Colonization of Germ-Free Piglets with Mucinolytic and Non-Mucinolytic Bifidobacterium boum Strains Isolated from the Intestine of Wild Boar and Their Interference with Salmonella Typhimurium
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Isolation of Wild Pig Indigenous Bifidobacteria
2.3. Identification and Characterization of Pig B. boum Strains
2.4. Mucinolytic Activity of B. boum Strains
2.5. Salmonella Typhimurium Strain LT2
2.6. Bacterial Suspensions
2.7. Gnotobiotic Piglets
2.8. Experimental Design
2.9. Clinical Signs
2.10. Bacterial Colonization of the GIT and Translocation
2.11. Blood Plasma
2.12. Goblet Cells in the Ileum and Colon
2.13. Total RNA Isolation and Reverse Transcription
2.14. Real-Time PCR
2.15. Luminex xMAP Technology
2.16. Statistical Analysis
3. Results
3.1. Isolation, Characterization, and Selection of Pig Indigenous Bifidobacterium boum Strains
3.2. Clinical Signs of Salmonellosis
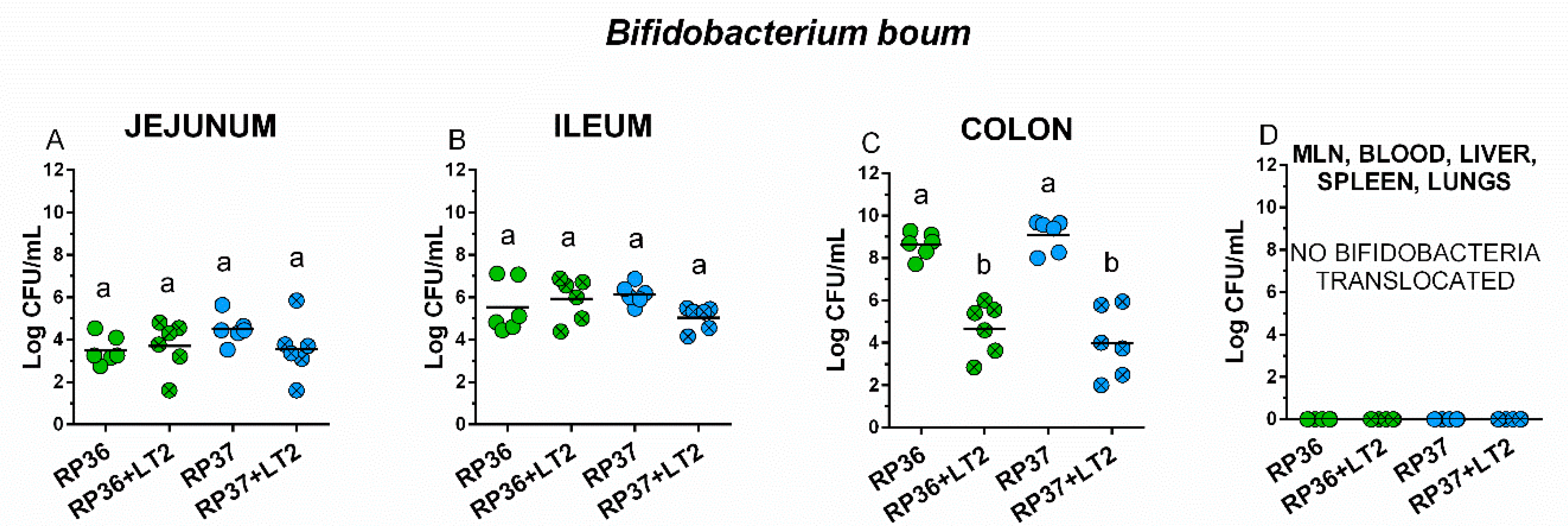
3.3. Colonization of the Intestine with Bifidobacterium boum RP36 and RP37 Strains, Their Translocation to Mesenteric Lymph Nodes, Blood, Liver, Spleen, and Lungs and Interference with Salmonella Typhimurium
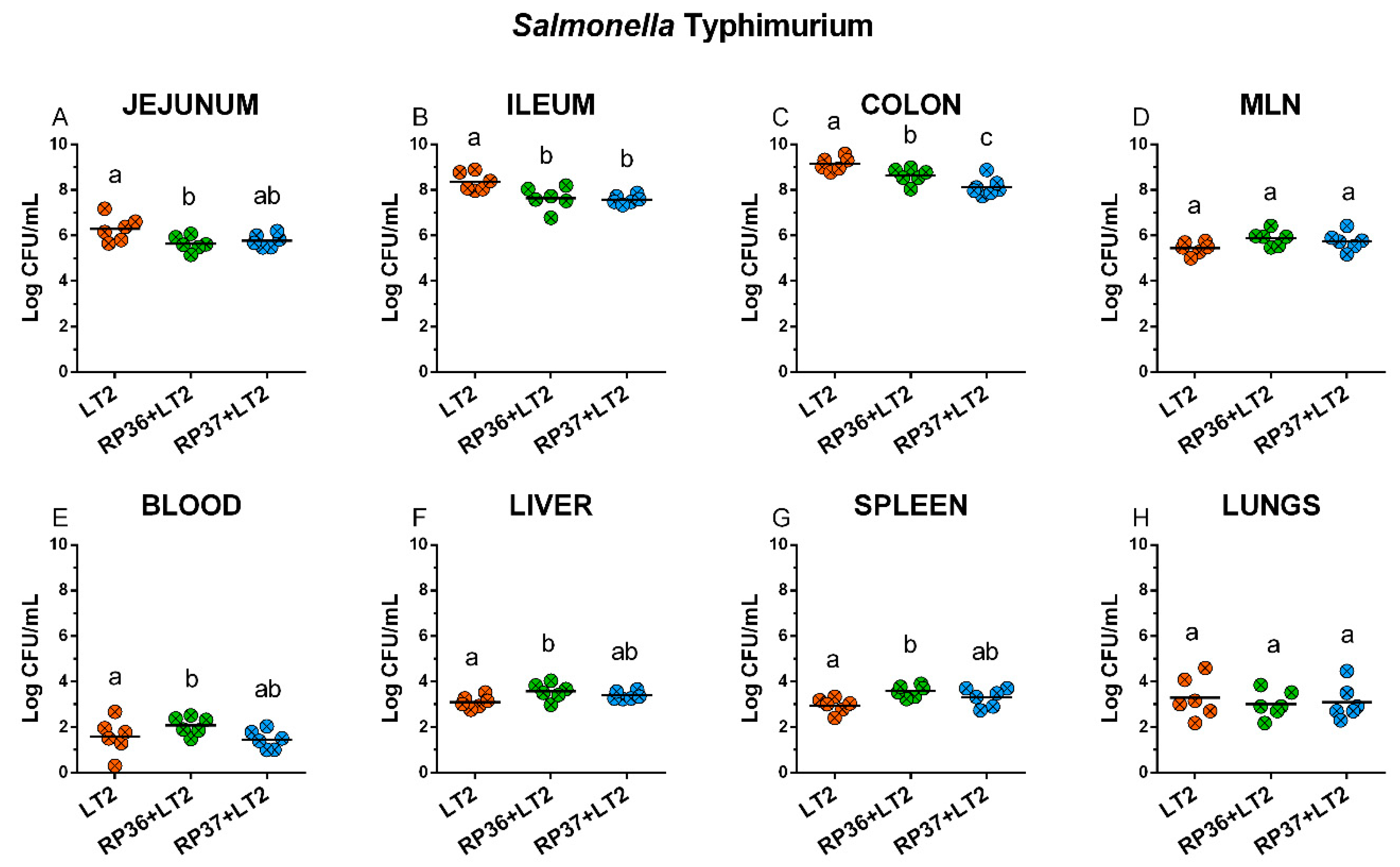
3.4. Colonization of the Intestine with Salmonella Typhimurium, Its Translocation to Mesenteric Mesenteric Lymph Nodes, Blood, Liver, Spleen, and Lungs, and Interference with Bifidobacterium boum RP36 and RP37 Strains
3.5. Density of Goblet Cells in the Ileum and Colon
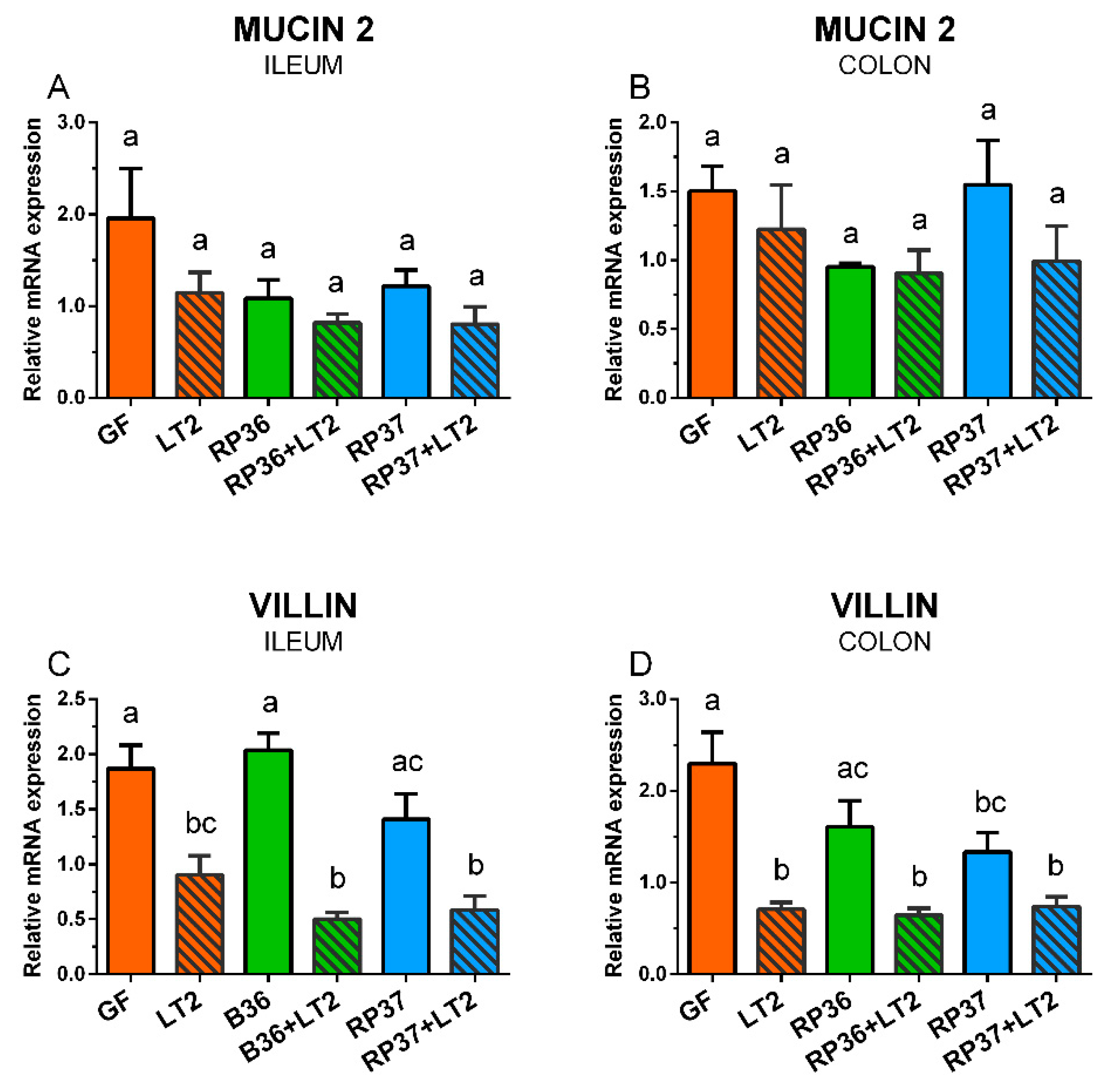
3.6. Expression of Mucin 2 and Villin mRNA in the Ileum and Colon
3.7. Expression of Tight Junction Protein mRNA in the Ileum and Colon
3.8. Plasmatic Levels of Inflammatory Cytokines IL-8, TNF-α, and IL-10
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sommer, F.; Backhed, F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Scarpellini, E.; Ianiro, G.; Attili, F.; Bassanelli, C.; De, S.A.; Gasbarrini, A. The human gut microbiota and virome: Potential therapeutic implications. Dig. Liver Dis. 2015, 47, 1007–1012. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Moissl-Eichinger, C.; Pausan, M.; Taffner, J.; Berg, G.; Bang, C.; Schmitz, R.A. Archaea are interactive components of complex microbiomes. Trends Microbiol. 2018, 26, 70–85. [Google Scholar] [CrossRef] [PubMed]
- Kapitan, M.; Niemiec, M.J.; Steimle, A.; Frick, J.S.; Jacobsen, I.D. Fungi as Part of the Microbiota and Interactions with Intestinal Bacteria. Curr. Top. Microbiol. Immunol. 2019, 422, 265–301. [Google Scholar] [CrossRef] [PubMed]
- Chabe, M.; Lokmer, A.; Segurel, L. Gut Protozoa: Friends or Foes of the Human Gut Microbiota? Trends Parasitol. 2017, 33, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Swain Ewald, H.A.; Ewald, P.W. Natural Selection, The Microbiome, and Public Health. Yale J. Biol. Med. 2018, 91, 445–455. [Google Scholar]
- Schroeder, B.O.; Backhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef]
- Turroni, F.; Milani, C.; Duranti, S.; Lugli, G.A.; Bernasconi, S.; Margolles, A.; Di Pierro, F.; van Sinderen, D.; Ventura, M. The infant gut microbiome as a microbial organ influencing host well-being. Ital. J. Pediatr. 2020, 46, 16. [Google Scholar] [CrossRef]
