Cysteine Residues in Helicobacter pylori Adhesin HopQ Are Required for CEACAM–HopQ Interaction and Subsequent CagA Translocation
Abstract
1. Introduction
2. Materials and Methods
2.1. H. pylori Strains and Generation of Mutant Strains
2.2. Cells
2.3. Infections and Western Blot for CagA Translocation
2.4. Pull Down Assays
2.5. Binding Assay (Flow Cytometry)
2.6. Immunofluorescence
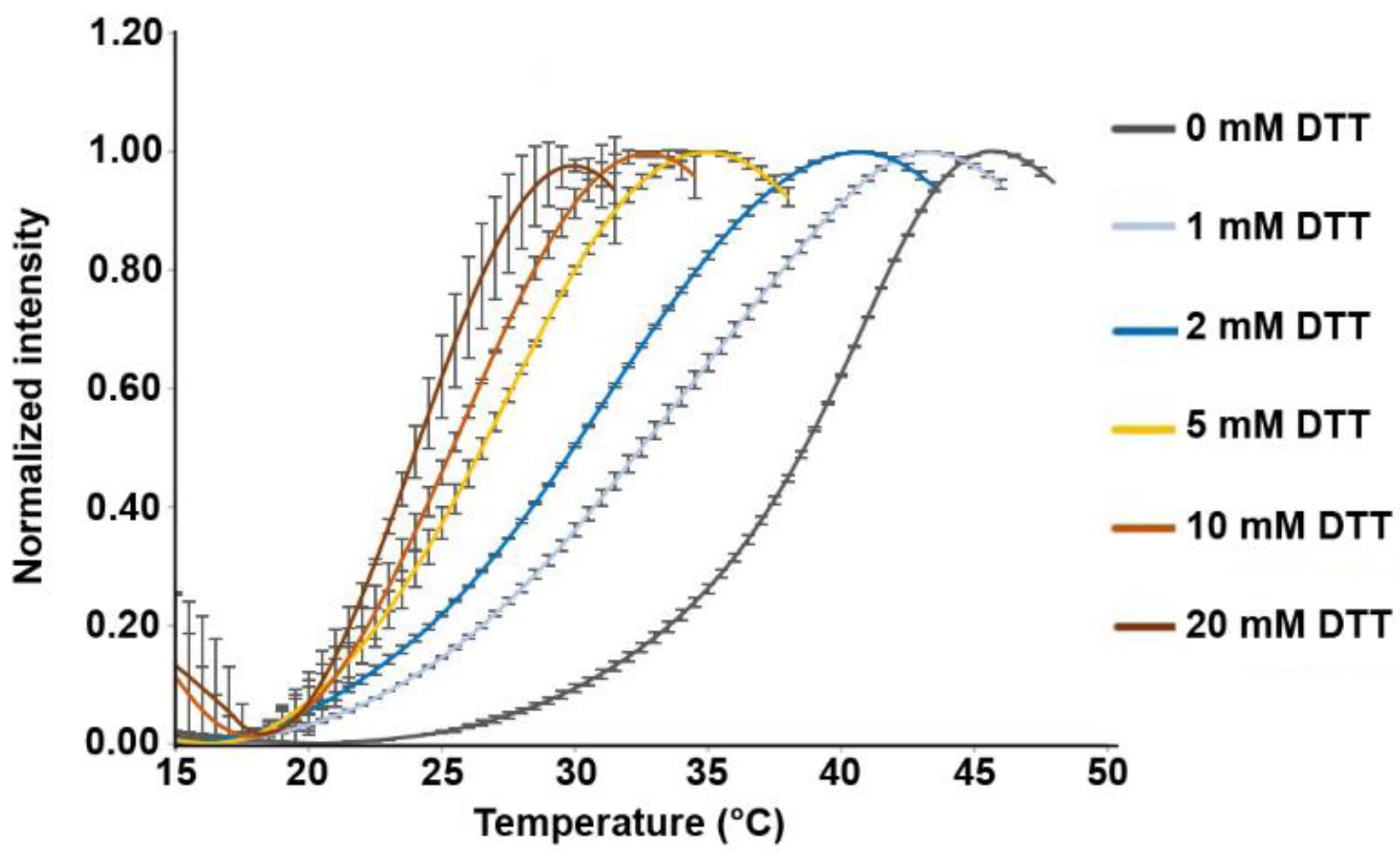
2.7. Thermal Denaturation Assay
2.8. CEACAM-GFP Binding Assay
2.9. H. pylori Outer Membrane Preparation
2.10. List of Antibodies Used
- Mouse anti-pTyr (1:300, PY99, Santa Cruz Biotechnology, Dallas, TX, USA);
- Rabbit anti-CagA serum (1:3000);
- Rabbit anti-GAPDH (1:1000, loading control, 14C10, Cell Signaling Technology, Danvers, MA, USA);
- Rabbit anti-alpha tubulin (1:1000, loading control, polyclonal, Cell Signaling Technology);
- SAB3 antibody (anti-CEACAM1/3/5 from B.B. Singer), 2.5 µg/mL;
- Mouse anti-HopQ serum (1:3000);
- Mouse anti-HpaA serum (1:3000);
- Mouse anti-gGT serum (1:3000).
