Different Dynamics of Bacterial and Fungal Communities in Hive-Stored Bee Bread and Their Possible Roles: A Case Study from Two Commercial Honey Bees in China
Abstract
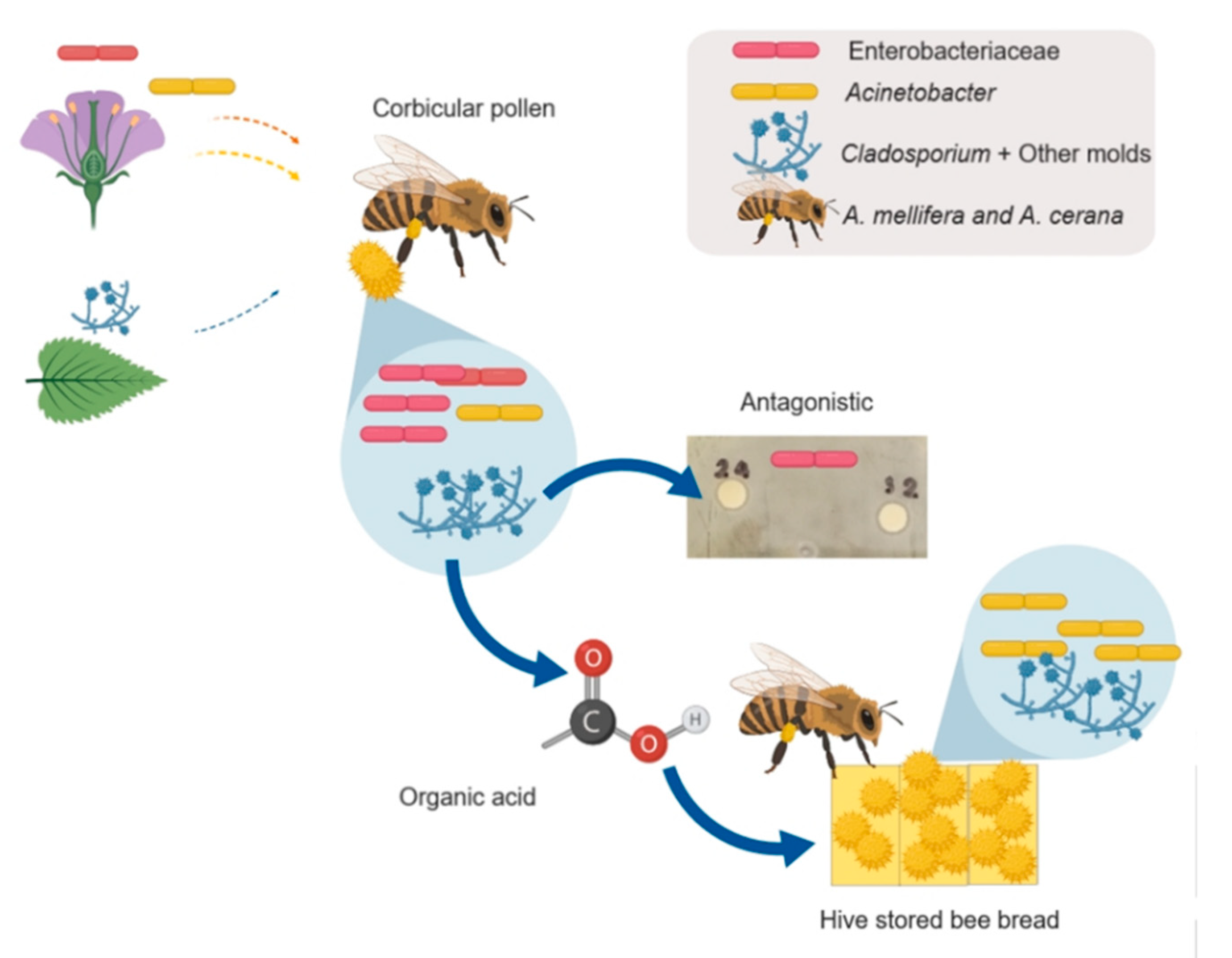
1. Introduction
2. Materials and Methods
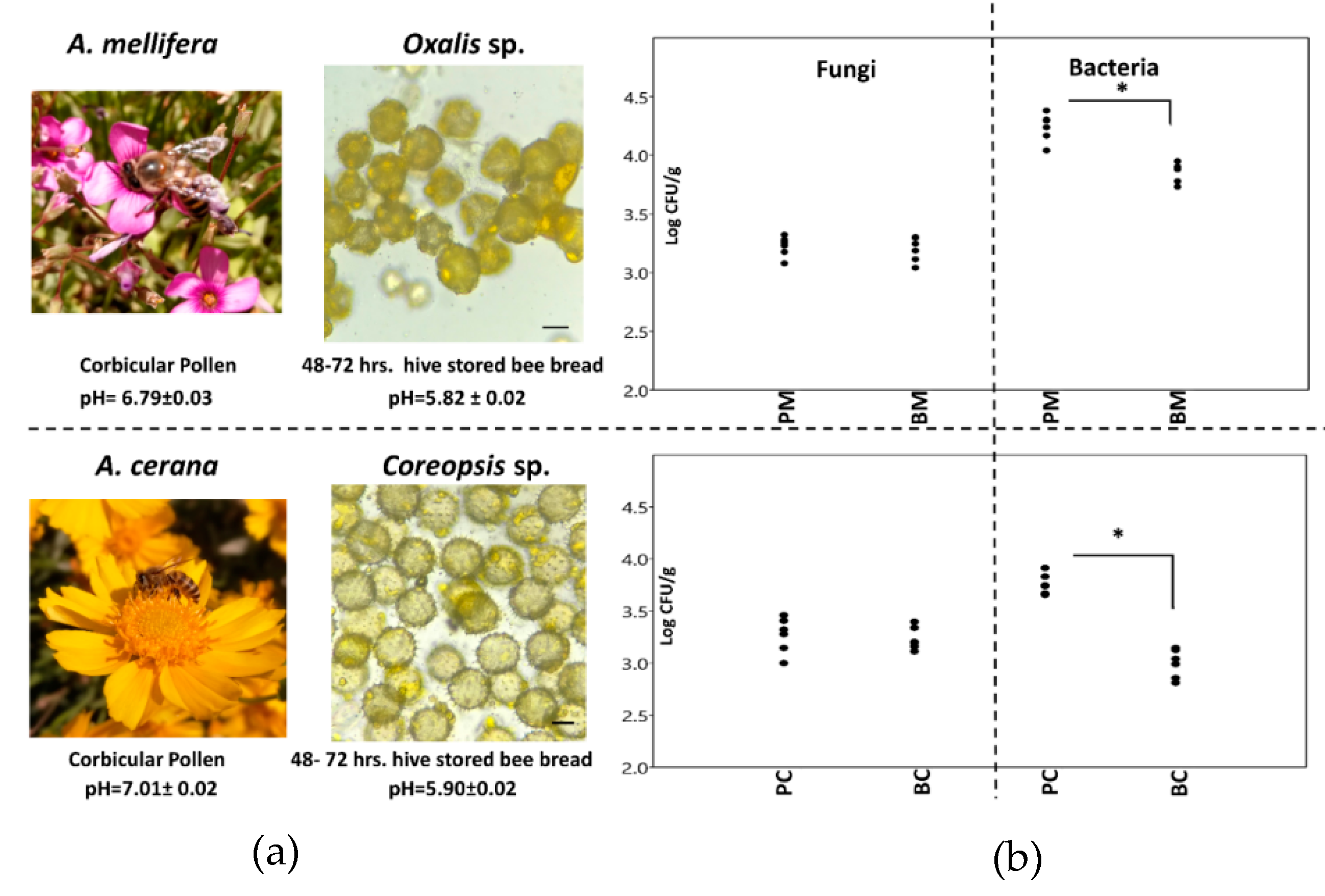
2.1. Corbicular and Hive Storage Pollen Collection
2.2. Pollen Identification
2.3. Microbe Number Determination
2.4. DNA Extraction
2.5. Amplicon Sequencing Analysis with Illumina Solexa
2.6. Microbial Isolate Identification
2.7. Organic Acid and Hydrolytic Enzyme Screening
2.8. The Antagonistic Inhibition of Filamentous Fungal Isolates with Association Bacteria in the Hive and During Chalkbrood Infection
2.9. Statistical Analysis
2.10. Sequence Deposition
3. Results
3.1. Pollen Majority and Colony-Forming Unit Numbers
3.2. The Quantitative Data of Illumina Solexa
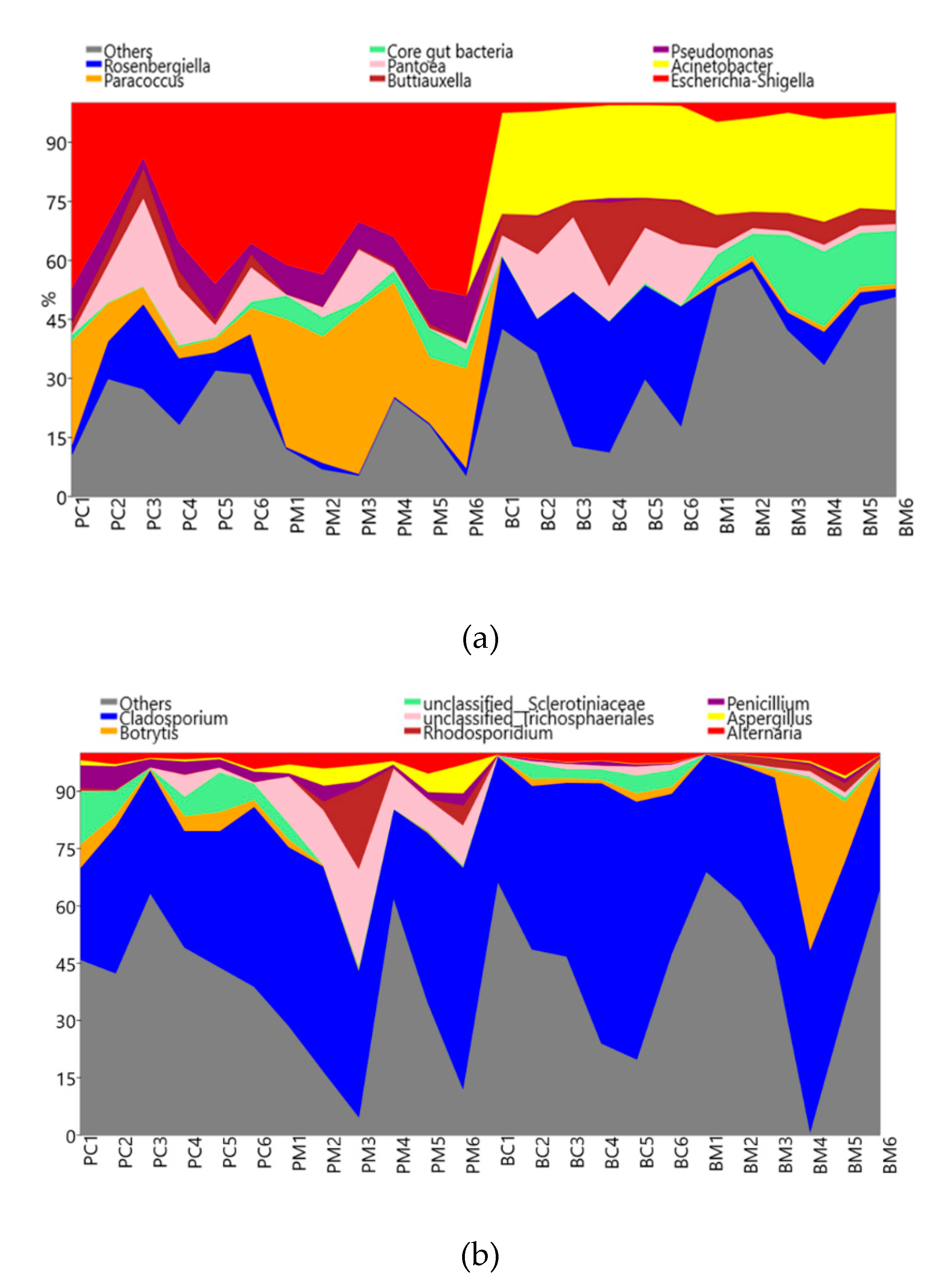
3.3. Microbial Diversity in Corbicular Pollen and Hive-Stored Bee Bread
3.4. Microbial Communities in Corbicular Pollen and Hive-Stored Bee Bread
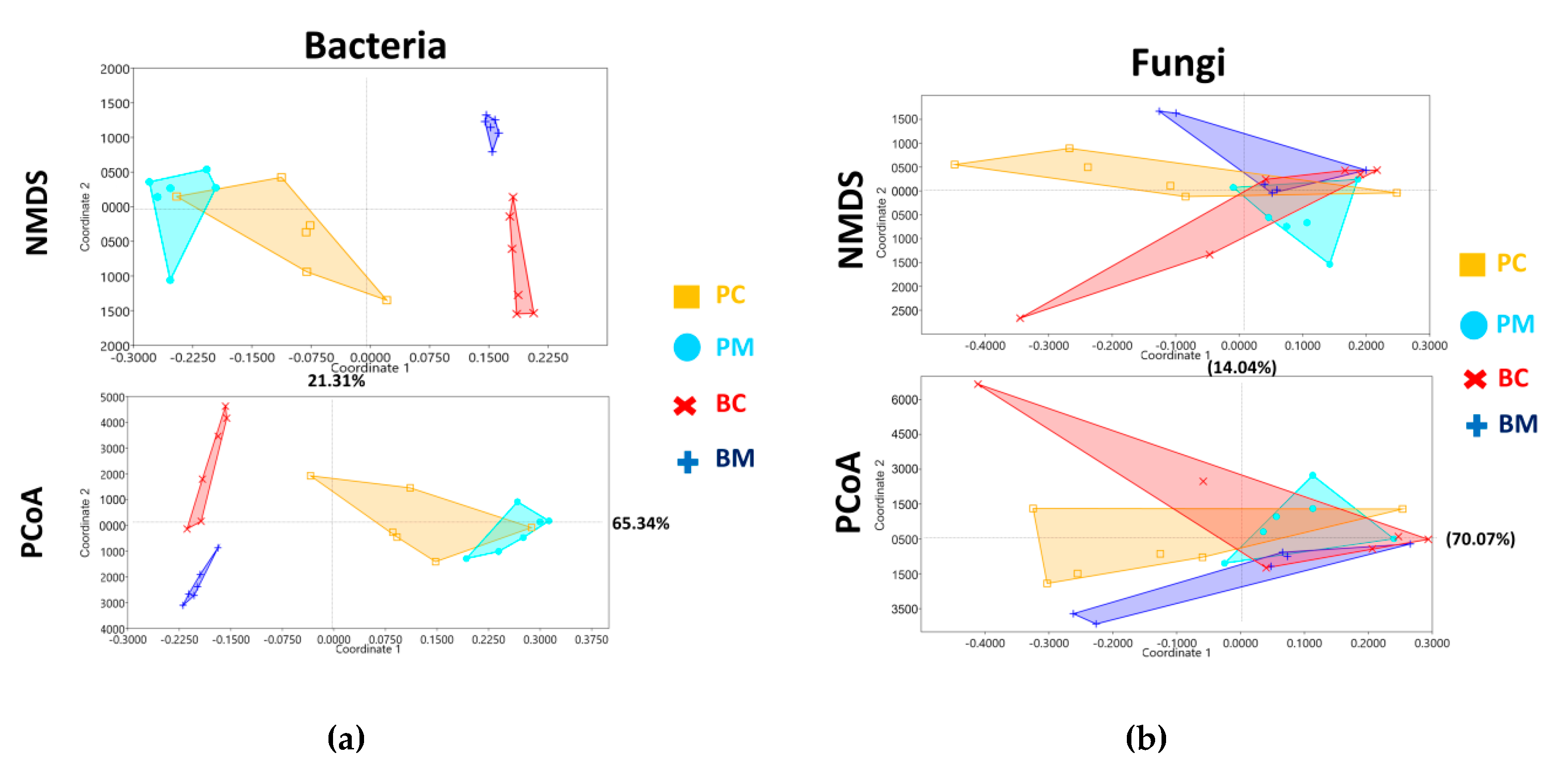
3.5. Beta Diversity and Bacterial Functional Prediction
3.6. Functional Prediction from 16S rRNA Gene Data via PICRUSTs Based on COGs
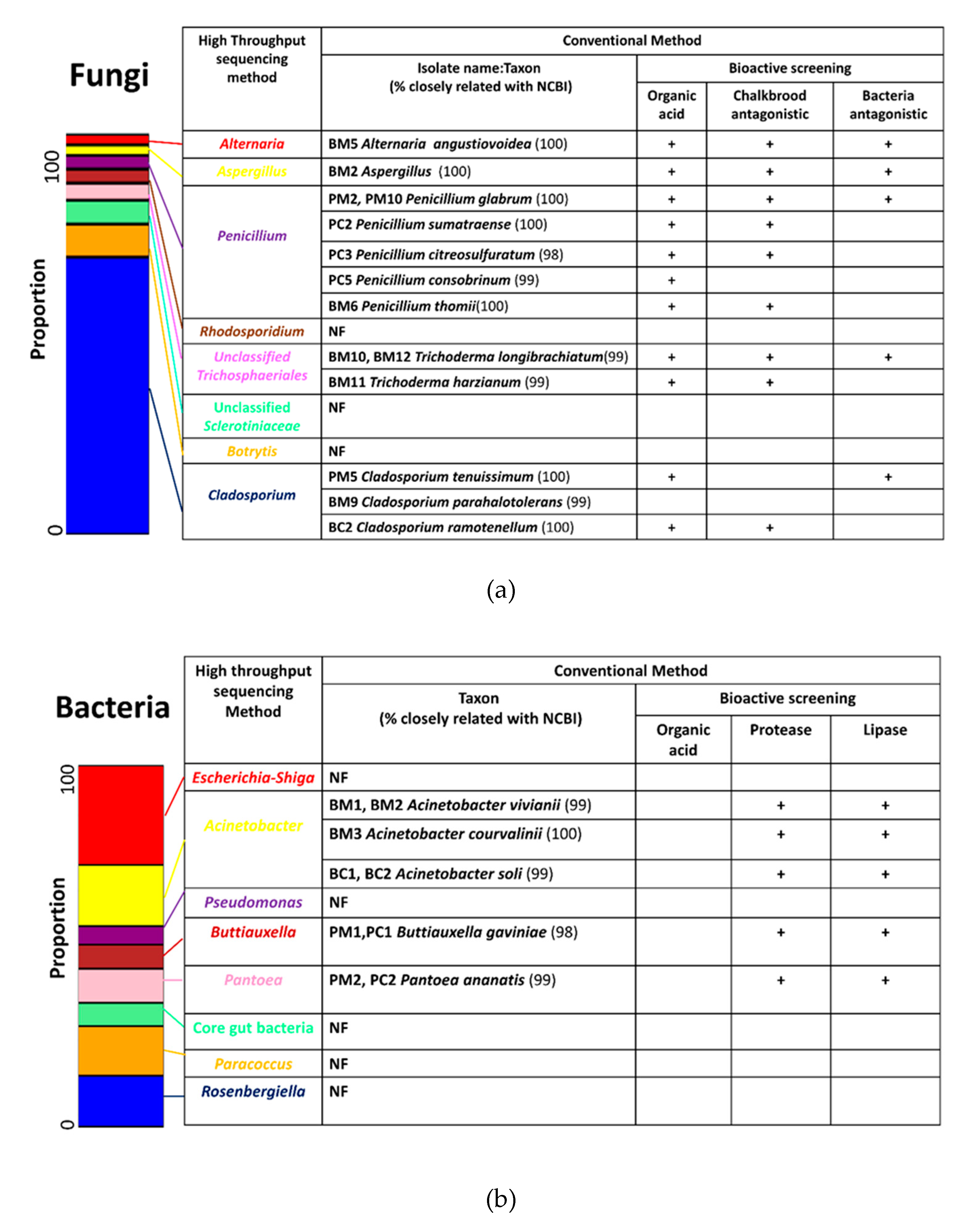
3.7. Isolates and Their Biological Activities
3.8. Cooperation between Conventional and High Throughput Sequencing Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Neumann, P.; Carreck, N.L. Honey bee colony losses. J. Apic. Res. 2010, 49, 1–6. [Google Scholar] [CrossRef]
- Goulson, D.; Nicholls, E.; Botías, C.; Rotheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef] [PubMed]