- Rutayisire, E.; Huang, K.; Liu, Y.; Tao, F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: A systematic review. BMC. Gastroenterol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Wang, M.; Radlowski, E.C.; Monaco, M.H.; Fahey, G.C., Jr.; Gaskins, H.R.; Donovan, S.M. Mode of delivery and early nutrition modulate microbial colonization and fermentation products in neonatal piglets. J. Nutr. 2013, 143, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Donovan, S.M. Evolution of the gut microbiome in infancy within an ecological context. Curr. Opin. Clin. Nutr. Metab. Care 2020, 23, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.C.; Dinsmoor, A.M.; Wang, M.; Donovan, S.M. Microbiome composition in pediatric populations from birth to adolescence: Impact of diet and prebiotic and probiotic interventions. Dig. Dis. Sci. 2020, 65, 706–722. [Google Scholar] [CrossRef]
- Trebichavsky, I.; Rada, V.; Splichalova, A.; Splichal, I. Cross-talk of human gut with bifidobacteria. Nutr. Rev. 2009, 67, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Peano, C.; Pass, D.A.; Foroni, E.; Severgnini, M.; Claesson, M.J.; Kerr, C.; Hourihane, J.; Murray, D.; Fuligni, F.; et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 2012, 7, e36957. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Milani, C.; Duranti, S.; Ferrario, C.; Lugli, G.A.; Mancabelli, L.; van Sinderen, D.; Ventura, M. Bifidobacteria and the infant gut: An example of co-evolution and natural selection. Cell Mol. Life Sci. 2018, 75, 103–118. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Barba-Vidal, E.; Castillejos, L.; Lopez-Colom, P.; Rivero, U.M.; Moreno Munoz, J.A.; Martin-Orue, S.M. Evaluation of the probiotic strain Bifidobacterium longum subsp. infantis CECT 7210 capacities to improve health status and fight digestive pathogens in a piglet model. Front. Microbiol. 2017, 8, 533. [Google Scholar] [CrossRef]
- Delcaru, C.; Alexandru, I.; Podgoreanu, P.; Cristea, V.C.; Bleotu, C.; Chifiriuc, M.C.; Bezirtzoglou, E.; Lazar, V. Antagonistic activities of some Bifidobacterium sp. strains isolated from resident infant gastrointestinal microbiota on Gram-negative enteric pathogens. Anaerobe 2016, 39, 39–44. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Ficara, M.; Pietrella, E.; Spada, C.; Della Casa, M.E.; Lucaccioni, L.; Iughetti, L.; Berardi, A. Changes of intestinal microbiota in early life. J. Matern. Fetal Neonatal Med. 2020, 33, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.; Mitchell, A.L.; Boland, M.; Forster, S.C.; Gloor, G.B.; Tarkowska, A.; Lawley, T.D.; Finn, R.D. A new genomic blueprint of the human gut microbiota. Nature 2019, 568, 499–504. [Google Scholar] [CrossRef]
- Nayfach, S.; Shi, Z.J.; Seshadri, R.; Pollard, K.S.; Kyrpides, N.C. New insights from uncultivated genomes of the global human gut microbiome. Nature 2019, 568, 505–510. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef]
- Allaire, J.M.; Crowley, S.M.; Law, H.T.; Chang, S.Y.; Ko, H.J.; Vallance, B.A. The intestinal epithelium: Central coordinator of mucosal immunity. Trends Immunol. 2018, 1. [Google Scholar] [CrossRef]