3. Results
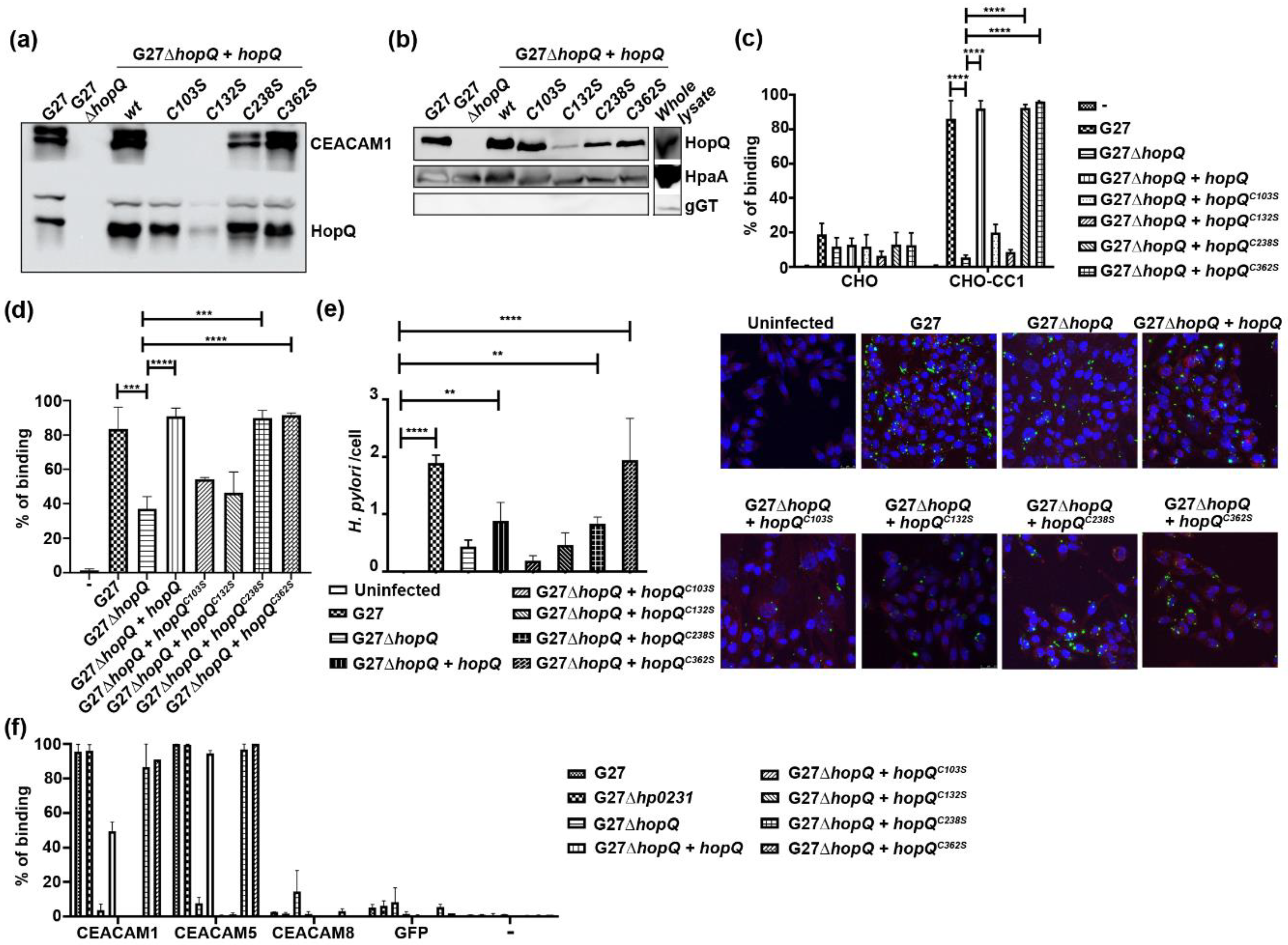
3.1. Disulfide Bridges in the HopQ Adhesin Domain are Essential for CEACAM1 Binding
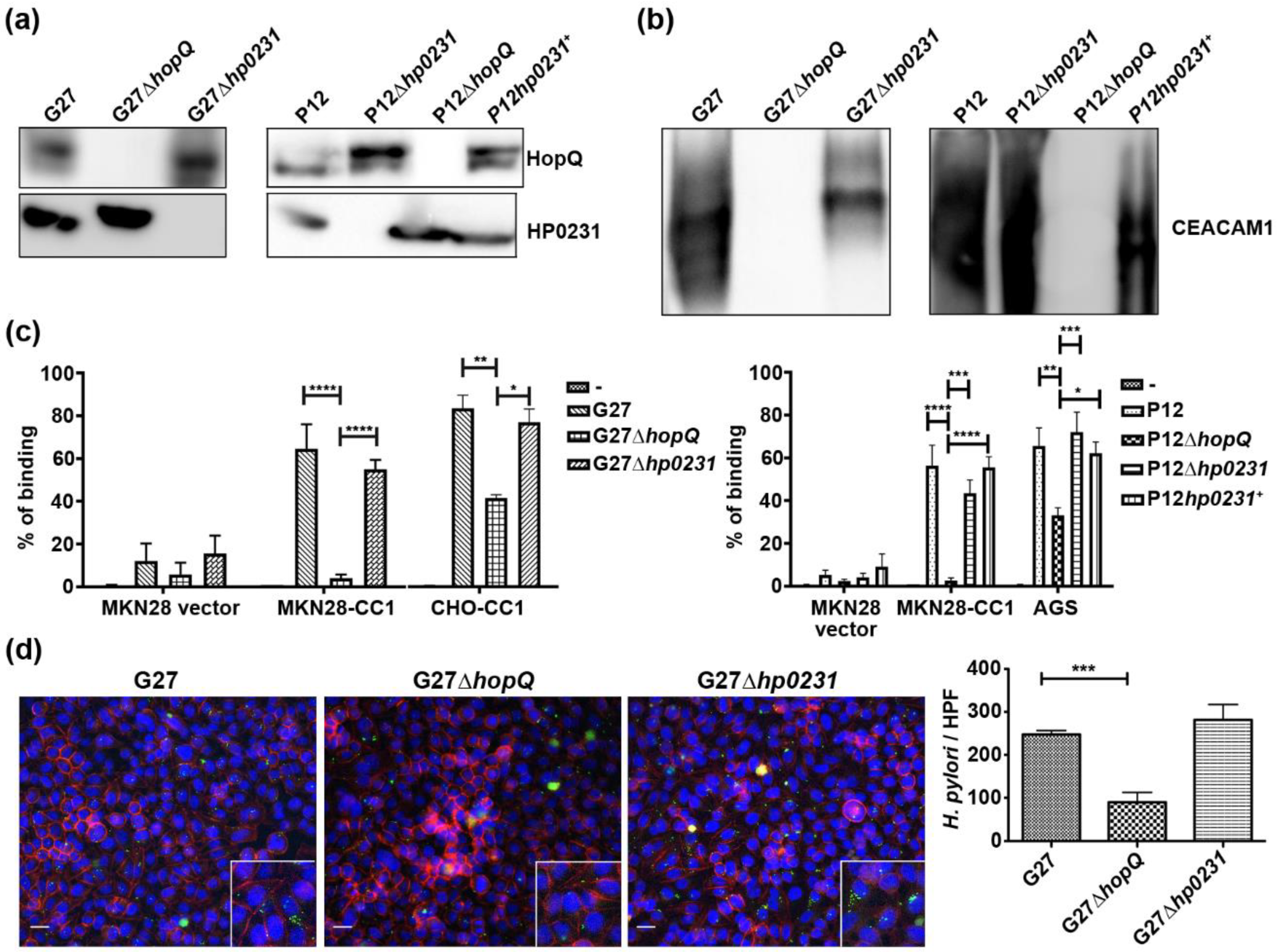
3.2. H. pylori Oxidoreductase HP0231 is Not Required for HopQ Binding to Gastric Cells
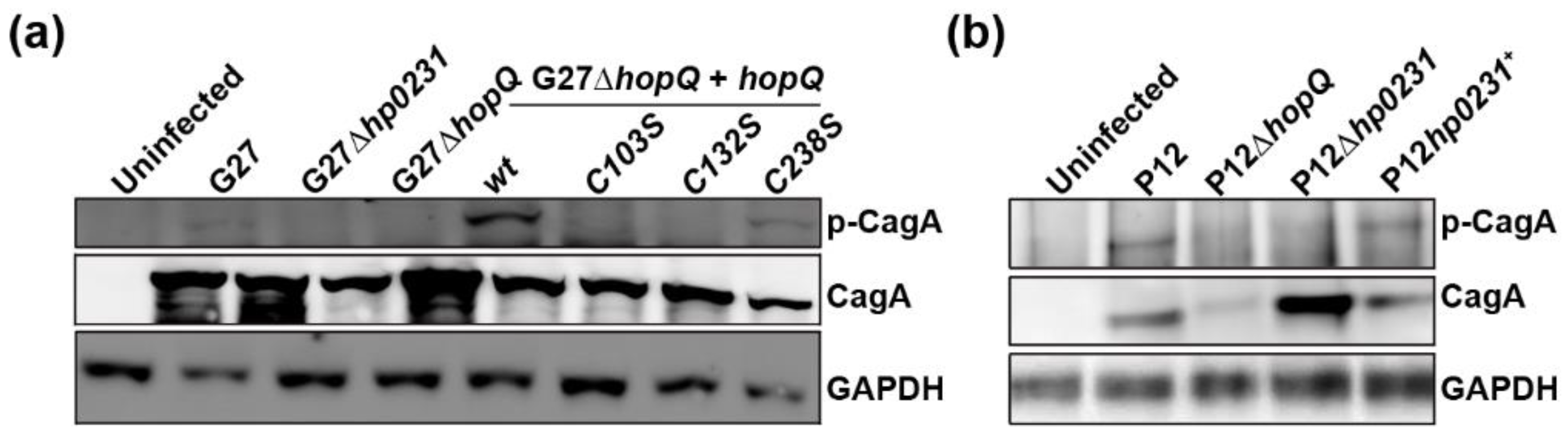
3.3. HopQ-CEACAM1 Mediated CagA Translocation is Dependent on the CL1 Disulfide
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ishaq, S.; Nunn, L. Helicobacter pylori and gastric cancer: A state of the art review. Gastroenterol. Hepatol. Bed Bench 2015, 8, S6–S14. [Google Scholar]
- Lindén, S.; Nordman, H.; Hedenbro, J.; Hurtig, M.; Borén, T.; Carlstedt, I. Strain- and blood group-dependent binding of Helicobacter pylori to human gastric MUC5AC glycoforms. Gastroenterology 2002, 123, 1923–1930. [Google Scholar] [CrossRef] [PubMed]
- Javaheri, A.; Kruse, T.; Moonens, K.; Mejías-Luque, R.; Debraekeleer, A.; Asche, C.I.; Tegtmeyer, N.; Kalali, B.; Bach, N.C.; Sieber, S.A.; et al. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat. Microbiol. 2016, 2, 16189. [Google Scholar] [CrossRef] [PubMed]
- Königer, V.; Holsten, L.; Harrison, U.; Busch, B.; Loell, E.; Zhao, Q.; Bonsor, D.A.; Roth, A.; Kengmo-Tchoupa, A.; Smith, S.I.; et al. Erratum: Helicobacter pylori exploits human CEACAMs via HopQ for adherence and translocation of CagA. Nat. Microbiol. 2016, 2, 16188. [Google Scholar] [CrossRef] [PubMed]
- Moonens, K.; Hamway, Y.; Neddermann, M.; Reschke, M.; Tegtmeyer, N.; Kruse, T.; Kammerer, R.; Mejías-Luque, R.; Singer, B.B.; Backert, S.; et al. Helicobacter pylori adhesin HopQ disrupts trans dimerization in human CEACAM s. EMBO J. 2018, 37, e98665. [Google Scholar] [CrossRef]