- Cox, L.; Yamanishi, S.; Sohn, J.; Alekseyenko, A.; Leung, J.; Cho, I.; Kim, S.; Li, H.; Gao, Z.; Mahana, D.; et al. Altering the Intestinal Microbiota during a Critical Developmental Window Has Lasting Metabolic Consequences. Cell 2014, 158, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.H.; Kim, S.H.; Lee, H.Y.; Bai, J.Y.; Nam, Y.D.; Bae, J.W.; Lee, D.G.; Shin, S.C.; Ha, E.M.; Lee, W.J. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 2008, 319, 777–782. [Google Scholar] [CrossRef]
- Wada-Katsumata, A.; Zurek, L.; Nalyanya, G.; Roelofs, W.L.; Schal, C. Gut bacteria mediate aggregation in the German cockroach. Proc. Natl. Acad. Sci. USA 2015, 112, 15678–15683. [Google Scholar] [CrossRef]
- Krych, L.; Hansen, C.H.; Hansen, A.K.; van den Berg, F.W.; Nielsen, D.S. Quantitatively different, yet qualitatively alike: A meta-analysis of the mouse core gut microbiome with a view towards the human gut microbiome. PLoS ONE 2013, 8, e62578. [Google Scholar] [CrossRef]
- Nguyen, T.L.; Vieira-Silva, S.; Liston, A.; Raes, J. How informative is the mouse for human gut microbiota research? Dis. Models Mech. 2015, 8, 1–16. [Google Scholar] [CrossRef]
- Huang, S.K.; Ye, K.T.; Huang, W.F.; Ying, B.H.; Su, X.; Lin, L.H.; Li, J.H.; Chen, Y.P.; Li, J.L.; Bao, X.L.; et al. Influence of Feeding Type and Nosema ceranae Infection on the Gut Microbiota of Apis cerana Workers. MSystems 2018, 3, e00177-18. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Annah, S.R.; Parthasarathy, R.; Adam, R.B.; Brendan, J.M.B.; Guillemin, K. Individual Members of the Microbiota Disproportionately Modulate Host Innate Immune Responses. Cell Host Microbe 2015, 18, 613–620. [Google Scholar]
- Yun, J.H.; Jung, M.J.; Kim, P.S.; Bae, J.W. Social status shapes the bacterial and fungal gut communities of the honey bee. Sci. Rep. 2018, 8, 2019. [Google Scholar] [CrossRef] [PubMed]
- Bonvehí, J.; Pesudo, F.; Pallarés, J. Quantitative determination of free amino acids in honeybee collected pollen using gas chromatography and spectrophotometry. Ann. Falsif. Expert Chim. 1991, 897, 153–166. [Google Scholar]