- Gunzel, D.; Fromm, M. Claudins and other tight junction proteins. Compr. Physiol. 2012, 2, 1819–1852. [Google Scholar] [CrossRef]
- Kim, C.H.; Kim, D.; Ha, Y.; Cho, K.D.; Lee, B.H.; Seo, I.W.; Kim, S.H.; Chae, C. Expression of mucins and trefoil factor family protein-1 in the colon of pigs naturally infected with Salmonella typhimurium. J. Comp. Pathol. 2009, 140, 38–42. [Google Scholar] [CrossRef]
- Linden, S.K.; Florin, T.H.; McGuckin, M.A. Mucin dynamics in intestinal bacterial infection. PLoS ONE 2008, 3, e3952. [Google Scholar] [CrossRef]
- Breschi, A.; Gingeras, T.R.; Guigo, R. Comparative transcriptomics in human and mouse. Nat. Rev. Genet. 2017, 18, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Feng, Q.; Liang, S.; Sonne, S.B.; Xia, Z.; Qiu, X.; Li, X.; Long, H.; Zhang, J.; Zhang, D.; et al. A catalog of the mouse gut metagenome. Nat. Biotechnol. 2015, 33, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Cavaillon, J.M.; Singer, M.; Skirecki, T. Sepsis therapies: Learning from 30 years of failure of translational research to propose new leads. EMBO Mol. Med. 2020, 12, e10128. [Google Scholar] [CrossRef] [PubMed]
- Burrin, D.; Sangild, P.T.; Stoll, B.; Thymann, T.; Buddington, R.; Marini, J.; Olutoye, O.; Shulman, R.J. Translational advances in pediatric nutrition and gastroenterology: New insights from pig models. Annu. Rev. Anim. Biosci. 2020, 8, 321–354. [Google Scholar] [CrossRef]
- Meurens, F.; Summerfield, A.; Nauwynck, H.; Saif, L.; Gerdts, V. The pig: A model for human infectious diseases. Trends Microbiol. 2012, 20, 50–57. [Google Scholar] [CrossRef]
- Waterhouse, A.; Leslie, D.C.; Bolgen, D.E.; Lightbown, S.; Dimitrakakis, N.; Cartwright, M.J.; Seiler, B.; Lightbown, K.; Smith, K.; Lombardo, P.; et al. Modified clinical monitoring assesment criteria for multi-organ failure during bacteremia and sepsis progression in a pig model. Adv. Crit. Care Med. 2018, 1, 2. [Google Scholar]
- Xiao, L.; Estelle, J.; Kiilerich, P.; Ramayo-Caldas, Y.; Xia, Z.; Feng, Q.; Liang, S.; Pedersen, A.O.; Kjeldsen, N.J.; Liu, C.; et al. A reference gene catalogue of the pig gut microbiome. Nat. Microbiol. 2016, 1, 16161. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Paim, F.C.; Kandasamy, S.; Alhamo, M.A.; Fischer, D.D.; Langel, S.N.; Deblais, L.; Kumar, A.; Chepngeno, J.; Shao, L.; et al. Protein malnutrition modifies innate immunity and gene expression by intestinal epithelial cells and human rotavirus infection in neonatal gnotobiotic pigs. mSphere 2017, 2. [Google Scholar] [CrossRef]
- Wang, M.; Donovan, S.M. Human microbiota-associated swine: Current progress and future opportunities. ILAR J. 2015, 56, 63–73. [Google Scholar] [CrossRef]
