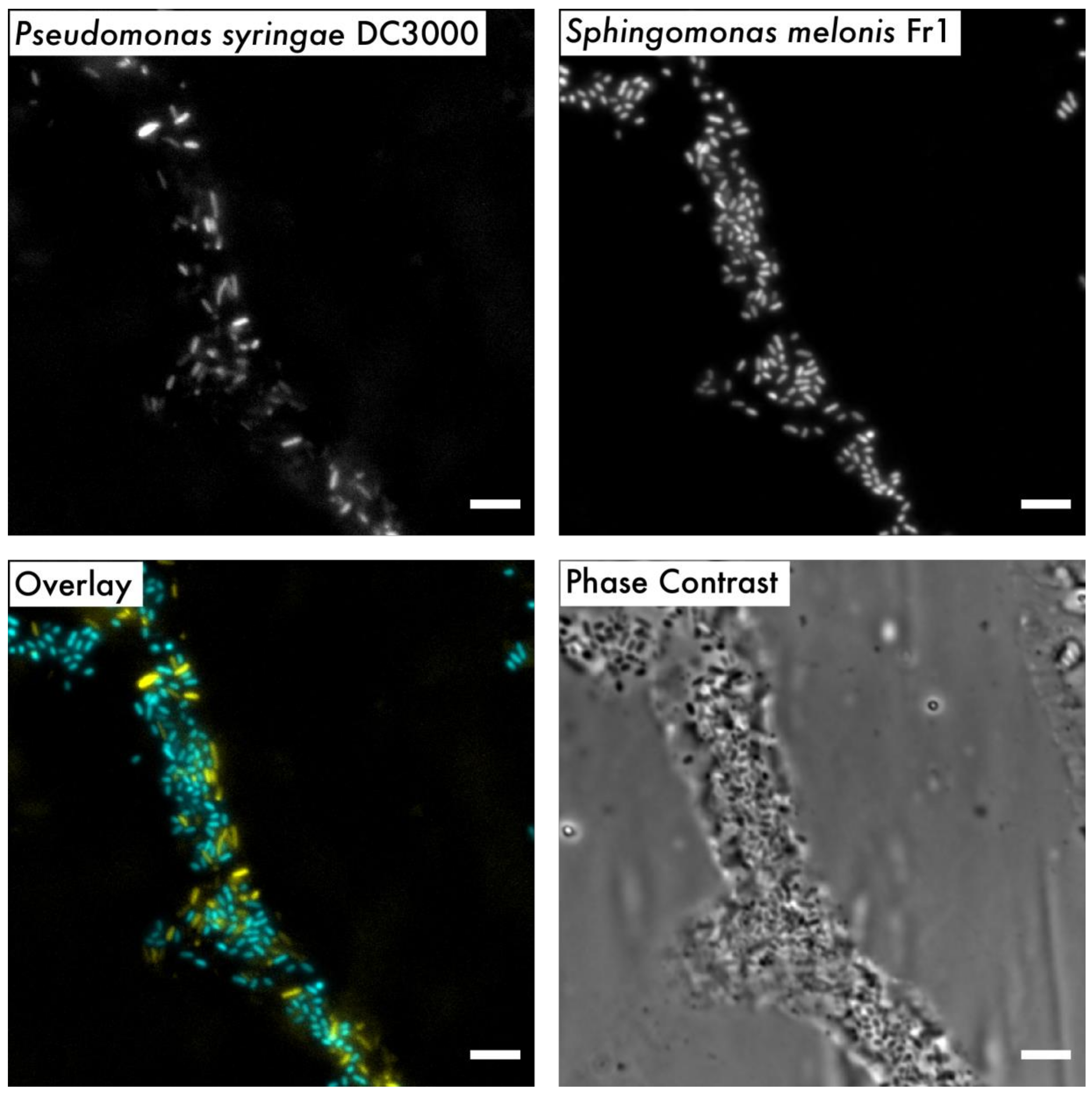
2.6. Microscopy and Image Processing
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Remus-Emsermann, M.N.P.; Schlechter, R.O. Phyllosphere microbiology: at the interface between microbial individuals and the plant host. New Phytol. 2018, 218, 1327–1333. [Google Scholar] [CrossRef] [PubMed]
- Schlechter, R.O.; Miebach, M.; Remus-Emsermann, M.N.P. Driving factors of epiphytic bacterial communities: A review. J. Adv. Res. 2019, 19, 57–65. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Ver Loren van Themaat, E.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cañizares, C.; Jorrín, B.; Poole, P.S.; Tkacz, A. Understanding the holobiont: the interdependence of plants and their microbiome. Curr. Opin. Microbiol. 2017, 38, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef]
- Chen, M.; Arato, M.; Borghi, L.; Nouri, E.; Reinhardt, D. Beneficial services of arbuscular mycorrhizal fungi - from ecology to application. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.; Zhu, H. Genetic and molecular mechanisms underlying symbiotic specificity in Legume-Rhizobium interactions. Front. Plant Sci. 2018, 9, 313. [Google Scholar] [CrossRef]
- Innerebner, G.; Knief, C.; Vorholt, J.A. Protection of Arabidobsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 2011, 77, 3202–3210. [Google Scholar] [CrossRef]
- Ritpitakphong, U.; Falquet, L.; Vimoltust, A.; Berger, A.; Métraux, J.-P.; L’Haridon, F. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 2016, 210, 1033–1043. [Google Scholar] [CrossRef]
- Berg, M.; Koskella, B. Nutrient- and dose-dependent microbiome-mediated protection against a plant pathogen. Curr. Biol. 2018, 28, 2487–2492. [Google Scholar] [CrossRef] [PubMed]
- Spaepen, S.; Vanderleyden, J.; Okon, Y. Chapter 7 plant growth-promoting actions of Rhizobacteria. Adv. Bot. Res. 2009, 51, 283–320, Academic Press. [Google Scholar]
- Lau, J.A.; Lennon, J.T. Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Natl. Acad. Sci. USA 2012, 109, 14058–14062. [Google Scholar] [CrossRef] [PubMed]
- Zengerer, V.; Schmid, M.; Bieri, M.; Müller, D.C.; Remus-Emsermann, M.N.P.; Ahrens, C.H.; Pelludat, C. Pseudomonas orientalis F9: A potent antagonist against phytopathogens with phytotoxic effect in the apple flower. Front. Microbiol. 2018, 9, 145. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Hart, M.; Liu, H. Paving the way from the lab to the field: using synthetic microbial consortia to produce high-quality crops. Front. Plant Sci. 2018, 9, 1467. [Google Scholar] [CrossRef] [PubMed]
- Sergaki, C.; Lagunas, B.; Lidbury, I.; Gifford, M.L.; Schäfer, P. Challenges and approaches in microbiome research: from fundamental to applied. Front. Plant Sci. 2018, 9, 1205. [Google Scholar] [CrossRef]
- Busby, P.E.; Soman, C.; Wagner, M.R.; Friesen, M.L.; Kremer, J.; Bennett, A.; Morsy, M.; Eisen, J.A.; Leach, J.E.; Dangl, J.L. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017, 15, e2001793. [Google Scholar] [CrossRef]