- Bonsor, D.A.; Zhao, Q.; Schmidinger, B.; Weiss, E.; Wang, J.; Deredge, D.; Beadenkopf, R.; Dow, B.; Fischer, W.; Beckett, D.; et al. The Helicobacter pylori adhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA. EMBO J. 2018, 37, e98664. [Google Scholar] [CrossRef]
- Zhong, Y.; Ander, F.; Kruse, T.; Schindele, F.; Jagusztyn-Krynicka, E.K.; Fischer, W.; Gerhard, M.; Mejías-Luque, R. Helicobacter pylori HP0231 influences bacterial virulence and is essential for gastric colonization. PLoS ONE 2016, 11, e0154643. [Google Scholar] [CrossRef]
- Moonens, K.; Gideonsson, P.; Subedi, S.; Bugaytsova, J.; Romaõ, E.; Mendez, M.; Nordén, J.; Fallah, M.; Rakhimova, L.; Shevtsova, A.; et al. Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori. Cell Host Microbe 2016, 19, 55–66. [Google Scholar] [CrossRef]
- Denoncin, K.; Collet, J.F. Disulfide bond formation in the bacterial periplasm: Major achievements and challenges ahead. Antioxid. Redox Signal. 2013, 19, 63–71. [Google Scholar] [CrossRef]
- Shouldice, S.R.; Heras, B.; Walden, P.M.; Totsika, M.; Schembri, M.A.; Martin, J.L. Structure and function of DsbA, a key bacterial oxidative folding catalyst. Antioxid. Redox Signal. 2011, 14, 1729–1760. [Google Scholar] [CrossRef]
- Berkmen, M. Production of disulfide-bonded proteins in Escherichia coli. Protein Expr. Purif. 2012, 82, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Lester, J.; Kichler, S.; Oickle, B.; Fairweather, S.; Oberc, A.; Chahal, J.; Ratnayake, D.; Creuzenet, C. Characterization of Helicobacter pyloriHP0231 (DsbK): Role in disulfide bond formation, redox homeostasis and production of Helicobacter cystein-rich protein HcpE. Mol. Microbiol. 2015, 96, 110–133. [Google Scholar] [CrossRef] [PubMed]