- Brugiroux, S.; Beutler, M.; Pfann, C.; Garzetti, D.; Ruscheweyh, H.J.; Ring, D.; Diehl, M.; Herp, S.; Lotscher, Y.; Hussain, S.; et al. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol. 2016, 2, 16215. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Sakamoto, K.; Seo, S.U.; Pickard, J.M.; Gillilland, M.G., III; Pudlo, N.A.; Hoostal, M.; Li, X.; Wang, T.D.; Feehley, T.; et al. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science 2017, 356, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Splichal, I.; Donovan, S.M.; Splichalova, Z.; Bunesova, V.N.; Vlkova, E.; Jenistova, V.; Killer, J.; Svejstil, R.; Skrivanova, E.; Splichalova, A. Colonization of germ-free piglets with commensal Lactobacillus amylovorus, Lactobacillus mucosae, and probiotic E. coli Nissle 1917 and their interference with Salmonella Typhimurium. Microorganisms 2019, 7, 273. [Google Scholar] [CrossRef] [PubMed]
- Baumler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef]
- Hurley, D.; McCusker, M.P.; Fanning, S.; Martins, M. Salmonella-host interactions—Modulation of the host innate immune system. Front. Immunol. 2014, 5, 481. [Google Scholar] [CrossRef]
- Campos, J.; Mourao, J.; Peixe, L.; Antunes, P. Non-typhoidal Salmonella in the pig production chain: A comprehensive analysis of Its impact on human health. Pathogens 2019, 8, 19. [Google Scholar] [CrossRef]
- Kaiser, P.; Hardt, W.D. Salmonella typhimurium diarrhea: Switching the mucosal epithelium from homeostasis to defense. Curr. Opin. Immunol. 2011, 23, 456–463. [Google Scholar] [CrossRef]
- Zhang, S.; Kingsley, R.A.; Santos, R.L.; Andrews-Polymenis, H.; Raffatellu, M.; Figueiredo, J.; Nunes, J.; Tsolis, R.M.; Adams, L.G.; Baumler, A.J. Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect. Immun. 2003, 71, 1–12. [Google Scholar] [CrossRef]
- Wen, S.C.; Best, E.; Nourse, C. Non-typhoidal Salmonella infections in children: Review of literature and recommendations for management. J. Paediatr. Child. Health 2017, 53, 936–941. [Google Scholar] [CrossRef]
- Rada, V.; Petr, J. A new selective medium for the isolation of glucose non-fermenting bifidobacteria from hen caeca. J. Microbiol. Methods 2000, 43, 127–132. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Killer, J.; Sedlacek, I.; Rada, V.; Havlik, J.; Kopecny, J. Reclassification of Bifidobacterium stercoris Kim et al. 2010 as a later heterotypic synonym of Bifidobacterium adolescentis. Int. J. Syst. Evol. Microbiol. 2013, 63, 4350–4353. [Google Scholar] [CrossRef] [PubMed]
- Pechar, R.; Rada, V.; Parafati, L.; Musilova, S.; Bunesova, V.; Vlkova, E.; Killer, J.; Mrazek, J.; Kmet, V.; Svejstil, R. Mupirocin-mucin agar for selective enumeration of Bifidobacterium bifidum. Int. J. Food Microbiol. 2014, 191, 32–35. [Google Scholar] [CrossRef]
- Splichal, I.; Rychlik, I.; Splichalova, I.; Karasova, D.; Splichalova, A. Toll-like receptor 4 signaling in the ileum and colon of gnotobiotic piglets infected with Salmonella Typhimurium or Its isogenic rfa mutants. Toxins 2020, 12, 545. [Google Scholar] [CrossRef]
- Mandel, L.; Travnicek, J. The minipig as a model in gnotobiology. Nahrung 1987, 31, 613–618. [Google Scholar] [CrossRef]
- Splichalova, A.; Slavikova, V.; Splichalova, Z.; Splichal, I. Preterm life in sterile conditions: A study on preterm, germ-free piglets. Front. Immunol. 2018, 9, 220. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Pechar, R.; Killer, J.; Mekadim, C.; Geigerova, M.; Rada, V. Classification of culturable bifidobacterial population from colonic samples of wild pigs (Sus scrofa) based on three molecular genetic methods. Curr. Microbiol. 2017, 74, 1324–1331. [Google Scholar] [CrossRef]
