Characterization of the Kenyan Honey Bee (Apis mellifera) Gut Microbiota: A First Look at Tropical and Sub-Saharan African Bee Associated Microbiomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Honey Bee Sample Collection and Preparation
2.2. DNA Extraction
2.3. 16S rRNA Gene Amplification and Sequencing
2.4. 16S rRNA Gut Community Analysis
2.5. qPCR Analysis
3. Results
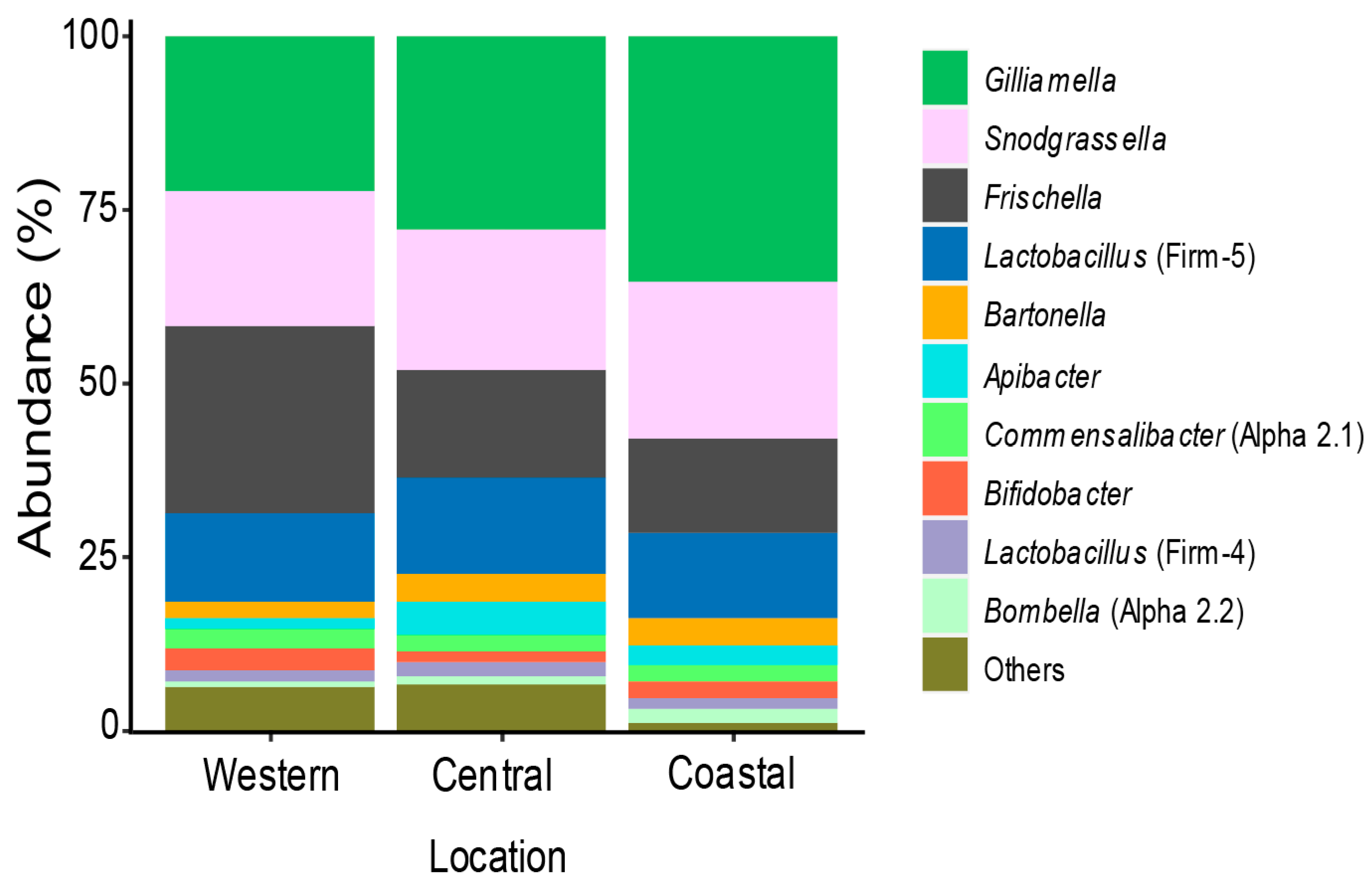
3.1. Bacterial Community Members Associated with Kenyan Apis Mellifera Gut
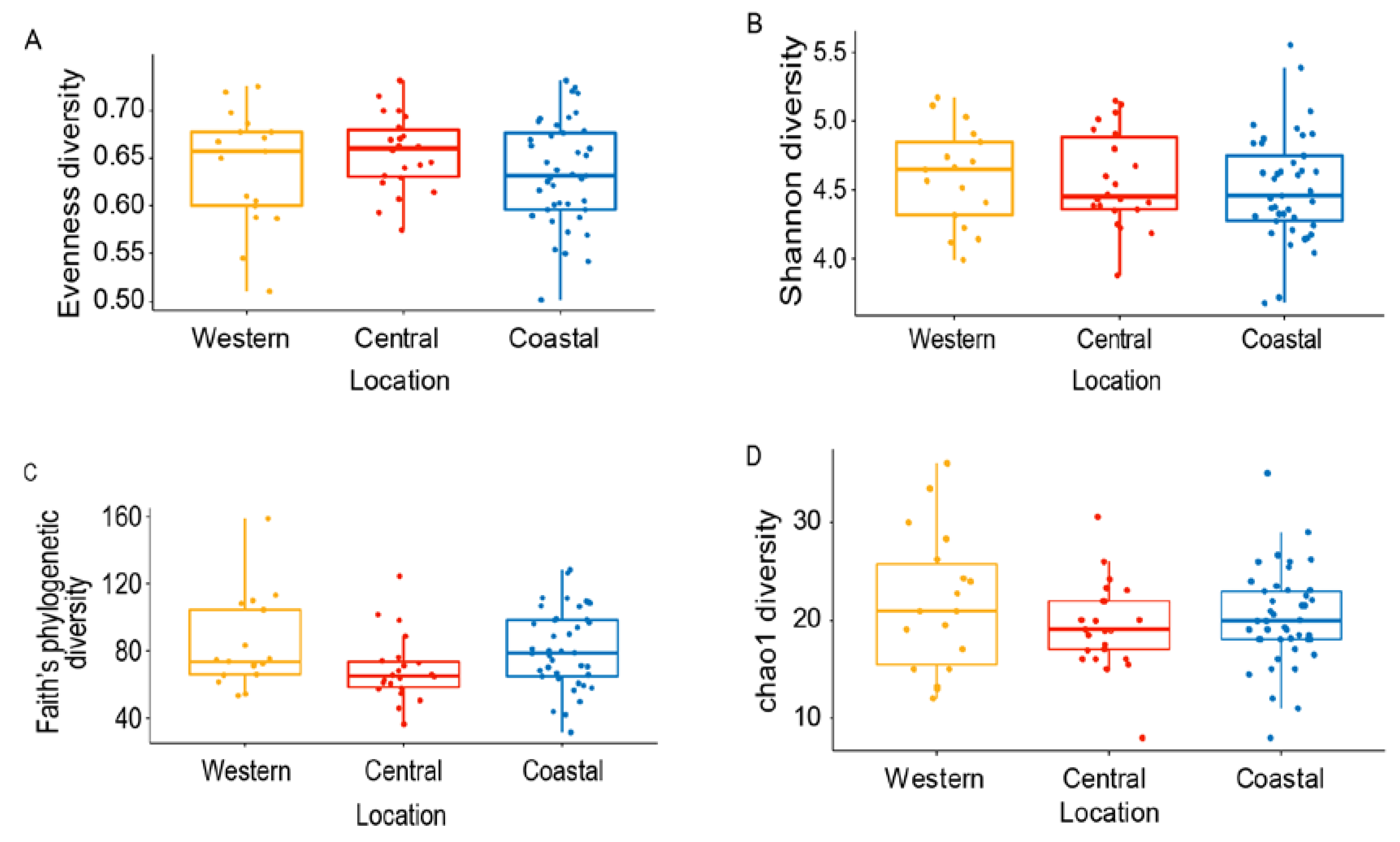
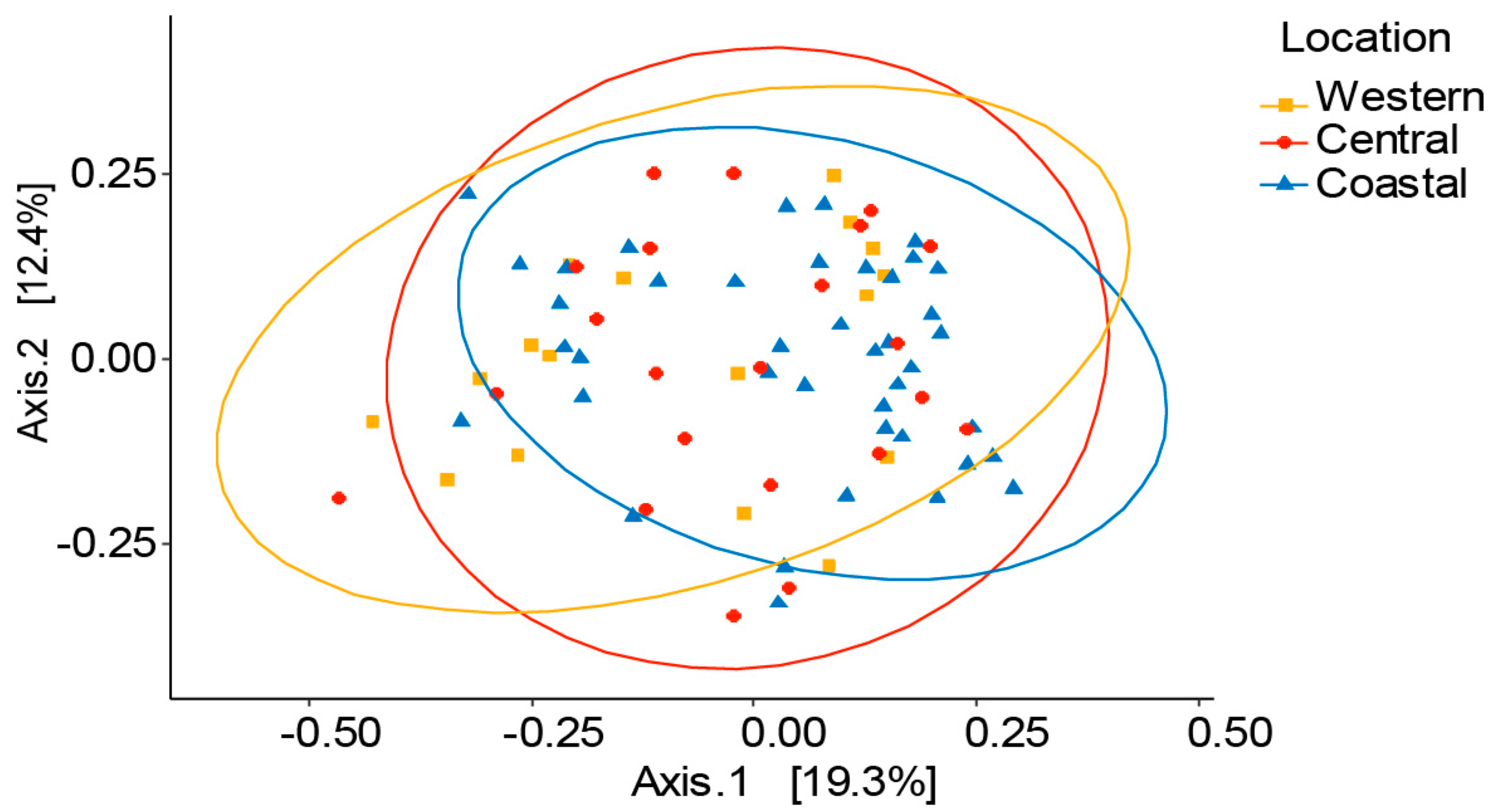
3.2. Variation of Honey Bee Gut Microbiota Across the Locations
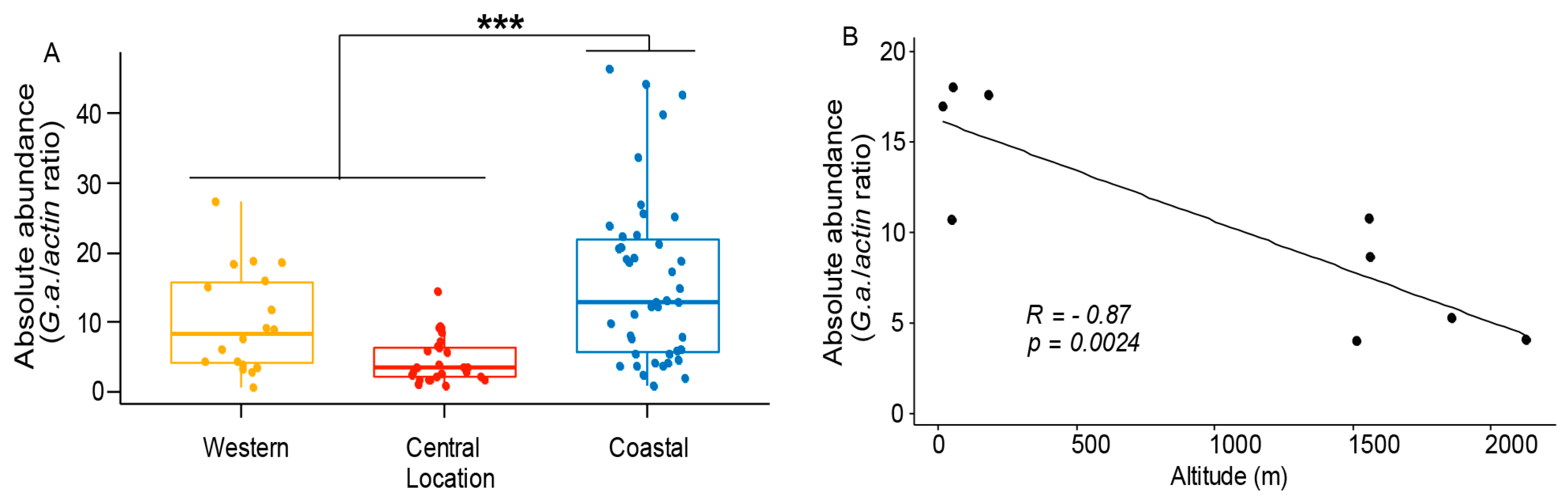
3.3. Gilliamela Abundance is Affected by the Local Environment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carroll, T.; Kinsella, J. Livelihood improvement and smallholder beekeeping in Kenya: The unrealised potential. Dev. Pract. 2013, 23, 332–345. [Google Scholar] [CrossRef]
- Gidey, Y. Assessment of beekeeping practices in Asgede Tsimbla district, Northern Ethiopia: Absconding, bee forage and bee pests. Afr. J. Agric. Res. 2012, 7, 1–5. [Google Scholar] [CrossRef]
- Kuboja, N.M.; Isinika, A.C.; Kilima, F.T.M. Determinants of economic efficiency among small-scale beekeepers in Tabora and Katavi regions, Tanzania: A stochastic profit frontier approach. Dev. Stud. Res. 2017, 4, 1–8. [Google Scholar] [CrossRef][Green Version]
- Ricketts, K.; Shackleton, C.M. Integrating livelihoods and forest conservation through beekeeping in northern KwaZulu-Natal. Dev. S. Afr. 2020, 37, 661–677. [Google Scholar] [CrossRef]
- Breeze, T.D.; Gallai, N.; Garibaldi, L.A.; Li, X.S. Economic Measures of Pollination Services: Shortcomings and Future Directions. Trends Ecol. Evol. 2016, 31, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Garibaldi, L.A.; Carvalheiro, L.G.; Vaissière, B.E.; Gemmill-Herren, B.; Hipólito, J.; Freitas, B.M.; Ngo, H.T.; Azzu, N.; Sáez, A.; Åström, J.; et al. Mutually beneficial pollinator diversity and crop yield outcomes in small and large farms. Science 2016, 351, 388–391. [Google Scholar] [CrossRef]