- Casteel, D.B. The behavior of the honey bee in pollen collection. U.S.D.A. Bur. Entomol. Bull. 1912, 121, 1–36. [Google Scholar]
- Foote, H.L. Possible use of microorganisms in synthetic bee bread production. Am. Bee J. 1957, 97, 476–478. [Google Scholar]
- Haydak, M.H. Pollen-pollen substitutes-bee bread. Am. Bee J. 1958, 98, 145–146. [Google Scholar]
- Chevtchik, V. Mikrobiologie pylového kvas’eni. Publ. Fac. Sci. Univ. Masaryk. 1950, 323, 103–130. [Google Scholar]
- Gilliam, M. Identification and roles of non-pathogenic microflora associated with honey bees1Mention of a proprietary product or company name does not constitute an endorsement of this product by the U.S. Department of Agriculture.1. FEMS Microbiol. Lett. 1997, 155, 1–10. [Google Scholar] [CrossRef]
- Gilliam, M.; Prest, D.; Lorenz, B. Microbiology of pollen and bee bread: Taxonomy and enzymology of molds. Apidologie 1989, 20, 53–68. [Google Scholar] [CrossRef]
- Martinson, V.G.; Danforth, B.N.; Minckley, R.L.; Rueppell, O.; Tingek, S.; Moran, N.A. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 2011, 20, 619–628. [Google Scholar] [CrossRef]
- Okunuki, K. Uber den Gaswechsel der Pollen. II. Acta Phytochim 2019, 11, 27–64. [Google Scholar]
- Pain, J.; Maugenet, J. Recherches biochimiques et physiologiques sur le pollen emmagsiné par les abeilles. Apidologie 1966, 9, 209–236. [Google Scholar] [CrossRef][Green Version]
- Sinpoo, C.; Williams, G.R.; Chantawannakul, P. Dynamics of fungal communities in corbicular pollen and bee bread. Chiang Mai J. Sci. 2017, 44, 1244–1256. [Google Scholar]
- Gilliam, M.; Wickerham, L.J.; Morton, H.L.; Martin, R.D. Yeasts isolated from honey bees, Apis mellifera, fed 2,4-D and antibiotics. J. Invertebr. Pathol. 1974, 24, 349–356. [Google Scholar] [CrossRef]
