Identification and Heterologous Expression of the Albucidin Gene Cluster from the Marine Strain Streptomyces Albus Subsp. Chlorinus NRRL B-24108
Abstract
1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
2.2. Isolation and Manipulation of DNA
2.3. Metabolite Extraction and Analysis
2.4. Chemical Mutagenesis
2.5. Albucidin Isolation and 1H-NMR Spectroscopy
2.6. Construction of the 1K1 BAC Derivatives
2.7. Genome Mining and Bioinformatics Analysis
3. Results and Discussion
3.1. Identification of the Albucidin Biosynthetic Gene Cluster
3.2. Identification of the Minimal Set of Albucidin Biosynthetic Genes
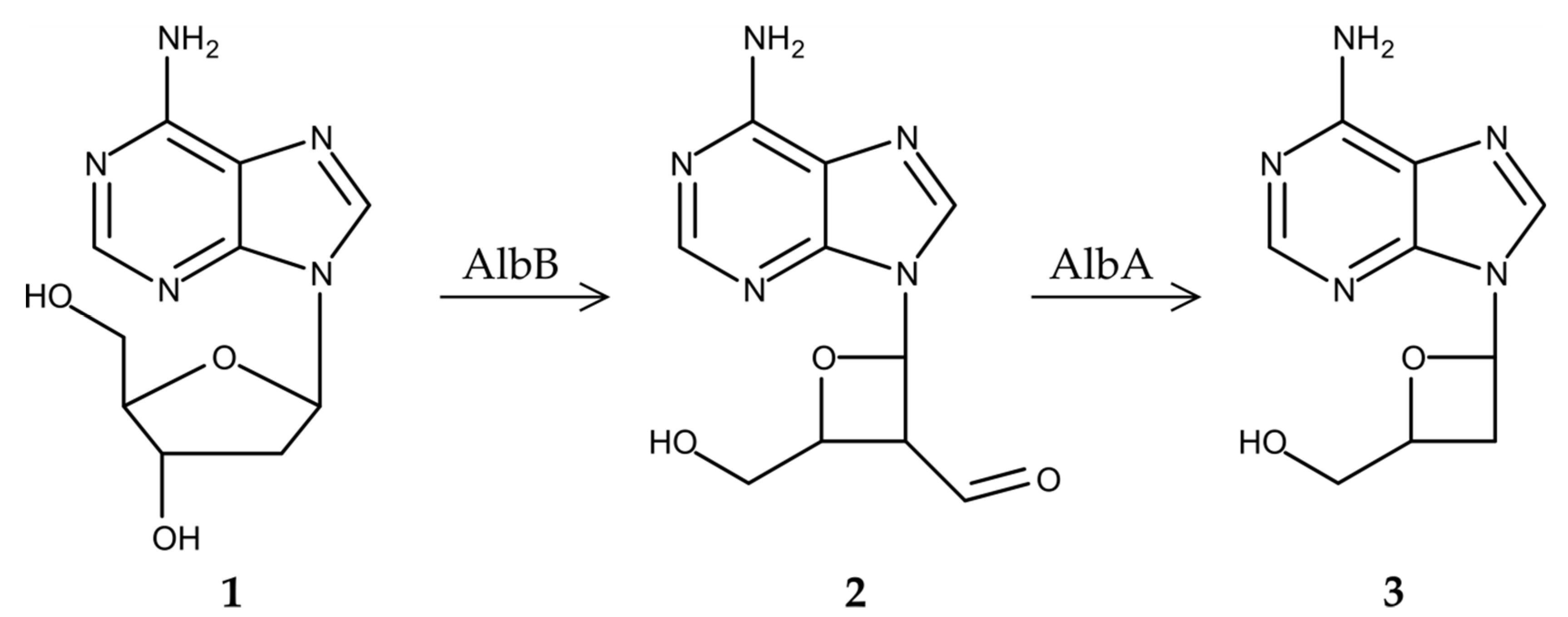
3.3. Proposed Biosynthetic Pathway of Albucidin
4. Patents
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Malakof, D.; Stokstad, E. Infographic: Pesticide Planet. Science 2013, 341, 730–731. [Google Scholar]
- Pimentel, D.; Zuniga, R.; Morrison, D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol. Econ. 2005, 52, 273–288. [Google Scholar] [CrossRef]
- Dayan, F.E.; Duke, S.O. Natural compounds as next-generation herbicides. Plant Physiol. 2014, 166, 1090–1105. [Google Scholar] [CrossRef]
- Duke, S.O. Why have no new herbicide modes of action appeared in recent years? Pest Manag. Sci. 2012, 68, 505–512. [Google Scholar] [CrossRef]
- Heap, I. Global perspective of herbicide-resistant weeds. Pest Manag. Sci. 2014, 70, 1306–1315. [Google Scholar] [CrossRef]
- Cantrell, C.L.; Dayan, F.E.; Duke, S.O. Natural products as sources for new pesticides. J. Nat. Prod. 2012, 75, 1231–1242. [Google Scholar] [CrossRef]
- Duke, S.O.; Stidham, M.A.; Dayan, F.E. A novel genomic approach to herbicide and herbicide mode of action discovery. Pest Manag. Sci. 2019, 75, 314–317. [Google Scholar] [CrossRef]
- Harvey, A.L. Natural products as a screening resource. Curr. Opin. Chem. Biol. 2007, 11, 480–484. [Google Scholar] [CrossRef]
- Koch, M.A.; Schuffenhauer, A.; Scheck, M.; Wetzel, S.; Casaulta, M.; Odermatt, A.; Ertl, P.; Waldmann, H. Charting biologically relevant chemical space: A structural classification of natural products (SCONP). Proc. Natl. Acad. Sci. USA 2005, 102, 17272–17277. [Google Scholar] [CrossRef]
- Hahn, D.R.; Graupner, P.R.; Chapin, E.; Gray, J.; Heim, D.; Gilbert, J.R.; Gerwick, B.C. Albucidin: A novel bleaching herbicide from Streptomyces albus subsp. chlorinus NRRL B-24108. J. Antibiot. 2009, 62, 191–194. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012. [Google Scholar]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Rebets, Y.; Ostash, B.; Luzhetskyy, A.; Hoffmeister, D.; Brana, A.; Mendez, C.; Salas, J.A.; Bechthold, A.; Fedorenko, V. Production of landomycins in Streptomyces globisporus 1912 and S. cyanogenus S136 is regulated by genes encoding putative transcriptional activators. FEMS Microbiol. Lett. 2003, 222, 149–153. [Google Scholar] [CrossRef]