- Kasina, M.; Kraemer, M.; Martius, C.; Wittmann, D. Diversity and activity density of bees visiting crop flowers in Kakamega, Western Kenya. J. Apic. Res. Bee World 2009, 48, 134–139. [Google Scholar] [CrossRef]
- Warui, M.; Gikungu, M.; Bosselmann, A.; Hansted, L. Pollination of Acacia woodlands and honey production by honey bees in Kitui, Kenya. Future Food J. Food Agric. Soc. 2018, 6, 40–50. [Google Scholar]
- Kasina, J.M.; Mburu, J.; Kraemer, M.; Holm-Mueller, K. Economic Benefit of Crop Pollination by Bees: A Case of Kakamega Small-Holder Farming in Western Kenya. J. Econ. Entomol. 2009, 102, 467–473. [Google Scholar] [CrossRef]
- Awino, O.I.; Skilton, R.; Muya, S.; Kabochi, S.; Kutima, H.; Kasina, M. Varroa mites, viruses and bacteria incidences in Kenyan domesticated honeybee colonies. East Afr. Agric. For. J. 2016, 8325. [Google Scholar] [CrossRef]
- Muli, E.; Patch, H.; Frazier, M.; Frazier, J.; Torto, B.; Baumgarten, T.; Kilonzo, J.; Kimani, J.N.; Mumoki, F.; Masiga, D.; et al. Evaluation of the distribution and impacts of parasites, pathogens, and pesticides on honey bee (Apis mellifera) populations in east Africa. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Moran, N.A.; Tran, P.; Gerardo, N.M. Symbiosis and insect diversification: An ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl. Environ. Microbiol. 2005, 71, 8802–8810. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, L.; Ferreira, Á.; Ashburner, M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008, 6, e1000002. [Google Scholar] [CrossRef] [PubMed]
- Hedges, L.M.; Brownlie, J.C.; O’Neill, S.L.; Johnson, K.N. Wolbachia and virus protection in insects. Science 2008, 322, 702. [Google Scholar] [CrossRef] [PubMed]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef]
- Harumoto, T.; Lemaitre, B. Male-killing toxin in a bacterial symbiont of Drosophila. Nature 2018, 557, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Dillon, R.J.; Vennard, C.T.; Charnley, A.K. Exploitation of gut bacteria in the locust. Nature 2000. [Google Scholar] [CrossRef]
- Wada-Katsumata, A.; Zurek, L.; Nalyanya, G.; Roelofs, W.L.; Zhang, A.; Schal, C. Gut bacteria mediate aggregation in the German cockroach. Proc. Natl. Acad. Sci. USA 2015, 112, 201504031. [Google Scholar] [CrossRef]