- Roszczenko, P.; Radomska, K.A.; Wywial, E.; Collet, J.F.; Jagusztyn-Krynicka, E.K. A Novel Insight into the Oxidoreductase Activity of Helicobacter pylori HP0231 Protein. PLoS ONE 2012, 7, e46563. [Google Scholar] [CrossRef]
- Roszczenko, P.; Grzeszczuk, M.; Kobierecka, P.; Wywial, E.; Urbanowicz, P.; Wincek, P.; Nowak, E.; Jagusztyn-Krynicka, E.K. Helicobacter pylori HP0377, a member of the Dsb family, is an untypical multifunctional CcmG that cooperates with dimeric thioldisulfide oxidase HP0231. BMC Microbiol. 2015, 15, 135. [Google Scholar] [CrossRef]
- Odenbreit, S.; Till, M.; Haas, R. Optimized BlaM-transposon shuttle mutagenesis of Helicobacter pylori allows the identification of novel genetic loci involved in bacterial virulence. Mol. Microbiol. 1996, 20, 361–373. [Google Scholar] [CrossRef]
- Xiang, Z.; Censini, S.; Bayeli, P.F.; Telford, J.L.; Figura, N.; Rappuoli, R.; Covacci, A. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect. Immun. 1995, 63, 94–98. [Google Scholar] [CrossRef]
- Romano, M.; Razandi, M.; Sekhon, S.; Krause, W.J.; Ivey, K.J. Human cell line for study of damage to gastric epithelial cells in vitro. J. Lab. Clin. Med. 1988, 111, 430–440. [Google Scholar]
- Müller, M.M.; Klaile, E.; Vorontsova, O.; Singer, B.B.; Öbrink, B. Homophilic adhesion and CEACAM1-S regulate dimerization of CEACAM1-L and recruitment of SHP-2 and c-Src. J. Cell Biol. 2009, 187, 569–581. [Google Scholar] [CrossRef]
- Tchoupa, A.K.; Lichtenegger, S.; Reidl, J.; Hauck, C.R. Outer membrane protein P1 is the CEACAM-binding adhesin of Haemophilus influenzae. Mol. Microbiol. 2015, 98, 440–455. [Google Scholar] [CrossRef]