- Killer, J.; Marounek, M. Fermentation of mucin by bifidobacteria from rectal samples of humans and rectal and intestinal samples of animals. Folia Microbiol. 2011, 56, 85–89. [Google Scholar] [CrossRef]
- Pechar, R.; Killer, J.; Salmonova, H.; Geigerova, M.; Svejstil, R.; Svec, P.; Sedlacek, I.; Rada, V.; Benada, O. Bifidobacterium apri sp. nov., a thermophilic actinobacterium isolated from the digestive tract of wild pigs (Sus scrofa). Int. J. Syst. Evol. Microbiol. 2017, 67, 2349–2356. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.D.; Kandasamy, S.; Paim, F.C.; Langel, S.N.; Alhamo, M.A.; Shao, L.; Chepngeno, J.; Miyazaki, A.; Huang, H.C.; Kumar, A.; et al. Protein Malnutrition Alters Tryptophan and Angiotensin-Converting Enzyme 2 Homeostasis and Adaptive Immune Responses in Human Rotavirus-Infected Gnotobiotic Pigs with Human Infant Fecal Microbiota Transplant. Clin. Vaccine Immunol. 2017, 24. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brunse, A.; Martin, L.; Rasmussen, T.S.; Christensen, L.; Skovsted, C.M.; Wiese, M.; Khakimov, B.; Pieper, R.; Nielsen, D.S.; Sangild, P.T.; et al. Effect of fecal microbiota transplantation route of administration on gut colonization and host response in preterm pigs. ISME J. 2019, 13, 720–733. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.N.; Jiang, P.; Frokiaer, H.; Heegaard, P.M.; Thymann, T.; Sangild, P.T. Delayed development of systemic immunity in preterm pigs as a model for preterm infants. Sci. Rep. 2016, 6, 36816. [Google Scholar] [CrossRef]
- Lamendella, R.; Santo Domingo, J.W.; Kelty, C.; Oerther, D.B. Bifidobacteria in feces and environmental waters. Appl. Environ. Microbiol. 2008, 74, 575–584. [Google Scholar] [CrossRef]
- Killer, J.; Mrazek, J.; Bunesova, V.; Havlik, J.; Koppova, I.; Benada, O.; Rada, V.; Kopecny, J.; Vlkova, E. Pseudoscardovia suis gen. nov., sp. nov., a new member of the family Bifidobacteriaceae isolated from the digestive tract of wild pigs (Sus scrofa). Syst. Appl. Microbiol. 2013, 36, 11–16. [Google Scholar] [CrossRef]
- Chattha, K.S.; Vlasova, A.N.; Kandasamy, S.; Rajashekara, G.; Saif, L.J. Divergent immunomodulating effects of probiotics on T cell responses to oral attenuated human rotavirus vaccine and virulent human rotavirus infection in a neonatal gnotobiotic piglet disease model. J. Immunol. 2013, 191, 2446–2456. [Google Scholar] [CrossRef]
- Splichalova, A.; Trebichavsky, I.; Rada, V.; Vlkova, E.; Sonnenborn, U.; Splichal, I. Interference of Bifidobacterium choerinum or Escherichia coli Nissle 1917 with Salmonella Typhimurium in gnotobiotic piglets correlates with cytokine patterns in blood and intestine. Clin. Exp. Immunol. 2011, 163, 242–249. [Google Scholar] [CrossRef]
- Abe, F.; Muto, M.; Yaeshima, T.; Iwatsuki, K.; Aihara, H.; Ohashi, Y.; Fujisawa, T. Safety evaluation of probiotic bifidobacteria by analysis of mucin degradation activity and translocation ability. Anaerobe 2010, 16, 131–136. [Google Scholar] [CrossRef]