- Vernier, C.L.; Chin, I.M.; Adu-Oppong, B.; Krupp, J.J.; Levine, J.D.; Dantas, G.; Ben-Shahar, Y. The gut microbiome defines social group membership in honey bee colonies. Sci. Adv. 2020, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Armstrong, T.-N. Antagonistic interactions between honey bee bacterial symbionts and implications for disease. BMC Ecol. 2006, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Jeyaprakash, A.; Hoy, M.A.; Allsopp, M.H. Bacterial diversity in worker adults of Apis mellifera capensis and Apis mellifera scutellata (Insecta: Hymenoptera) assessed using 16S rRNA sequences. J. Invertebr. Pathol. 2003, 84, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Kešnerová, L.; Mars, R.A.T.; Ellegaard, K.M.; Troilo, M.; Sauer, U.; Engel, P. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 2017, 15, e2003467. [Google Scholar] [CrossRef] [PubMed]
- Raymann, K.; Moran, N.A. The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef]
- Schwarz, R.S.; Moran, N.A.; Evans, J.D. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc. Natl. Acad. Sci. USA 2016, 113, 9345–9350. [Google Scholar] [CrossRef]
- Zheng, H.; Nishida, A.; Kwong, W.K.; Koch, H.; Engel, P.; Steele, M.I.; Moran, N.A. Metabolism of Toxic Sugars by Strains of the Bee Gut Symbiont Gilliamella apicola. MBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Bonilla-Rosso, G.; Engel, P. Functional roles and metabolic niches in the honey bee gut microbiota. Curr. Opin. Microbiol. 2018, 43, 69–76. [Google Scholar] [CrossRef]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.-W.; Jia, E.; Soh, Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic microbiome evolution in social bees. Sci. Adv. 2017. [Google Scholar] [CrossRef]
- Koch, H.; Abrol, D.P.; Li, J.; Schmid-Hempel, P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol. Ecol. 2013, 22, 2028–2044. [Google Scholar] [CrossRef]