- Kuespert, K.; Weibel, S.; Hauck, C.R. Profiling of bacterial adhesin--host receptor recognition by soluble immunoglobulin superfamily domains. J. Microbiol. Methods 2007, 68, 478–485. [Google Scholar] [CrossRef]
- Kuespert, K.; Hauck, C.R. Characterizing host receptor recognition by individual bacterial pathogens. Methods Mol. Biol. 2009, 470, 57–65. [Google Scholar] [PubMed]
- Muturi, H.T.; Dreesen, J.D.; Nilewski, E.; Jastrow, H.; Giebel, B.; Ergun, S.; Singer, B.B. Tumor and Endothelial Cell-Derived Microvesicles Carry Distinct CEACAMs and Influence T-Cell Behavior. PLoS ONE 2013, 8, e74654. [Google Scholar] [CrossRef] [PubMed]
- Grzeszczuk, M.J.; Bocian-Ostrzycka, K.M.; Banaś, A.M.; Roszczenko-Jasinska, P.; Malinowska, A.; Stralova, H.; Haas, R.; Meyer, T.F.; Jagusztyn-Krynicka, E.K. Thioloxidoreductase HP0231 of Helicobacter pylori impacts HopQ-dependent CagA translocation. Int. J. Med. Microbiol. 2018, 308, 977–985. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Zhou, M.; He, Y.; Wan, Y.; Bai, W.; Tao, S.; Ren, Y.; Zhang, X.; Xu, J.; Liu, J.; et al. Efficient Inhibition of Hepatitis B Virus Infection by a preS1-binding Peptide. Sci. Rep. 2016, 6, 29391. [Google Scholar] [CrossRef]
- Melnik, L.I.; Garry, R.F.; Morris, C.A. Peptide inhibition of human cytomegalovirus infection. Virol. J. 2011, 8, 76. [Google Scholar] [CrossRef]
- Rajik, M.; Omar, A.R.; Ideris, A.; Hassan, S.S.; Yusoff, K. A novel peptide inhibits the influenza virus replication by preventing the viral attachment to the host cells. Int. J. Biol. Sci. 2009, 5, 543–548. [Google Scholar] [CrossRef]
- Krepstakies, M.; Lucifora, J.; Nagel, C.-H.; Zeisel, M.B.; Holstermann, B.; Hohenberg, H.; Kowalski, I.; Gutsmann, T.; Baumert, T.F.; Brandenburg, K.; et al. A New Class of Synthetic Peptide Inhibitors Blocks Attachment and Entry of Human Pathogenic Viruses. J. Infect. Dis. 2012, 205, 1654–1664. [Google Scholar] [CrossRef]