- Rodríguez Estévez, M.; Myronovskyi, M.; Gummerlich, N.; Nadmid, S.; Luzhetskyy, A. Heterologous Expression of the Nybomycin Gene Cluster from the Marine Strain Streptomyces albus subsp. chlorinus NRRL B-24108. Mar. Drugs 2018, 16, 435. [Google Scholar]
- Mazodier, P.; Petter, R.; Thompson, C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J. Bacteriol. 1989, 171, 3583–3585. [Google Scholar] [CrossRef] [PubMed]
- Myronovskyi, M.; Brötz, E.; Rosenkränzer, B.; Manderscheid, N.; Tokovenko, B.; Rebets, Y.; Luzhetskyy, A. Generation of new compounds through unbalanced transcription of landomycin A cluster. Appl. Microbiol. Biotechnol. 2016, 100, 9175–9186. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Medema, M.H.; Kottmann, R.; Lee, S.Y.; Weber, T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2017, 45, D555–D559. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Myronovskyi, M.; Rosenkränzer, B.; Nadmid, S.; Pujic, P.; Normand, P.; Luzhetskyy, A. Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metab. Eng. 2018, 49, 316–324. [Google Scholar] [CrossRef]
- Rückert, C.; Albersmeier, A.; Busche, T.; Jaenicke, S.; Winkler, A.; Friðjónsson, Ó.H.; Hreggviðsson, G.Ó.; Lambert, C.; Badcock, D.; Bernaerts, K.; et al. Complete genome sequence of Streptomyces lividans TK24. J. Biotechnol. 2015, 199, 21–22. [Google Scholar] [CrossRef]
- Siegl, T.; Tokovenko, B.; Myronovskyi, M.; Luzhetskyy, A. Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab. Eng. 2013, 19, 98–106. [Google Scholar] [CrossRef]
- Shimada, N.; Hasegawa, S.; Harada, T.; Tomisawa, T.; Fujii, A.; Takita, T. Oxetanocin, a novel nucleoside from bacteria. J. Antibiot. 1986, 39, 1623–1625. [Google Scholar] [CrossRef]
- Morita, M.; Tomita, K.; Ishizawa, M.; Takagi, K.; Kawamura, F.; Takahashi, H.; Morino, T. Cloning of oxetanocin A biosynthetic and resistance genes that reside on a plasmid of Bacillus megaterium strain NK84-0128. Biosci. Biotechnol. Biochem. 1999, 63, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Bridwell-Rabb, J.; Zhong, A.; Sun, H.G.; Drennan, C.L.; Liu, H.-W. A B12-dependent radical SAM enzyme involved in oxetanocin A biosynthesis. Nature 2017, 544, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Bridwell-Rabb, J.; Kang, G.; Zhong, A.; Liu, H.-W.; Drennan, C.L. An HD domain phosphohydrolase active site tailored for oxetanocin-A biosynthesis. Proc. Natl. Acad. Sci. USA 2016, 113, 13750–13755. [Google Scholar] [CrossRef] [PubMed]
- Grant, S.G.; Jessee, J.; Bloom, F.R.; Hanahan, D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 1990, 87, 4645–4649. [Google Scholar] [CrossRef]
- Flett, F.; Mersinias, V.; Smith, C.P. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 1997, 155, 223–229. [Google Scholar] [CrossRef]


| Gene | Locus Tag 1 | Putative Function |