- Martinson, V.G.; Danforth, B.N.; Minckley, R.L.; Rueppell, O.; Tingek, S.; Moran, N.A. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 2011, 20, 619–628. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: Description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a memb. Int. J. Syst. Evol. Microbiol. 2013, 63, 2008–2018. [Google Scholar] [CrossRef]
- Kwong, W.K.; Engel, P.; Koch, H.; Moran, N.A. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc. Natl. Acad. Sci. USA 2014, 111, 11509–11514. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Martinson, V.G.; Moran, N.A. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. USA 2012. [Google Scholar] [CrossRef]
- Zheng, H.; Perreau, J.; Elijah Powell, J.; Han, B.; Zhang, Z.; Kwong, W.K.; Tringe, S.G.; Moran, N.A. Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc. Natl. Acad. Sci. USA 2019, 116, 25909–25916. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Ellegaard, K.M.; Tamarit, D.; Javelind, E.; Olofsson, T.C.; Andersson, S.G.E.; Vásquez, A. Extensive intra-phylotype diversity in lactobacilli and bifidobacteria from the honeybee gut. BMC Genom. 2015, 16, 284. [Google Scholar] [CrossRef]
- Moran, N.A.; Hansen, A.K.; Powell, J.E.; Sabree, Z.L. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE 2012, 7, e36393. [Google Scholar] [CrossRef]
- Olofsson, T.C.; Vásquez, A. Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. Curr. Microbiol. 2008, 57, 356–363. [Google Scholar] [CrossRef]
- Vásquez, A.; Forsgren, E.; Fries, I.; Paxton, R.J.; Flaberg, E.; Szekely, L.; Olofsson, T.C. Symbionts as Major Modulators of Insect Health: Lactic Acid Bacteria and Honeybees. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Audisio, M.C. Gram-Positive Bacteria with Probiotic Potential for the Apis mellifera L. Honey Bee: The Experience in the Northwest of Argentina. Probiotics Antimicrob. Proteins 2017, 9, 22–31. [Google Scholar] [CrossRef]
- Evans, J.D.; Lopez, D.L. Bacterial probiotics induce an immune response in the honey bee (Hymenoptera: Apidae). J. Econ. Entomol. 2004, 97, 752–756. [Google Scholar] [CrossRef]
- Forsgren, E.; Olofsson, T.C.; Vásquez, A.; Fries, I. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 2010, 41, 99–108. [Google Scholar] [CrossRef]