- De Palma, A.; Abrahamczyk, S.; Aizen, M.A.; Albrecht, M.; Basset, Y.; Bates, A.; Blake, R.J.; Boutin, C.; Bugter, R.; Connop, S.; et al. Predicting bee community responses to land-use changes: Effects of geographic and taxonomic biases. Sci. Rep. 2016, 6, 31153. [Google Scholar] [CrossRef]
- Anderson, K.; Sheehan, T.; Mott, B.; Maes, P.; Snyder, L.; Schwan, M.; Walton, A.; Jones, B.; Corby-Harris, V. Microbial Ecology of the Hive and Pollination Landscape: Bacterial Associates from Floral Nectar, the Alimentary Tract and Stored Food of Honey Bees (Apis mellifera). PLoS ONE 2013, 8, e83125. [Google Scholar] [CrossRef]
- Anderson, K.E.; Carroll, M.J.; Sheehan, T.; Mott, B.M.; Corby-Harris, V. Hive-stored pollen of honey bees: Many lines of evidence are consistent with pollen preservation, not nutrient conversion. Mol. Ecol. 2014, 23, 5904–5917. [Google Scholar] [CrossRef]
- Herbert, EW., Jr.; Shimanuki, H. Chemical composition and nutritive value of bee-collected and bee stored pollen. Apidologie 1978, 9, 33–40. [Google Scholar] [CrossRef]
- Loper, G.M.; Standifer, L.N.; Thompson, M.J.; Gilliam, M. Biochemistry and microbiology of bee-collected almond (prunus dulcis) pollen and bee bread. I-Fatty Acids, Sterols, Vitamins and Minerals. Apidologie 1980, 11, 63–73. [Google Scholar] [CrossRef]
- Anderson, K.; Sheehan, T.H.; Eckholm, B.; Mott, B.M.; Degrandi-Hoffman, G. An emerging paradigm of colony health: Microbial balance of the honey bee and hive (Apis mellifera). Insectes Sociaux 2011, 58, 431–444. [Google Scholar] [CrossRef]
- Vasquez, A.; Olofsson, T.C. The honey crop-the holy grail when antibiotics fail. Microbiol. Today 2011, 38, 226–229. [Google Scholar]
- DeGrandi-Hoffman, G.; Eckholm, B.J.; Huang, M.H. A comparison of bee bread made by Africanized and European honey bees (Apis mellifera) and its effects on hemolymph protein titers. Apidologie 2013, 44, 52–63. [Google Scholar] [CrossRef]