- Delmotte, N.; Knief, C.; Chaffron, S.; Innerebner, G.; Roschitzki, B.; Schlapbach, R.; von Mering, C.; Vorholt, J.A. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. USA 2009, 106, 16428–16433. [Google Scholar] [CrossRef]
- Knief, C.; Delmotte, N.; Chaffron, S.; Stark, M.; Innerebner, G.; Wassmann, R.; von Mering, C.; Vorholt, J.A. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012, 6, 1378–1390. [Google Scholar] [CrossRef]
- Levy, A.; Salas Gonzalez, I.; Mittelviefhaus, M.; Clingenpeel, S.; Herrera Paredes, S.; Miao, J.; Wang, K.; Devescovi, G.; Stillman, K.; Monteiro, F.; et al. Genomic features of bacterial adaptation to plants. Nat. Genet. 2018, 50, 138–150. [Google Scholar] [CrossRef]
- Turner, T.R.; Ramakrishnan, K.; Walshaw, J.; Heavens, D.; Alston, M.; Swarbreck, D.; Osbourn, A.; Grant, A.; Poole, P.S. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 2013, 7, 2248–2258. [Google Scholar] [CrossRef] [PubMed]
- Makiola, A.; Dickie, I.A.; Holdaway, R.J.; Wood, J.R.; Orwin, K.H.; Glare, T.R. Land use is a determinant of plant pathogen alpha- but not beta-diversity. Mol. Ecol. 2019, 28, 3786–3798. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, J.A.; Vogel, C.; Carlström, C.I.; Müller, D.B. Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 2017, 22, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Qin, Y.; Bai, Y. Reductionist synthetic community approaches in root microbiome research. Curr. Opin. Microbiol. 2019, 49, 97–102. [Google Scholar] [CrossRef]
- Carlström, C.I.; Field, C.M.; Bortfeld-Miller, M.; Müller, B.; Sunagawa, S.; Vorholt, J.A. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nat. Ecol. Evol. 2019, 3, 1445–1454. [Google Scholar] [CrossRef]
- Stockwell, V.O.; Johnson, K.B.; Sugar, D.; Loper, J.E. Control of fire blight by Pseudomonas fluorescens A506 and Pantoea vagans C9-1 applied as single strains and mixed inocula. Phytopathology 2010, 100, 1330–1339. [Google Scholar] [CrossRef]
- Stockwell, V.O.; Johnson, K.B.; Sugar, D.; Loper, J.E. Mechanistically compatible mixtures of bacterial antagonists improve biological control of fire blight of pear. Phytopathology 2011, 101, 113–123. [Google Scholar] [CrossRef]
- Berens, M.L.; Wolinska, K.W.; Spaepen, S.; Ziegler, J.; Nobori, T.; Nair, A.; Krüler, V.; Winkelmüller, T.M.; Wang, Y.; Mine, A.; et al. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. USA 2019, 116, 2364–2373. [Google Scholar] [CrossRef]
- Finkel, O.M.; Salas-González, I.; Castrillo, G.; Spaepen, S.; Law, T.F.; Teixeira, P.J.P.L.; Jones, C.D.; Dangl, J.L. The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biol. 2019, 17, e3000534. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Andrew, D.R.; Fitak, R.R.; Munguia-Vega, A.; Racolta, A.; Martinson, V.G.; Dontsova, K. Abiotic factors shape microbial diversity in Sonoran Desert soils. Appl. Environ. Microbiol. 2012, 78, 7527–7537. [Google Scholar] [CrossRef]