- Anderson, K.E.; Sheehan, T.H.; Mott, B.M.; Maes, P.; Snyder, L.; Schwan, M.R.; Walton, A.; Jones, B.M.; Corby-Harris, V. Microbial ecology of the hive and pollination landscape: Bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Engel, P.; Bartlett, K.D.; Moran, A. The Bacterium Frischella perrara Causes Scab Formation in the Gut of its Honeybee Host. Mbio 2015, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Emery, O.; Schmidt, K.; Engel, P. Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera). Mol. Ecol. 2017. [Google Scholar] [CrossRef]
- Kešnerová, L.; Moritz, R.; Engel, P. Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. 2016, 66, 414–421. [Google Scholar] [CrossRef]
- Siozios, S.; Moran, J.; Chege, M.; Hurst, G.D.D.; Paredes, J.C. Complete Reference Genome Assembly for Commensalibacter sp. Strain AMU001, an Acetic Acid Bacterium Isolated from the Gut of Honey Bees. Microbiol. Resour. Announc. 2019, 8, 1–2. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Apibacter adventoris gen. nov., sp. nov., a member of the phylum Bacteroidetes isolated from honey bees. Int. J. Syst. Evol. Microbiol. 2016, 66, 1323–1329. [Google Scholar] [CrossRef]
- Kwong, W.K.; Steele, M.I.; Moran, N.A. Genome Sequences of Apibacter spp., Gut Symbionts of Asian Honey Bees. Genome Biol. Evol. 2018, 10, 1174–1179. [Google Scholar] [CrossRef]
- Praet, J.; Aerts, M.; de Brandt, E.; Meeus, I.; Smagghe, G.; Vandamme, P. Apibacter mensalis sp. Nov.: A rare member of the bumblebee gut microbiota. Int. J. Syst. Evol. Microbiol. 2016, 66, 1645–1651. [Google Scholar] [CrossRef]
- Mockler, B.K.; Kwong, W.K.; Moran, N.A.; Koch, H. Microbiome Structure Influences Infection by the Parasite Crithidia bombi in Bumble Bees. Appl. Environ. Microbiol. 2018, 84, 1–11. [Google Scholar] [CrossRef]
- Ellegaard, K.M.; Engel, P. Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun. 2019, 10, 446. [Google Scholar] [CrossRef]
- Engel, P.; Stepanauskas, R.; Moran, N.A. Hidden Diversity in Honey Bee Gut Symbionts Detected by Single-Cell Genomics. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Bleau, N.; Bouslama, S.; Giovenazzo, P. Dynamics of the Honeybee (Apis mellifera) Gut Microbiota Throughout the Overwintering Period in Canada. Microorganisms 2020, 8, 1146. [Google Scholar] [CrossRef] [PubMed]