- Mattila, H.R.; Rios, D.; Walker-Sperling, V.E.; Roeselers, G.; Newton, I.L.G. Characterization of the active microbiotas associated with honey bees reveals healthier and broader communities when colonies are genetically diverse. PLoS ONE 2012, 7, e32962. [Google Scholar] [CrossRef]
- Lee, F.J.; Rusch, D.B.; Stewart, F.J.; Mattila, H.R.; Newton, I.L. Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ. Microbiol. 2015, 17, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Breed, M.; Mizrahi, A.; Lensky, Y. Bee Products: Properties, Applications, and Apitherapy. Bioscience 1998, 48. [Google Scholar] [CrossRef]
- Chen, Y.C. Apicultural in China; China Agriculture Press: Beijing, China, 1993. [Google Scholar]
- Jun, G.; Wu, J.; Chen, Y.; Evans, J.; Dai, R.; Luo, W.; Li, J. Characterization of gut bacteria at different developmental stages of Asian honey bees, Apis Cerana. J. Invertebr. Pathol. 2015, 127. [Google Scholar]
- Wang, C.; Huang, Y.; Li, L.; Guo, J.; Wu, Z.; Deng, Y.; Dai, L.; Ma, S. Lactobacillus panisapium sp. nov. from honeybee Apis cerana bee bread. Int. J. Syst. Evol. Microbiol. 2018, 68, 703–708. [Google Scholar] [CrossRef]
- Lee, C.K.; Barbier, B.A.; Bottos, E.M.; McDonald, I.R.; Cary, S.C. The Inter-Valley Soil Comparative Survey: The ecology of Dry Valley edaphic microbial communities. ISME J. 2012, 6, 1046–1057. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes--application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.; Bahram, M.; Bates, S.; Bruns, T.; Bengtsson-Palme, J.; Callaghan, T.; et al. Towards a unified paradigm for sequence-based identification of Fungi. Mol. Ecol. 2013, 22. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Goodfellow, E.S., Ed.; John Wiley & Sons: New York, NY, USA, 1991; pp. 115–176. [Google Scholar]
- Hammer, O.; PAST (Paleontological Statistics). Natural History Museum, University of Oslo. 2011. Available online: http://folk. uio.no/ohammer/past/index.html (accessed on 9 April 2019).