- Tao, Y.; Xie, Z.; Chen, W.; Glazebrook, J.; Chang, H.-S.; Han, B.; Zhu, T.; Zou, G.; Katagiri, F. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 2003, 15, 317–330. [Google Scholar] [CrossRef]
- Vogel, C.; Bodenhausen, N.; Gruissem, W.; Vorholt, J.A. The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. New Phytol. 2016, 212, 192–207. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Innerebner, G.; Zingg, J.; Guder, J.; Vorholt, J.A. Forward genetic in planta screen for identification of plant-protective traits of Sphingomonas sp. strain Fr1 against Pseudomonas syringae DC3000. Appl. Environ. Microbiol. 2012, 78, 5529–5535. [Google Scholar] [CrossRef] [PubMed]
- Henry, A.; Doucette, W.; Norton, J.; Jones, S.; Chard, J.; Bugbee, B. An axenic plant culture system for optimal growth in long-term studies. J. Environ. Qual. 2006, 35, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Gunning, T.; Cahill, D.M. A soil-free plant growth system to facilitate analysis of plant pathogen interactions in roots. J. Phytopathol. 2009, 157, 497–501. [Google Scholar] [CrossRef]
- Koch, K.; Hartmann, K.D.; Schreiber, L.; Barthlott, W.; Neinhuis, C. Influences of air humidity during the cultivation of plants on wax chemical composition, morphology and leaf surface wettability. Environ. Exp. Bot. 2006, 56, 1–9. [Google Scholar] [CrossRef]
- Kosma, D.K.; Bourdenx, B.; Bernard, A.; Parsons, E.P.; Lü, S.; Joubès, J.; Jenks, M.A. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 2009, 151, 1918–1929. [Google Scholar] [CrossRef] [PubMed]
- Reisberg, E.E.; Hildebrandt, U.; Riederer, M.; Hentschel, U. Distinct phyllosphere bacterial communities on Arabidopsis wax mutant leaves. PLoS ONE 2013, 8, e78613. [Google Scholar] [CrossRef]
- Beattie, G.A. Water relations in the interaction of foliar bacterial pathogens with plants. Annu. Rev. Phytopathol. 2011, 49, 533–555. [Google Scholar] [CrossRef] [PubMed]
- Shaw, L.J.; Beaton, Y.; Glover, L.A.; Killham, K.; Meharg, A.A. Re-inoculation of autoclaved soil as a non-sterile treatment for xenobiotic sorption and biodegradation studies. Appl. Soil Ecol. 1999, 11, 217–226. [Google Scholar] [CrossRef]
- Bank, T.L.; Kukkadapu, R.K.; Madden, A.S.; Ginder-Vogel, M.A.; Baldwin, M.E.; Jardine, P.M. Effects of gamma-sterilization on the physico-chemical properties of natural sediments. Chem. Geol. 2008, 251, 1–7. [Google Scholar] [CrossRef]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Glavina del Rio, T.; Jones, C.D.; Tringe, S.G.; et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.-X.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X.; et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef]
- Adams, C.; Jacobson, A.; Bugbee, B. Ceramic aggregate sorption and desorption chemistry: Implications for use as a component of soilless media. J. Plant Nutr. 2014, 37, 1345–1357. [Google Scholar] [CrossRef]