|---|---|---|
| 1 | SACHL2_05539 | Hypothetical protein |
| 2 | SACHL2_05538 | ABC transporter |
| 3 | SACHL2_05537 | Transcriptional regulatory protein LiaR |
| 4 | SACHL2_05536 | Hypothetical protein |
| 5 | SACHL2_05535 | Hypothetical protein |
| 6 | SACHL2_05534 | beta-lactamase/D-alanine carboxypeptidase |
| 7 | SACHL2_05533 | Chondramide synthase |
| 8 | SACHL2_05532 | Hypothetical protein |
| 9 | SACHL2_05531 | Hypothetical protein |
| 10 | SACHL2_05530 | Thymidylate kinase |
| 11 | SACHL2_05529 | Pyruvate, phosphate dikinase |
| 12 | SACHL2_05528 | Hypothetical protein |
| 13 | SACHL2_05527 | Hypothetical protein |
| 14 | SACHL2_05526 | Hypothetical protein |
| 15; albA | SACHL2_05525 | Biotin synthase, radical SAM protein |
| 16; albB | SACHL2_05524 | Radical SAM protein |
| 17; albC | SACHL2_05523 | Ribonucleoside-triphosphate reductase |
| 18 | SACHL2_05522 | Tyrocidine synthase 3 |
| 19 | SACHL2_05521 | Plipastatin synthase, subunit A |
| 20 | SACHL2_05520 | Acyl carrier protein |
| 21 | SACHL2_05519 | Demethylmenaquinone methyltransferase |
| 22 | SACHL2_05518 | Linear gramicidin synthase, subunit D |
| 23 | SACHL2_05517 | Hypothetical protein |
| 24 | SACHL2_05516 | Acyl carrier protein |
| 25 | SACHL2_05515 | Fatty-acid--CoA ligase |
| 26 | SACHL2_05514 | Ribonucleotide-diphosphate reductase |
| 27 | SACHL2_05513 | Hypothetical protein |
| 28 | SACHL2_05512 | Tyrocidine synthase 3 |
| 29 | SACHL2_05511 | Hypothetical protein |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Myronovskyi, M.; Rosenkränzer, B.; Stierhof, M.; Petzke, L.; Seiser, T.; Luzhetskyy, A. Identification and Heterologous Expression of the Albucidin Gene Cluster from the Marine Strain Streptomyces Albus Subsp. Chlorinus NRRL B-24108. Microorganisms 2020, 8, 237. https://doi.org/10.3390/microorganisms8020237
Myronovskyi M, Rosenkränzer B, Stierhof M, Petzke L, Seiser T, Luzhetskyy A. Identification and Heterologous Expression of the Albucidin Gene Cluster from the Marine Strain Streptomyces Albus Subsp. Chlorinus NRRL B-24108. Microorganisms. 2020; 8(2):237. https://doi.org/10.3390/microorganisms8020237
Chicago/Turabian StyleMyronovskyi, Maksym, Birgit Rosenkränzer, Marc Stierhof, Lutz Petzke, Tobias Seiser, and Andriy Luzhetskyy. 2020. "Identification and Heterologous Expression of the Albucidin Gene Cluster from the Marine Strain Streptomyces Albus Subsp. Chlorinus NRRL B-24108" Microorganisms 8, no. 2: 237. https://doi.org/10.3390/microorganisms8020237
APA StyleMyronovskyi, M., Rosenkränzer, B., Stierhof, M., Petzke, L., Seiser, T., & Luzhetskyy, A. (2020). Identification and Heterologous Expression of the Albucidin Gene Cluster from the Marine Strain Streptomyces Albus Subsp. Chlorinus NRRL B-24108. Microorganisms, 8(2), 237. https://doi.org/10.3390/microorganisms8020237