- Arguello, H.; Estelle, J.; Zaldivar-Lopez, S.; Jimenez-Marin, A.; Carvajal, A.; Lopez-Bascon, M.A.; Crispie, F.; O’Sullivan, O.; Cotter, P.D.; Priego-Capote, F.; et al. Early Salmonella Typhimurium infection in pigs disrupts microbiome composition and functionality principally at the ileum mucosa. Sci. Rep. 2018, 8, 7788. [Google Scholar] [CrossRef]
- McClelland, M.; Sanderson, K.E.; Spieth, J.; Clifton, S.W.; Latreille, P.; Courtney, L.; Porwollik, S.; Ali, J.; Dante, M.; Du, F.; et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 2001, 413, 852–856. [Google Scholar] [CrossRef] [PubMed]
- Clarke, R.C.; Gyles, C.L. Virulence of wild and mutant strains of Salmonella typhimurium in ligated intestinal segments of calves, pigs, and rabbits. Am. J. Vet. Res. 1987, 48, 504–510. [Google Scholar] [PubMed]
- Perez-Lopez, A.; Behnsen, J.; Nuccio, S.P.; Raffatellu, M. Mucosal immunity to pathogenic intestinal bacteria. Nat. Rev. Immunol. 2016, 16, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.; Phillipson, M.; Petersson, J.; Velcich, A.; Holm, L.; Hansson, G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 15064–15069. [Google Scholar] [CrossRef] [PubMed]
- Linden, S.K.; Sutton, P.; Karlsson, N.G.; Korolik, V.; McGuckin, M.A. Mucins in the mucosal barrier to infection. Mucosal. Immunol. 2008, 1, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Griffiths, G.; Repnik, U.; Hornef, M. Seeing is understanding: Salmonella’s way to penetrate the intestinal epithelium. Int. J. Med. Microbiol. 2018, 308, 97–106. [Google Scholar] [CrossRef]
- Viswanathan, V.K.; Hodges, K.; Hecht, G. Enteric infection meets intestinal function: How bacterial pathogens cause diarrhoea. Nat. Rev. Microbiol. 2009, 7, 110–119. [Google Scholar] [CrossRef]
- West, A.B.; Isaac, C.A.; Carboni, J.M.; Morrow, J.S.; Mooseker, M.S.; Barwick, K.W. Localization of villin, a cytoskeletal protein specific to microvilli, in human ileum and colon and in colonic neoplasms. Gastroenterology 1988, 94, 343–352. [Google Scholar] [CrossRef]
- Lhocine, N.; Arena, E.T.; Bomme, P.; Ubelmann, F.; Prevost, M.C.; Robine, S.; Sansonetti, P.J. Apical invasion of intestinal epithelial cells by Salmonella typhimurium requires villin to remodel the brush border actin cytoskeleton. Cell Host Microbe 2015, 17, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Galen, J.E.; Buskirk, A.D.; Tennant, S.M.; Pasetti, M.F. Live attenuated human Salmonella vaccine candidates: Tracking the pathogen in natural infection and stimulation of host immunity. EcoSal Plus 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Gunzel, D.; Yu, A.S. Claudins and the modulation of tight junction permeability. Physiol. Rev. 2013, 93, 525–569. [Google Scholar] [CrossRef] [PubMed]
- Kiela, P.R.; Ghishan, F.K. Physiology of intestinal absorption and secretion. Best. Pract. Res. Clin. Gastroenterol. 2016, 30, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Al-Sadi, R.; Khatib, K.; Guo, S.; Ye, D.; Youssef, M.; Ma, T. Occludin regulates macromolecule flux across the intestinal epithelial tight junction barrier. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G1054–G1064. [Google Scholar] [CrossRef] [PubMed]