- Ming, D.W.; Allen, E.R. Use of natural zeolites in agronomy, horticulture and environmental soil remediation. Rev. Mineral. Geochem. 2001, 45, 619–654. [Google Scholar] [CrossRef]
- Iskander, A.L.; Khald, E.M.; Sheta, A.S. Zinc and manganese sorption behavior by natural zeolite and bentonite. Sci. Ann. Univ. Agric. Sci. Vet. Med. 2011, 56, 43–48. [Google Scholar] [CrossRef]
- ZeoponiX Inc. Research and Application. Available online: http://zeoponix.com/?page_id=94 (accessed on 8 January 2020).
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef]
- Peyraud, R.; Kiefer, P.; Christen, P.; Massou, S.; Portais, J.-C.; Vorholt, J.A. Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. Proc. Natl. Acad. Sci. USA 2009, 106, 4846–4851. [Google Scholar] [CrossRef]
- Jameson, P.E.; Morris, R.O. Zeatin-like cytokinins in yeast: detection by immunological methods. J. Plant Physiol. 1989, 135, 385–390. [Google Scholar] [CrossRef]
- Schlechter, R.O.; Jun, H.; Bernach, M.; Oso, S.; Boyd, E.; Muñoz-Lintz, D.A.; Dobson, R.C.J.; Remus, D.M.; Remus-Emsermann, M.N.P. Chromatic Bacteria - a broad host-range plasmid and chromosomal insertion toolbox for fluorescent protein expression in bacteria. Front. Microbiol. 2018, 9, 3052. [Google Scholar] [CrossRef] [PubMed]
- Buonaurio, R.; Stravato, V.M.; Kosako, Y.; Fujiwara, N.; Naka, T.; Kobayashi, K.; Cappelli, C.; Yabuuchi, E. Sphingomonas melonis sp. nov., a novel pathogen that causes brown spots on yellow Spanish melon fruits. Int. J. Syst. Evol. Microbiol. 2002, 52, 2081–2087. [Google Scholar] [PubMed]
- Cuppels, D.A. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 1986, 51, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Huggett, J.F.; Sanchez, E. Good Practice Guide for the Application of Quantitative PCR (qPCR); LGC: Teddington, UK, 2013. [Google Scholar]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef]
- Kim, Y.; Tsuda, K.; Igarashi, D.; Hillmer, R.A.; Sakakibara, H.; Myers, C.L.; Katagiri, F. Mechanisms underlying robustness and tunability in a plant immune signaling network. Cell Host Microbe 2014, 15, 84–94. [Google Scholar] [CrossRef]
- Remus-Emsermann, M.N.P.; Lücker, S.; Müller, D.B.; Potthoff, E.; Daims, H.; Vorholt, J.A. Spatial distribution analyses of natural phyllosphere-colonizing bacteria on Arabidopsis thaliana revealed by fluorescence in situ hybridization. Environ. Microbiol. 2014, 16, 2329–2340. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Roué, J.; Chauvet, H.; Brunel-Michac, N.; Bizet, F.; Moulia, B.; Badel, E.; Legué, V. Root cap size and shape influence responses to the physical strength of the growth medium in Arabidopsis thaliana primary roots. J. Exp. Bot. 2019. [Google Scholar] [CrossRef]
- Eapen, D.; Barroso, M.L.; Ponce, G.; Campos, M.E.; Cassab, G.I. Hydrotropism: Root growth responses to water. Trends Plant Sci. 2005, 10, 44–50. [Google Scholar] [CrossRef]