- Bosmans, L.; Pozo, M.I.; Verreth, C.; Crauwels, S.; Wäckers, F.; Jacquemyn, H.; Lievens, B. Hibernation leads to altered gut communities in Bumblebee queens (Bombus terrestris). Insects 2018, 9, 188. [Google Scholar] [CrossRef]
- Brosi, B.J.; Daily, G.C.; Shih, T.M.; Oviedo, F.; Durán, G. The effects of forest fragmentation on bee communities in tropical countryside. J. Appl. Ecol. 2008, 45, 773–783. [Google Scholar] [CrossRef]
- Jones, J.C.; Fruciano, C.; Hildebrand, F.; Al Toufalilia, H.; Balfour, N.J.; Bork, P.; Engel, P.; Ratnieks, F.L.W.; Hughes, W.O.H. Gut microbiota composition is associated with environmental landscape in honey bees. Ecol. Evol. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Kešnerová, L.; Emery, O.; Troilo, M.; Liberti, J.; Erkosar, B.; Engel, P. Gut microbiota structure differs between honeybees in winter and summer. ISME J. 2019, 14, 801–814. [Google Scholar] [CrossRef]
- Mattila, H.R.; Rios, D.; Walker-Sperling, V.E.; Roeselers, G.; Newton, I.L.G. Characterization of the active microbiotas associated with honey bees reveals healthier and broader communities when colonies are genetically diverse. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Rothman, J.A.; Carroll, M.J.; Meikle, W.G.; Anderson, K.E.; McFrederick, Q.S. Longitudinal Effects of Supplemental Forage on the Honey Bee (Apis mellifera) Microbiota and Inter- and Intra-Colony Variability. Microb. Ecol. 2018, 76, 814–824. [Google Scholar] [CrossRef]
- D’Alvise, P.; Böhme, F.; Codrea, M.C.; Seitz, A.; Nahnsen, S.; Binzer, M.; Rosenkranz, P.; Hasselmann, M. The impact of winter feed type on intestinal microbiota and parasites in honey bees. Apidologie 2018, 49, 252–264. [Google Scholar] [CrossRef]
- Wang, H.; Liu, C.; Liu, Z.; Wang, Y.; Ma, L.; Xu, B. The different dietary sugars modulate the composition of the gut microbiota in honeybee during overwintering. BMC Microbiol. 2020, 20, 61. [Google Scholar] [CrossRef] [PubMed]
- Wallberg, A.; Schöning, C.; Webster, M.T.; Hasselmann, M. Two extended haplotype blocks are associated with adaptation to high altitude habitats in East African honey bees. PLoS Genet. 2017. [Google Scholar] [CrossRef]