- Edelblum, K.L.; Shen, L.; Weber, C.R.; Marchiando, A.M.; Clay, B.S.; Wang, Y.; Prinz, I.; Malissen, B.; Sperling, A.I.; Turner, J.R. Dynamic migration of gammadelta intraepithelial lymphocytes requires occludin. Proc. Natl. Acad. Sci. USA 2012, 109, 7097–7102. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Zhang, F.; He, Y.; Wang, X.; Jin, X.; Zhang, P.; Bi, D. Enterococcus faecium HDRsEf1 elevates the intestinal barrier defense against enterotoxigenic Escherichia coli and regulates occludin expression via activation of TLR-2 and PI3K signalling pathways. Lett. Appl. Microbiol. 2018, 67, 520–527. [Google Scholar] [CrossRef]
- Kohler, H.; Sakaguchi, T.; Hurley, B.P.; Kase, B.A.; Reinecker, H.C.; McCormick, B.A. Salmonella enterica serovar Typhimurium regulates intercellular junction proteins and facilitates transepithelial neutrophil and bacterial passage. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G178–G187. [Google Scholar] [CrossRef]
- Loetscher, Y.; Wieser, A.; Lengefeld, J.; Kaiser, P.; Schubert, S.; Heikenwalder, M.; Hardt, W.D.; Stecher, B. Salmonella transiently reside in luminal neutrophils in the inflamed gut. PLoS ONE 2012, 7, e34812. [Google Scholar] [CrossRef]
- Foster, N.; Lovell, M.A.; Marston, K.L.; Hulme, S.D.; Frost, A.J.; Bland, P.; Barrow, P.A. Rapid protection of gnotobiotic pigs against experimental salmonellosis following induction of polymorphonuclear leukocytes by avirulent Salmonella enterica. Infect. Immun. 2003, 71, 2182–2191. [Google Scholar] [CrossRef]
- Splichal, I.; Trebichavsky, I.; Splichalova, A.; Barrow, P.A. Protection of gnotobiotic pigs against Salmonella enterica serotype Typhimurium by rough mutant of the same serotype is accompanied by the change of local and systemic cytokine response. Vet. Immunol. Immunopathol. 2005, 103, 155–161. [Google Scholar] [CrossRef]
- Splichalova, A.; Splichalova, Z.; Karasova, D.; Rychlik, I.; Trevisi, P.; Sinkora, M.; Splichal, I. Impact of the lipopolysaccharide chemotype of Salmonella enterica serovar Typhimurium on virulence in gnotobiotic piglets. Toxins 2019, 11, 534. [Google Scholar] [CrossRef]
- Nevola, J.J.; Laux, D.C.; Cohen, P.S. In vivo colonization of the mouse large intestine and in vitro penetration of intestinal mucus by an avirulent smooth strain of Salmonella typhimurium and its lipopolysaccharide-deficient mutant. Infect. Immun. 1987, 55, 2884–2890. [Google Scholar] [CrossRef]
- Dinarello, C.A. Historical insights into cytokines. Eur. J. Immunol. 2007, 37 (Suppl. 1), S34–S45. [Google Scholar] [CrossRef]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Cole, E.; Gillespie, S.; Vulliamy, P.; Brohi, K. Multiple organ dysfunction after trauma. Br. J. Surg. 2020, 107, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Pierrakos, C.; Vincent, J.L. Sepsis biomarkers: A review. Crit. Care 2010, 14, R15. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, H.; Zhou, J.; Zhong, Y.; Ali, M.M.; McGuire, F.; Nagarkatti, P.S.; Nagarkatti, M. Role of cytokines as a double-edged sword in sepsis. In Vivo 2013, 27, 669–684. [Google Scholar] [PubMed]