- Rellán-Álvarez, R.; Lobet, G.; Lindner, H.; Pradier, P.-L.; Sebastian, J.; Yee, M.-C.; Geng, Y.; Trontin, C.; LaRue, T.; Schrager-Lavelle, A.; et al. GLO-Roots: An imaging platform enabling multidimensional characterization of soil-grown root systems. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Remus-Emsermann, M.N.P.; Tecon, R.; Kowalchuk, G.A.; Leveau, J.H.J. Variation in local carrying capacity and the individual fate of bacterial colonizers in the phyllosphere. ISME J. 2012, 6, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Remus-Emsermann, M.N.P.; Pelludat, C.; Gisler, P. Conjugation dynamics of self-transmissible and mobilisable plasmids into E. coli O157: H7 on Arabidopsis thaliana rosettes. bioRxiv 2018. [Google Scholar] [CrossRef]
- Kniskern, J.M.; Traw, M.B.; Bergelson, J. Salicylic acid and jasmonic acid signaling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana. Mol. Plant. Microbe. Interact. 2007, 20, 1512–1522. [Google Scholar] [CrossRef] [PubMed]
- Reisberg, E.E.; Hildebrandt, U.; Riederer, M.; Hentschel, U. Phyllosphere bacterial communities of trichome-bearing and trichomeless Arabidopsis thaliana leaves. Anton. Leeuw. Int. J. G 2012, 101, 551–560. [Google Scholar] [CrossRef]
- Burch, A.Y.; Do, P.T.; Sbodio, A.; Suslow, T.V.; Lindow, S.E. High-level culturability of epiphytic bacteria and frequency of biosurfactant producers on leaves. Appl. Environ. Microbiol. 2016, 82, 5997–6009. [Google Scholar] [CrossRef]
- Gekenidis, M.-T.; Gossin, D.; Schmelcher, M.; Schöner, U.; Remus-Emsermann, M.N.P.; Drissner, D. Dynamics of culturable mesophilic bacterial communities of three fresh herbs and their production environment. J. Appl. Microbiol. 2017, 123, 916–932. [Google Scholar] [CrossRef]
- Rastogi, G.; Sbodio, A.; Tech, J.J.; Suslow, T.V.; Coaker, G.L.; Leveau, J.H.J. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012, 6, 1812–1822. [Google Scholar] [CrossRef]
- Kremer, J.M.; Paasch, B.C.; Rhodes, D.; Thireault, C.; Froehlich, J.E.; Schulze-Lefert, P.; Tiedje, J.M.; He, S.Y. FlowPot axenic plant growth system for microbiota research. bioRxiv 2018, 254953. [Google Scholar] [CrossRef]
- Van der Ent, S.; Van Hulten, M.; Pozo, M.J.; Czechowski, T.; Udvardi, M.K.; Pieterse, C.M.J.; Ton, J. Priming of plant innate immunity by rhizobacteria and β-aminobutyric acid: differences and similarities in regulation. New Phytol. 2009, 183, 419–431. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Leeman, M.; Van Oorschot, M.M.P.; Van der Sluis, I.; Schippers, B.; Bakker, P. Dose-response relationships in biological control of Fusarium wilt of radish by Pseudomonas spp. Phytopathology 1995, 85, 1075–1080. [Google Scholar] [CrossRef]