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Abou-Shaara, H.F. The foraging behaviour of honey bees, Apis mellifera: A review. Veterinární Med. 2014, 59, 1–10. [Google Scholar] [CrossRef]
- Alqarni, A. Tolerance of Summer Temperature in Imported and Indigenous Honeybee Apis mellifera L. Races in Central Saudi Arabia. Saudi J. Biol. Sci. 2006, 13, 123–127. [Google Scholar]
- Joshi, N.; Joshi, P. Foraging Behaviour of Apis Spp. on Apple Flowers in a Subtropical Environment. NY Sci. J. 2010, 3, 71–76. [Google Scholar]
- Mohammad, M.; Edriss, M.; Basiri, M. Analysis of Colony and Morphological Characters in Honey Bees (Apis mellifera meda). Pak. J. Biol. Sci. 2006, 9, 2685–2688. [Google Scholar]
- Oldroyd, B.; Rinderer, T.; Wongsiri, S. Pollen resource partitioning by Apis dorsata, A cerana, A andreniformis and A florea in Thailand. J. Apic. Res. 1992, 31, 3–7. [Google Scholar] [CrossRef]
- Hendry, G.A.F. Evolutionary origins and natural functions of fructans—a climatological, biogeographic and mechanistic appraisal. New Phytol. 2006, 123, 3–14. [Google Scholar] [CrossRef]
- Laere, A.V.; Ende, W.V.D. Inulin metabolism in dicots: Chicory as a model system. Plant Cell Environ. 2002, 25, 803–813. [Google Scholar] [CrossRef]
- Lenaerts, M.; Pozo, M.I.; Wäckers, F.; Ende, W.V.D.; Jacquemyn, H.; Lievens, B. Impact of microbial communities on floral nectar chemistry: Potential implications for biological control of pest insects. Basic Appl. Ecol. 2016, 17, 189–198. [Google Scholar] [CrossRef]
- Lokoschus, F.S.; Keularts, J.L.W. A futher function of the medibular gland of the worker honey bee: Production of substance inhibiting pollen germination. Z. Bienenforch 1968, 9, 333–343. [Google Scholar]
- Egorova, A.I. Preservative microflora in stored pollen. Veterinaruya 1971, 8, 40–41. [Google Scholar]
- Ariizumi, T.; Toriyama, K. Genetic Regulation of Sporopollenin Synthesis and Pollen Exine Development. Annu. Rev. Plant Biol. 2010, 62, 437–460. [Google Scholar] [CrossRef] [PubMed]
- Blackmore, S.; Wortley, A.H.; Skvarla, J.J.; Rowley, J.R. Pollen wall development in flowering plants. New Phytol. 2007, 174, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Sanchez-Moreiras, A.M.; Abel, C.; Sohrabi, R.; Lee, S.; Gershenzon, J.; Tholl, D. The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. New Phytol. 2012, 193, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.; Thornburg, R. Is the nectar redox cycle a floral defense against microbial attack? Trends Plant Sci. 2004, 9, 320–324. [Google Scholar] [CrossRef]
- Harper, A.D.; Stalnaker, S.H.; Wells, L.; Darvill, A.; Thornburg, R.; York, W.S. Interaction of Nectarin 4 with a fungal protein triggers a microbial surveillance and defense mechanism in nectar. Phytochemistry 2010, 71, 1963–1969. [Google Scholar] [CrossRef]
- Remus-Emsermann, M.N.P.; Tecon, R.; Kowalchuk, G.A.; Leveau, J.H.J. Variation in local carrying capacity and the individual fate of bacterial colonizers in the phyllosphere. ISME J. Multidiscip. J. Microb. Ecol. 2012, 6, 756–765. [Google Scholar] [CrossRef]
- Nicolson, S.W.; Human, H. Chemical composition of the ‘low quality’ pollen of sunflower (Helianthus annuus, Asteraceae). Apidologie 2013, 44, 144–152. [Google Scholar] [CrossRef]
- Rousk, J.; Brookes, P.C.; Bååth, E. Contrasting Soil pH Effects on Fungal and Bacterial Growth Suggest Functional Redundancy in Carbon Mineralization. Appl. Environ. Microbiol. 2009, 75, 1589–1596. [Google Scholar] [CrossRef]
- Tate, R.L. Microbial biomass measurements in acidic soil: Effect of fungal:bacterial activity ratios and soil amendment. Soil Sci. 1991, 152, 220–225. [Google Scholar] [CrossRef]
- Longworthy, T.A. Microbial life in extreme pH value. In Microbial Life in Extreme Environment; Academic Press: London, UK, 1978; pp. 279–315. [Google Scholar]
- Hesse, S.J.A.; Ruijter, G.J.G.; Dijkema, C.; Visser, J. Intracellular pH homeostasis in the filamentous fungus Aspergillus niger. Eur. J. Biochem. 2002, 269, 3485–3494. [Google Scholar] [CrossRef]
- Slonczewski, J.L.; Rosen, B.P.; Alger, J.R.; Macnab, R.M. pH Homeostasis in Escherichia coli: Measurement by 31P Nuclear Magnetic Resonance of Methylphosphonate and Phosphate. Proc. Natl. Acad. Sci. USA 1981, 78, 6271–6275. [Google Scholar] [CrossRef] [PubMed]
- Telias, A.; White, J.R.; Pahl, D.M.; Ottesen, A.R.; Walsh, C.S. Bacterial community diversity and variation in spray water sources and the tomato fruit surface. BMC Microbiol. 2011, 11, 81. [Google Scholar] [CrossRef]
- Fridman, S.; Izhaki, I.; Gerchman, Y.; Halpern, M. Bacterial communities in floral nectar. Environ. Microbiol. Rep. 2012, 4, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Pérez, S.; Herrera, C.M.; Vega, C.D. Zooming-in on floral nectar: A first exploration of nectar-associated bacteria in wild plant communities. FEMS Microbiol. Ecol. 2012, 80, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Junker, R.; Loewel, C.; Gross, R.; Dötterl, S.; Keller, A.; Blüthgen, N. Composition of epiphytic bacterial communities differs on petals and leaves. Plant Biol. 2011, 13, 918–924. [Google Scholar] [CrossRef] [PubMed]
- Ercolani, G.L. Distribution of epiphytic bacteria on olive leaves and the influence of leaf age and sampling time. Microb. Ecol. 1991, 21, 35–48. [Google Scholar] [CrossRef]
- Thompson, I.P.; Bailey, M.J.; Fenlon, J.S.; Fermor, T.R.; Lilley, A.K.; Lynch, J.M.; McCormack, P.J.; McQuilken, M.P.; Purdy, K.J.; Rainey, P.B.; et al. Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar beet (Beta vulgaris). Plant Soil 1993, 150, 177–191. [Google Scholar] [CrossRef]
- Krimm, U.; Abanda-Nkpwatt, D.; Schwab, W.; Schreiber, L. Epiphytic microorganisms on strawberry plants (Fragaria ananassa cv. Elsanta): Identification of bacterial isolates and analysis of their interaction with leaf surfaces. FEMS Microbiol. Ecol. 2005, 53, 483–492. [Google Scholar] [CrossRef]