- Splichal, I.; Splichalova, A. Experimental enteric bacterial infections in pigs. J. Infect. Dis. 2018, 218, 504–505. [Google Scholar] [CrossRef] [PubMed]
- Splichalova, A.; Splichal, I.; Sonnenborn, U.; Rada, V. A modified MacConkey agar for selective enumeration of necrotoxigenic E. coli O55 and probiotic E. coli Nissle 1917. J. Microbiol. Methods 2014, 104, 82–86. [Google Scholar] [CrossRef]
- Splichalova, A.; Splichal, I. Local and systemic occurrences of HMGB1 in gnotobiotic piglets infected with E. coli O55 are related to bacterial translocation and inflammatory cytokines. Cytokine 2012, 60, 597–600. [Google Scholar] [CrossRef]
- Gogos, C.A.; Drosou, E.; Bassaris, H.P.; Skoutelis, A. Pro- versus anti-inflammatory cytokine profile in patients with severe sepsis: A marker for prognosis and future therapeutic options. J. Infect. Dis. 2000, 181, 176–180. [Google Scholar] [CrossRef]





| Gene | 5′-forward primer-3′ | 5′-reverse primer-3′ | #LNA Probe |
|---|---|---|---|
| BACT 1 | TCCCTGGAGAAGAGCTACGA | AAGAGCGCCTCTGGACAC | 9 |
| CYPA 2 | CCTGAAGCATACGGGTCCT | AAAGACCACATGTTTGCCATC | 48 |
| VILLIN | GCATGAAGAAGGTGGAGACC | ACGTTCCTCTTGCCCTTGA | 42 |
| CLD-1 3 | CACCACTTTGCAAGCAACC | TGGCCACAAAGATGGCTATT | 3 |
| CLD-2 4 | CTCGCGCCAAAGACAGAG | ATGAAGATTCCACGCAACG | 77 |
| MUC2 5 OCLN 6 | CCTGCTGCAAGGAGTATCG AAAGAGCTCTCTCGACTGGATAAA | TCTCGATGTGGGTGTAGGTG AGCAGCAGCCATGTACTCTTC | 20 42 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Splichalova, A.; Pechar, R.; Killer, J.; Splichalova, Z.; Bunesova, V.N.; Vlkova, E.; Salmonova, H.S.; Splichal, I. Colonization of Germ-Free Piglets with Mucinolytic and Non-Mucinolytic Bifidobacterium boum Strains Isolated from the Intestine of Wild Boar and Their Interference with Salmonella Typhimurium. Microorganisms 2020, 8, 2002. https://doi.org/10.3390/microorganisms8122002
Splichalova A, Pechar R, Killer J, Splichalova Z, Bunesova VN, Vlkova E, Salmonova HS, Splichal I. Colonization of Germ-Free Piglets with Mucinolytic and Non-Mucinolytic Bifidobacterium boum Strains Isolated from the Intestine of Wild Boar and Their Interference with Salmonella Typhimurium. Microorganisms. 2020; 8(12):2002. https://doi.org/10.3390/microorganisms8122002
Chicago/Turabian StyleSplichalova, Alla, Radko Pechar, Jiri Killer, Zdislava Splichalova, Vera Neuzil Bunesova, Eva Vlkova, Hana Subrtova Salmonova, and Igor Splichal. 2020. "Colonization of Germ-Free Piglets with Mucinolytic and Non-Mucinolytic Bifidobacterium boum Strains Isolated from the Intestine of Wild Boar and Their Interference with Salmonella Typhimurium" Microorganisms 8, no. 12: 2002. https://doi.org/10.3390/microorganisms8122002
APA StyleSplichalova, A., Pechar, R., Killer, J., Splichalova, Z., Bunesova, V. N., Vlkova, E., Salmonova, H. S., & Splichal, I. (2020). Colonization of Germ-Free Piglets with Mucinolytic and Non-Mucinolytic Bifidobacterium boum Strains Isolated from the Intestine of Wild Boar and Their Interference with Salmonella Typhimurium. Microorganisms, 8(12), 2002. https://doi.org/10.3390/microorganisms8122002