- McMenamin, A.; Mumoki, F.; Frazier, M.; Kilonzo, J.; Mweu, B.; Baumgarten, T.; Patch, H.; Torto, B.; Masiga, D.; Tumlinson, J.; et al. The impact of hive type on the behavior and health of honey bee colonies (Apis mellifera) in Kenya. Apidologie 2017. [Google Scholar] [CrossRef]
- Engel, P.; James, R.R.; Koga, R.; Kwong, W.K.; Mcfrederick, Q.S.; Moran, N.A. Standard methods for research on Apis mellifera gut symbionts. J. Apic. Res. 2013, 524, 1–24. [Google Scholar] [CrossRef]
- Apprill, A.; Mcnally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST plus: Architecture and applications. BMC Bioinform. 2009, 10, 1. [Google Scholar] [CrossRef]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef]
- Sabree, Z.L.; Hansen, A.K.; Moran, N.A. Independent studies using deep sequencing resolve the same set of core bacterial species dominating gut communities of honey bees. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Ma, W.H.; Zheng, X.; Li, L.; Shen, J.; Li, W.; Gao, Y. Changes in the gut microbiota of honey bees associated with jujube flower disease. Ecotoxicol. Environ. Saf. 2020, 198, 110616. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.-H.; Hong, I.-P.; Bok, J.-I.; Kim, B.-Y.; Song, J.; Weon, H.-Y. Pyrosequencing analysis of the bacterial communities in the guts of honey bees Apis cerana and Apis mellifera in Korea. J. Microbiol. 2012, 50, 735–745. [Google Scholar] [CrossRef]
- Khan, K.A.; Ansari, M.J.; Al-Ghamdi, A.; Nuru, A.; Harakeh, S.; Iqbal, J. Investigation of gut microbial communities associated with indigenous honey bee (Apis mellifera jemenitica) from two different eco-regions of Saudi Arabia. Saudi J. Biol. Sci. 2017, 24, 1061–1068. [Google Scholar] [CrossRef]
- Sepulveda, J.; Moeller, A.H. The Effects of Temperature on Animal Gut Microbiomes. Front. Microbiol. 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Ludvigsen, J.; Rangberg, A.; Avershina, E.; Sekelja, M.; Kreibich, C.; Amdam, G.; Rudi, K. Shifts in the Midgut/Pyloric Microbiota Composition within a Honey Bee Apiary throughout a Season. Microbes Environ. 2015, 30, 235–244. [Google Scholar] [CrossRef]