- Sánchez-Barinas, C.D.; Ocampo, M.; Tabares, L.; Bermúdez, M.; Patarroyo, M.A.; Patarroyo, M.E. Specific Binding Peptides from Rv3632: A Strategy for Blocking Mycobacterium tuberculosis Entry to Target Cells? Biomed Res. Int. 2019, 2019, 8680935. [Google Scholar] [CrossRef]
- Aldeghaither, D.; Smaglo, B.G.; Weiner, L.M. Beyond peptides and mAbs—Current status and future perspectives for biotherapeutics with novel constructs. J. Clin. Pharmacol. 2015, 55, S4–S20. [Google Scholar] [CrossRef]
- Siontorou, C.G. Nanobodies as novel agents for disease diagnosis and therapy. Int. J. Nanomed. 2013, 8, 4215–4227. [Google Scholar] [CrossRef]
- Chiu, J.; Hogg, P.J. Allosteric disulfides: Sophisticated molecular structures enabling flexible protein regulation. J. Biol. Chem. 2019, 294, 2949–2960. [Google Scholar] [CrossRef] [PubMed]
- Hallenbeck, K.K.; Turner, D.M.; Renslo, A.R.; Arkin, M.R. Targeting Non-Catalytic Cysteine Residues Through Structure-Guided Drug Discovery. Curr. Top. Med. Chem. 2017, 17, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.; Burke, B.; Thaï, R.; Dey, A.K.; Combes, O.; Heyd, B.; Geonnotti, A.R.; Montefiori, D.C.; Kan, E.; Lian, Y.; et al. Stabilization of HIV-1 envelope in the CD4-bound conformation through specific cross-linking of a CD4 mimetic. J. Biol. Chem. 2011, 286, 21706–21716. [Google Scholar] [CrossRef]
- Cerutti, N.; Mendelow, B.V.; Napier, G.B.; Papathanasopoulos, M.A.; Killick, M.; Khati, M.; Stevens, W.; Capovilla, A. Stabilization of HIV-1 gp120-CD4 receptor complex through targeted interchain disulfide exchange. J. Biol. Chem. 2010, 285, 25743–25752. [Google Scholar] [CrossRef] [PubMed]