- Xu, X.Q.; Pan, S.Q. An Agrobacterium catalase is a virulence factor involved in tumorigenesis. Mol. Microbiol. 2000, 35, 407–414. [Google Scholar] [CrossRef]
- Danhorn, T.; Fuqua, C. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 2007, 61, 401–422. [Google Scholar] [CrossRef]
- Barak, J.; Liang, A. Role of Soil, Crop Debris, and a Plant Pathogen in Salmonella enterica Contamination of Tomato Plants. PLoS ONE 2008, 3, e1657. [Google Scholar] [CrossRef] [PubMed]
- Meric, G.; Kemsley, E.K.; Falush, D.; Saggers, E.J.; Lucchini, S. Phylogenetic distribution of traits associated with plant colonization in Escherichia coli. Environ. Microbiol. 2013, 15, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Ferenci, T.; Galbiati, H.F.; Betteridge, T.; Phan, K.; Spira, B. The constancy of global regulation across a species: The concentrations of ppGpp and RpoS are strain-specific in Escherichia coli. BMC Microbiol. 2011, 11, 62. [Google Scholar] [CrossRef] [PubMed]
- Axtell, C.; Beattie, G. Construction and Characterization of a proU-gfp Transcriptional Fusion That Measures Water Availability in a Microbial Habitat. Appl. Environ. Microbiol. 2002, 68, 4604–4612. [Google Scholar] [CrossRef]
- Davidson, C.J.; White, A.P.; Surette, M.G. Evolutionary loss of the rdar morphotype in Salmonella as a result of high mutation rates during laboratory passage. ISME J. 2008, 2, 293–307. [Google Scholar] [CrossRef]
- Kyle, J.L.; Parker, C.T.; Goudeau, D.; Brandl, M.T. Transcriptome analysis of Escherichia coli O157:H7 exposed to lysates of lettuce leaves. Appl. Environ. Microbiol. 2010, 76, 1375–1387. [Google Scholar] [CrossRef]
- Etchells, J.L.; Fabian, F.W.; Jones, I.D. The aerobacter fermentation of cucumbers during salting. Tech. Bull. Mich. State Coll. 1945, 200, 1–56. [Google Scholar]
- Andersson, R. Characteristics of the bacterial flora isolated during spontaneous lactic acid fermentation of carrots and red beets. Lebensm. Wiss. Technol. 1984, 17, 282–286. [Google Scholar]
- Daeschel, M.A.; Andersson, R.E.; Fleming, H.P. Microbial ecology of fermenting plant materials & nbsp. FEMS Microbiol. Lett. 2006, 46, 357–367. [Google Scholar]
- Nicolson, S.W.; Fleming, P.A. Nectar as food for birds: The physiological consequences of drinking dilute sugar solutions. Plant Syst. Evol. 2003, 238, 139–153. [Google Scholar] [CrossRef]
- Donkersley, P.; Rhodes, G.; Pickup, R.W.; Jones, K.C.; Wilson, K. Bacterial communities associated with honeybee food stores are correlated with land use. Ecol. Evol. 2018, 8, 4743–4756. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Pérez, S.; Lievens, B.; Jacquemyn, H.; Herrera, C. Acinetobacter nectaris sp. nov. and Acinetobacter boissieri sp. nov. isolated from floral nectar of wild Mediterranean insect-pollinated plants. Int. J. Syst. Evol. Microbiol. 2013, 63, 1532–1539. [Google Scholar] [CrossRef]
- Kim, P.S.; Shin, N.R.; Kim, J.Y.; Yun, J.H.; Hyun, D.W.; Bae, J.W. Acinetobacter apis sp. nov. isolated from the intestinal tract of a honey bee, Apis mellifera. J. Microbiol. 2014, 52, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Leroy, P.D.; Sabri, A.; Heuskin, S.; Thonart, P.; Lognay, G.; Verheggen, F.J.; Francis, F.; Brostaux, Y.; Felton, G.W.; Haubruge, E. Microorganisms from aphid honeydew attract and enhance the efficacy of natural enemies. Nat. Commun. 2011, 2, 348. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Manda, R.; Gupta, S.; Kumar, S.; Kumar, V. Isolation and characterization of osmotolerant bacteria from Thar Desert of Western Rajasthan (India). Rev. De Biol. Trop. 2013, 61, 1551–1562. [Google Scholar] [CrossRef][Green Version]
- Hrenovic, J.; Ivankovic, T. Survival of Escherichia coli and Acinetobacter junii at various concentrations of sodium chloride. EurAsian J. Biosci. 2009, 3, 144–151. [Google Scholar] [CrossRef]
- Yang, J.R.; Wang, Y.; Chen, H.; Lyu, Y.K. Ammonium removal characteristics of an acid-resistant bacterium Acinetobacter sp. JR1 from pharmaceutical wastewater capable of heterotrophic nitrification-aerobic denitrification. Bioresour. Technol. 2019, 274, 56–64. [Google Scholar] [CrossRef]
- Disayathanoowat, T.; Young, J.P.W.; Helgason, T.; Chantawannakul, P. T-RFLP analysis of bacterial communities in the midguts of Apis mellifera and Apis cerana honey bees in Thailand. FEMS Microbiol. Ecol. 2011, 79, 273–281. [Google Scholar] [CrossRef][Green Version]
- Ahn, J.H.; Hong, I.P.; Bok, J.I.; Kim, B.Y.; Song, J.; Weon, H.Y. Pyrosequencing analysis of the bacterial communities in the guts of honey bees Apis cerana and Apis mellifera in Korea. J. Microbiol. 2012, 50, 735–745. [Google Scholar] [CrossRef]
- Lee, V.T.; Schneewind, O. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 2001, 15, 1725–1752. [Google Scholar] [CrossRef]
- King, L.B.; Pangburn, M.K.; McDaniel, L.S. Serine protease PKF of Acinetobacter baumannii results in serum resistance and suppression of biofilm formation. J. Infect. Dis. 2013, 207, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.; Slamti, L.; Avci, F.; Pier, G.; Maira-Litrán, T. The pgaABCD Locus of Acinetobacter baumannii Encodes the Production of Poly- -1-6-N-Acetylglucosamine, Which Is Critical for Biofilm Formation. J. Bacteriol. 2009, 191, 5953–5963. [Google Scholar] [CrossRef] [PubMed]
- Pour, N.K.; Dusane, D.H.; Dhakephalkar, P.K.; Zamin, F.R.; Zinjarde, S.S.; Chopade, B.A. Biofilm formation by Acinetobacter baumannii strains isolated from urinary tract infection and urinary catheters. FEMS Immunol. Med. Microbiol. 2011, 62, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Djeribi, R.; Bouchloukh, W.; Jouenne, T.; Menaa, B. Characterization of bacterial biofilms formed on urinary catheters. Am. J. Infect. Control 2012, 40, 854–859. [Google Scholar] [CrossRef] [PubMed]