- Motta, E.V.S.; Raymann, K.; Moran, N.A. Glyphosate perturbs the gut microbiota of honey bees. Proc. Natl. Acad. Sci. USA 2018, 115, 10305–10310. [Google Scholar] [CrossRef]
- Rouzé, R.; Moné, A.; Delbac, F.; Belzunces, L.; Blot, N. The honeybee gut microbiota is altered after chronic exposure to different families of insecticides and infection by Nosema ceranae. Microbes Environ. 2019, 34, 226–233. [Google Scholar] [CrossRef]
- Maes, P.W.; Rodrigues, P.A.P.; Oliver, R.; Mott, B.M.; Anderson, K.E. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera). Mol. Ecol. 2016, 25, 5439–5450. [Google Scholar] [CrossRef]
- Kakumanu, M.L.; Reeves, A.M.; Anderson, T.D.; Rodrigues, R.R.; Williams, M.A. Honey bee gut microbiome is altered by in-hive pesticide exposures. Front. Microbiol. 2016, 7, 1–11. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tola, Y.H.; Waweru, J.W.; Hurst, G.D.D.; Slippers, B.; Paredes, J.C. Characterization of the Kenyan Honey Bee (Apis mellifera) Gut Microbiota: A First Look at Tropical and Sub-Saharan African Bee Associated Microbiomes. Microorganisms 2020, 8, 1721. https://doi.org/10.3390/microorganisms8111721
Tola YH, Waweru JW, Hurst GDD, Slippers B, Paredes JC. Characterization of the Kenyan Honey Bee (Apis mellifera) Gut Microbiota: A First Look at Tropical and Sub-Saharan African Bee Associated Microbiomes. Microorganisms. 2020; 8(11):1721. https://doi.org/10.3390/microorganisms8111721
Chicago/Turabian StyleTola, Yosef Hamba, Jacqueline Wahura Waweru, Gregory D. D. Hurst, Bernard Slippers, and Juan C. Paredes. 2020. "Characterization of the Kenyan Honey Bee (Apis mellifera) Gut Microbiota: A First Look at Tropical and Sub-Saharan African Bee Associated Microbiomes" Microorganisms 8, no. 11: 1721. https://doi.org/10.3390/microorganisms8111721
APA StyleTola, Y. H., Waweru, J. W., Hurst, G. D. D., Slippers, B., & Paredes, J. C. (2020). Characterization of the Kenyan Honey Bee (Apis mellifera) Gut Microbiota: A First Look at Tropical and Sub-Saharan African Bee Associated Microbiomes. Microorganisms, 8(11), 1721. https://doi.org/10.3390/microorganisms8111721