- Tegtmeyer, N.; Harrer, A.; Schmitt, V.; Singer, B.B.; Backert, S. Expression of CEACAM1 or CEACAM5 in AZ-521 cells restores the type IV secretion deficiency for translocation of CagA by Helicobacter pylori. Cell. Microbiol. 2019, 21, e12965. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamway, Y.; Taxauer, K.; Moonens, K.; Neumeyer, V.; Fischer, W.; Schmitt, V.; Singer, B.B.; Remaut, H.; Gerhard, M.; Mejías-Luque, R. Cysteine Residues in Helicobacter pylori Adhesin HopQ Are Required for CEACAM–HopQ Interaction and Subsequent CagA Translocation. Microorganisms 2020, 8, 465. https://doi.org/10.3390/microorganisms8040465
Hamway Y, Taxauer K, Moonens K, Neumeyer V, Fischer W, Schmitt V, Singer BB, Remaut H, Gerhard M, Mejías-Luque R. Cysteine Residues in Helicobacter pylori Adhesin HopQ Are Required for CEACAM–HopQ Interaction and Subsequent CagA Translocation. Microorganisms. 2020; 8(4):465. https://doi.org/10.3390/microorganisms8040465
Chicago/Turabian StyleHamway, Youssef, Karin Taxauer, Kristof Moonens, Victoria Neumeyer, Wolfgang Fischer, Verena Schmitt, Bernhard B. Singer, Han Remaut, Markus Gerhard, and Raquel Mejías-Luque. 2020. "Cysteine Residues in Helicobacter pylori Adhesin HopQ Are Required for CEACAM–HopQ Interaction and Subsequent CagA Translocation" Microorganisms 8, no. 4: 465. https://doi.org/10.3390/microorganisms8040465
APA StyleHamway, Y., Taxauer, K., Moonens, K., Neumeyer, V., Fischer, W., Schmitt, V., Singer, B. B., Remaut, H., Gerhard, M., & Mejías-Luque, R. (2020). Cysteine Residues in Helicobacter pylori Adhesin HopQ Are Required for CEACAM–HopQ Interaction and Subsequent CagA Translocation. Microorganisms, 8(4), 465. https://doi.org/10.3390/microorganisms8040465