- Medrela-Kuder, E. Seasonal variations in the occurrence of culturable airborne fungi in outdoor and indoor air in Cracow. Int. Biodeterior. Biodegrad. 2003, 52, 203–205. [Google Scholar] [CrossRef]
- Bensch, K.; Braun, U.; Groenewald, J.Z.; Crous, P. The genus Cladosporium. Stud. Mycol. 2012, 72, 1–401. [Google Scholar] [CrossRef]
- Aleklett, K.; Hart, M.; Shade, A. The microbial ecology of flowers: An emerging frontier in phyllosphere research 1. Botany 2014, 92, 281–289. [Google Scholar] [CrossRef]
- Lagowska, B. The biological control perspective of scale insects (Homoptera, Coccinea) on ornamental plants in glasshouses. Wiad. Entomol. 1995, 14, 5–10. [Google Scholar]
- Vallejo, L.F.; Soto, S.U.; Bernal, U. Identification of pathogenic fungi for Lutzomyia sp. (Diptera: Psychodidae), vectors of Lei hmaniasis. Revista Colombia. Entomology 1996, 22, 13–17. [Google Scholar]
- Han, B.; Zhang, H.; Lin, C. Epizootic infection and spatial pattern within epizootic peak period of Cladosporium Sp. to the population of Aleurocanthus Spiniferus. Entomol. J. E. China 1997, 40, 66–70. [Google Scholar]
- Abdel-Baky, N.; Arafat, S.N.; Abdel-Salam, A. Three Cladosporium spp. As Promising Biological Control Candidates for Controlling Whiteflies (Bemisia spp.) In Egypt. Pak. J. Biol. Sci. 1998, 1, 188–195. [Google Scholar]
- Aizenberg-Gershtein, Y.; Izhaki, I.; Halpern, M. Do Honeybees Shape the Bacterial Community Composition in Floral Nectar? PLoS ONE 2013, 8, e67556. [Google Scholar] [CrossRef] [PubMed]
- Martinson, E.; Herre, E.A.; Machado, C.A.; Arnold, A.E. Culture-Free Survey Reveals Diverse and Distinctive Fungal Communities Associated with Developing Figs (Ficus spp.) in Panama. Micro Ecol. 2012, 64, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Heydenreich, B.; Bellinghausen, I.; Konig, B.; Becker, W.M.; Grabbe, S.; Petersen, A.; Saloga, J. Gram-positive bacteria on grass pollen exhibit adjuvant activity inducing inflammatory T cell responses. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2012, 42, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.; Robbins, E.I. Acidophilic and acid-tolerant fungi and yeasts. Hydrobiologia 2000, 433, 91–109. [Google Scholar] [CrossRef]
- Bridžiuvienė, D.; Levinskaitė, L. Fungal tolerance towards copper-based wood preservatives. Biologija 2007, 53. [Google Scholar]
- Zalar, P.; de Hoog, G.S.; Schroers, H.J.; Crous, P.W.; Groenewald, J.Z.; Gunde-Cimerman, N. Phylogeny and ecology of the ubiquitous saprobe Cladosporium sphaerospermum, with descriptions of seven new species from hypersaline environments. Stud. Mycol. 2007, 58, 157–183. [Google Scholar] [CrossRef]
- Moubasher, A.H.; Ismail, M.A.; Hussein, N.A.; Gouda, H.A. Osmophilic/osmotolerant and halophilic/halotolerant mycobiota of soil of Wadi El-Natrun regions, Egypt. J. Basic Appl. Mycol. 2015, 6, 27–42. [Google Scholar]
- Ma, R.; Chen, Q.; Fan, Y.; Wang, Q.; Yao, B. Six new soil-inhabiting Cladosporium species from plateaus in China. Mycologia 2017, 109, 1–17. [Google Scholar] [CrossRef]
- Horner, K.J.; Anagnostropoulos, G.D. Combined effects of water activity, pH and temperature on the growth and spoilage potential of fungi. J. Appl. Bacteriol. 1973, 36, 427–436. [Google Scholar] [CrossRef]
- El-Kady, I.A.; Moubasher, M.H.; Mostafa, M.E. Accumulation of sugar alcohols by filamentous fungi. Folia Microbiol. 1995, 40, 481–486. [Google Scholar] [CrossRef]
- Klungness, L.M.; Peng, Y. A scanning electron microscopic study of pollen loads collected and stored by honeybees. J. Apic. Res. 1983, 22, 264–271. [Google Scholar] [CrossRef]
- Howell, C.R. Mechanisms Employed by Trichoderma Species in the Biological Control of Plant Diseases: The History and Evolution of Current Concepts. Plant Dis. 2003, 87, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Bunbury-Blanchette, A.; Walker, A. Trichoderma species show biocontrol potential in dual culture and greenhouse bioassays against Fusarium basal rot of onion. Biol. Control 2018, 130, 127–135. [Google Scholar] [CrossRef]
- Harman, G.E. Myths and Dogmas of Biocontrol Changes in Perceptions Derived from Research on Trichoderma harzinum T-22. Plant Dis. 2000, 84, 377–393. [Google Scholar] [CrossRef]
- Howell, C.R.; Hanson, L.E.; Stipanovic, R.D.; Puckhaber, L.S. Induction of Terpenoid Synthesis in Cotton Roots and Control of Rhizoctonia solani by Seed Treatment with Trichoderma virens. Phytopathology 2000, 90, 248–252. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Disayathanoowat, T.; Li, H.; Supapimon, N.; Suwannarach, N.; Lumyong, S.; Chantawannakul, P.; Guo, J. Different Dynamics of Bacterial and Fungal Communities in Hive-Stored Bee Bread and Their Possible Roles: A Case Study from Two Commercial Honey Bees in China. Microorganisms 2020, 8, 264. https://doi.org/10.3390/microorganisms8020264
Disayathanoowat T, Li H, Supapimon N, Suwannarach N, Lumyong S, Chantawannakul P, Guo J. Different Dynamics of Bacterial and Fungal Communities in Hive-Stored Bee Bread and Their Possible Roles: A Case Study from Two Commercial Honey Bees in China. Microorganisms. 2020; 8(2):264. https://doi.org/10.3390/microorganisms8020264
Chicago/Turabian StyleDisayathanoowat, Terd, HuanYuan Li, Natapon Supapimon, Nakarin Suwannarach, Saisamorn Lumyong, Panuwan Chantawannakul, and Jun Guo. 2020. "Different Dynamics of Bacterial and Fungal Communities in Hive-Stored Bee Bread and Their Possible Roles: A Case Study from Two Commercial Honey Bees in China" Microorganisms 8, no. 2: 264. https://doi.org/10.3390/microorganisms8020264
APA StyleDisayathanoowat, T., Li, H., Supapimon, N., Suwannarach, N., Lumyong, S., Chantawannakul, P., & Guo, J. (2020). Different Dynamics of Bacterial and Fungal Communities in Hive-Stored Bee Bread and Their Possible Roles: A Case Study from Two Commercial Honey Bees in China. Microorganisms, 8(2), 264. https://doi.org/10.3390/microorganisms8020264