- Washington, E.J.; Mukhtar, M.S.; Finkel, O.M.; Wan, L.; Banfield, M.J.; Kieber, J.J.; Dangl, J.L. Pseudomonas syringae type III effector HopAF1 suppresses plant immunity by targeting methionine recycling to block ethylene induction. Proc. Natl. Acad. Sci. USA 2016, 113, E3577–E3586. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.M.; Leveau, J.H.J. Microbiology of the phyllosphere: a playground for testing ecological concepts. Oecologia 2012, 168, 621–629. [Google Scholar] [CrossRef]
- Leveau, J.H.; Lindow, S.E. Appetite of an epiphyte: Quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. USA 2001, 98, 3446–3453. [Google Scholar] [CrossRef]
- Monier, J.-M.; Lindow, S.E. Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl. Environ. Microbiol. 2004, 70, 346–355. [Google Scholar] [CrossRef]
- Sandhu, A.; Halverson, L.J.; Beattie, G.A. Bacterial degradation of airborne phenol in the phyllosphere. Environ. Microbiol. 2007, 9, 383–392. [Google Scholar] [CrossRef]
| Bacterial Strain | Reference | Fluorescence |
|---|---|---|
| Sphingomonas melonis Fr1 | [54] | n.a. |
| Pseudomonas syringae DC3000 | [55] | n.a. |
| Sphingomonas melonis Fr1::mTurquoise2 | This work | mTurquoise2 (cyan) |
| Pseudomonas syringae DC3000::mScarlet-I | This work | mScarlet-I (red) |
| Template | Reference | 5′ Primer | 3′ Primer |
|---|---|---|---|
| At2g41230 (ET marker) | This study | cgaaccgtccgtacatacataa | ttgcacgaaactaaaactaaaagc |
| PR1 (SA marker) | This study | gatgtgccaaagtgaggtgtaa | ttcacataattcccacgagga |
| At3g50280 (JA marker) | This study | ccttcgctggtcgtcttaac | cagagccatcaggtcgaaga |
| At2g28390 (reference gene) | This study | ggattttcagctactcttcaagcta | tcctgccttgactaagttgaca |
| At4g26410 (reference gene) | This study | cgtccacaaagctgaatgtg | cgaagtcatggaagccactt |
| At1g13320 (reference gene) | This study | tgctcagatgagggagagtg | caccagctgaaagtcgctta |
| EF-1α (reference gene) | This study | cttggtgtcaagcagatgattt | cgtacctagccttggagtatttg |
| Actin2 (reference gene) | [57] | cttgcaccaagcagcatgaa | ccgatccagacactgtacttcctt |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miebach, M.; Schlechter, R.O.; Clemens, J.; Jameson, P.E.; Remus-Emsermann, M.N.P. Litterbox—A gnotobiotic Zeolite-Clay System to Investigate Arabidopsis–Microbe Interactions. Microorganisms 2020, 8, 464. https://doi.org/10.3390/microorganisms8040464
Miebach M, Schlechter RO, Clemens J, Jameson PE, Remus-Emsermann MNP. Litterbox—A gnotobiotic Zeolite-Clay System to Investigate Arabidopsis–Microbe Interactions. Microorganisms. 2020; 8(4):464. https://doi.org/10.3390/microorganisms8040464
Chicago/Turabian StyleMiebach, Moritz, Rudolf O. Schlechter, John Clemens, Paula E. Jameson, and Mitja N.P. Remus-Emsermann. 2020. "Litterbox—A gnotobiotic Zeolite-Clay System to Investigate Arabidopsis–Microbe Interactions" Microorganisms 8, no. 4: 464. https://doi.org/10.3390/microorganisms8040464
APA StyleMiebach, M., Schlechter, R. O., Clemens, J., Jameson, P. E., & Remus-Emsermann, M. N. P. (2020). Litterbox—A gnotobiotic Zeolite-Clay System to Investigate Arabidopsis–Microbe Interactions. Microorganisms, 8(4), 464. https://doi.org/10.3390/microorganisms8040464