Inter- and Intra-Species Diversity of Lactic Acid Bacteria in Apis mellifera ligustica Colonies
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Processing
2.3. Bacterial Isolation
2.4. Genotypic Characterization
2.4.1. DNA Extraction
2.4.2. Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis (PCR-DGGE) Analysis
2.4.3. DNA Sequencing and Data Processing
2.4.4. RAPD-PCR Analysis
2.5. Biochemical Characterization
3. Results and Discussion
3.1. Genetic Characterization of LAB
3.1.1. Species Diversity (Species Identification)
3.1.2. Species Distribution
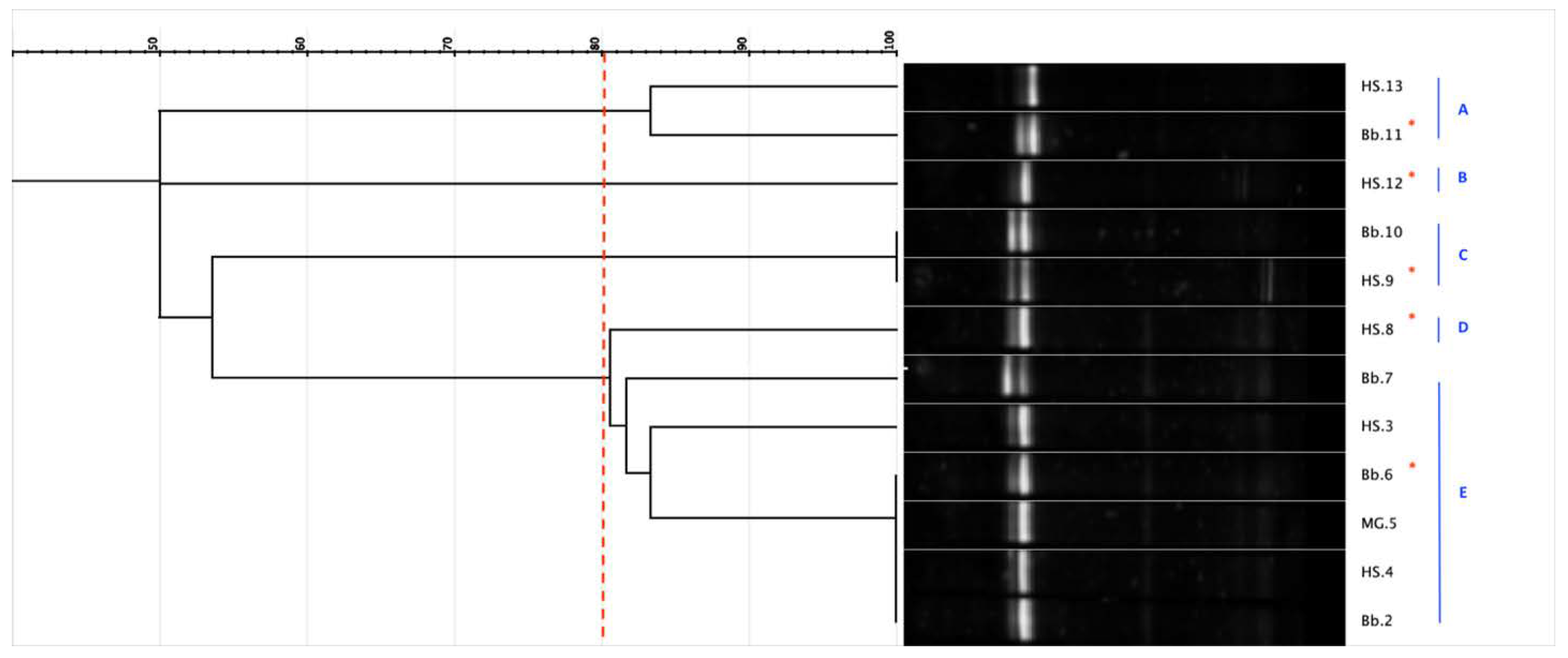
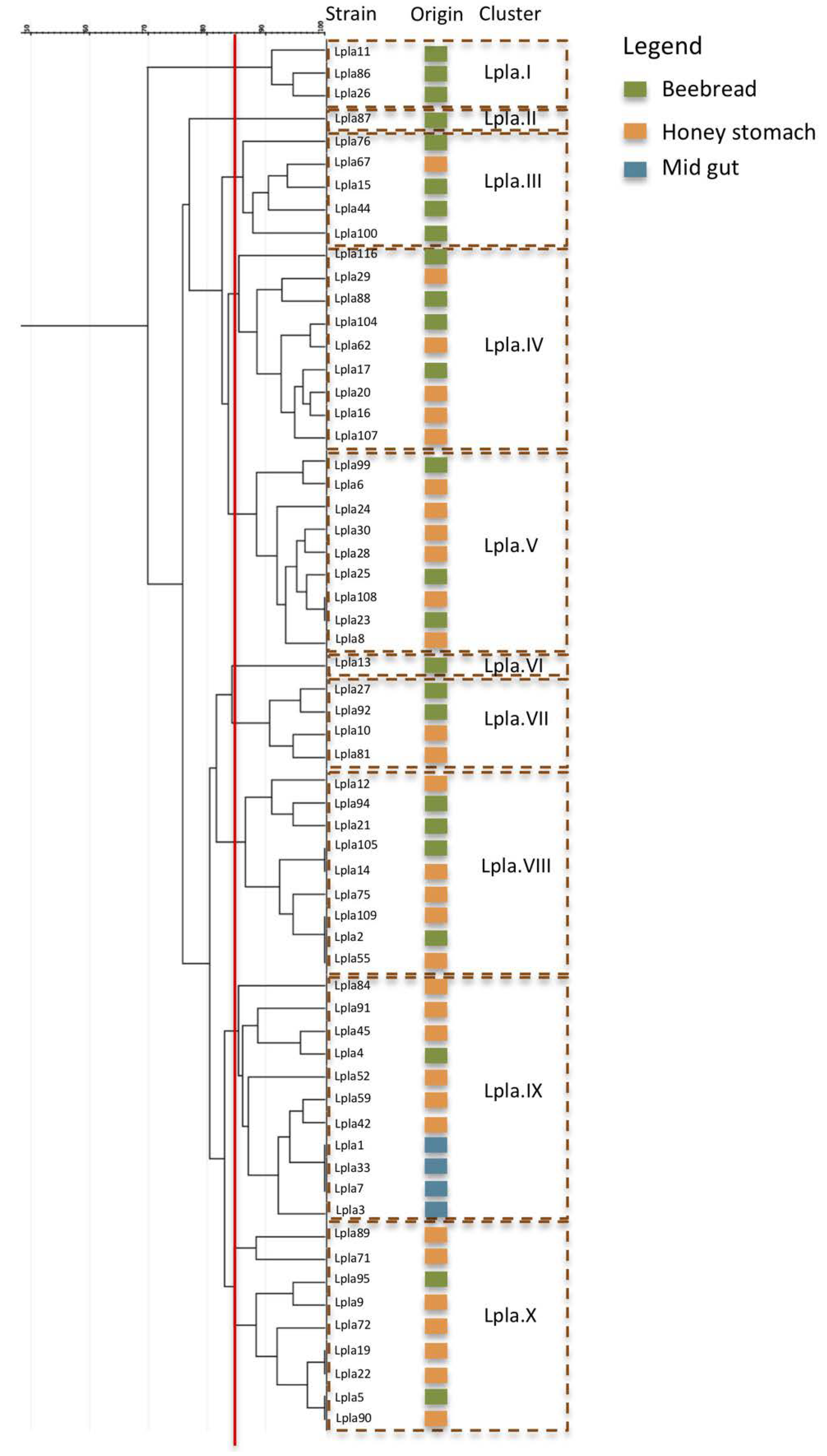
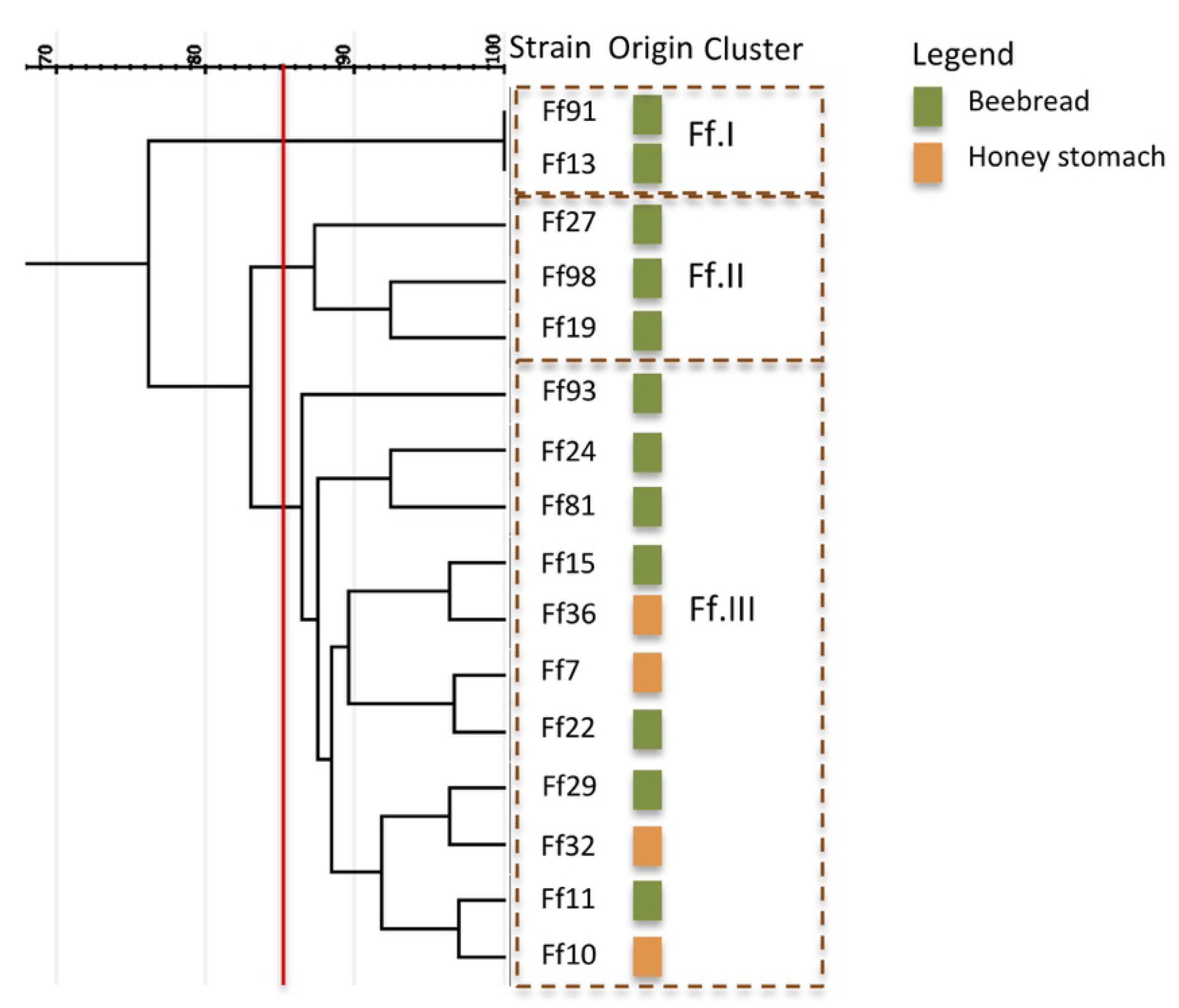
3.1.3. Strain Diversity
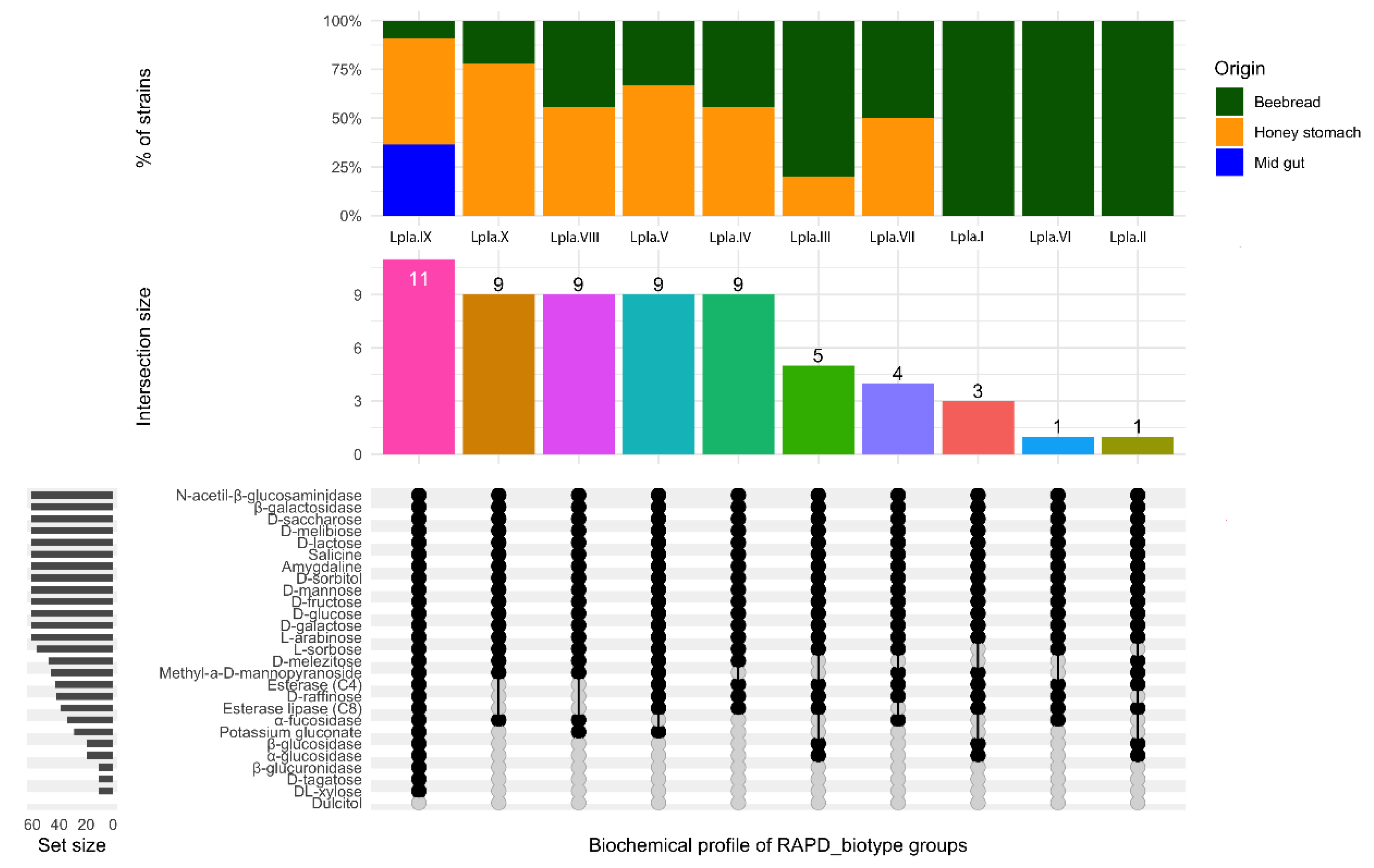
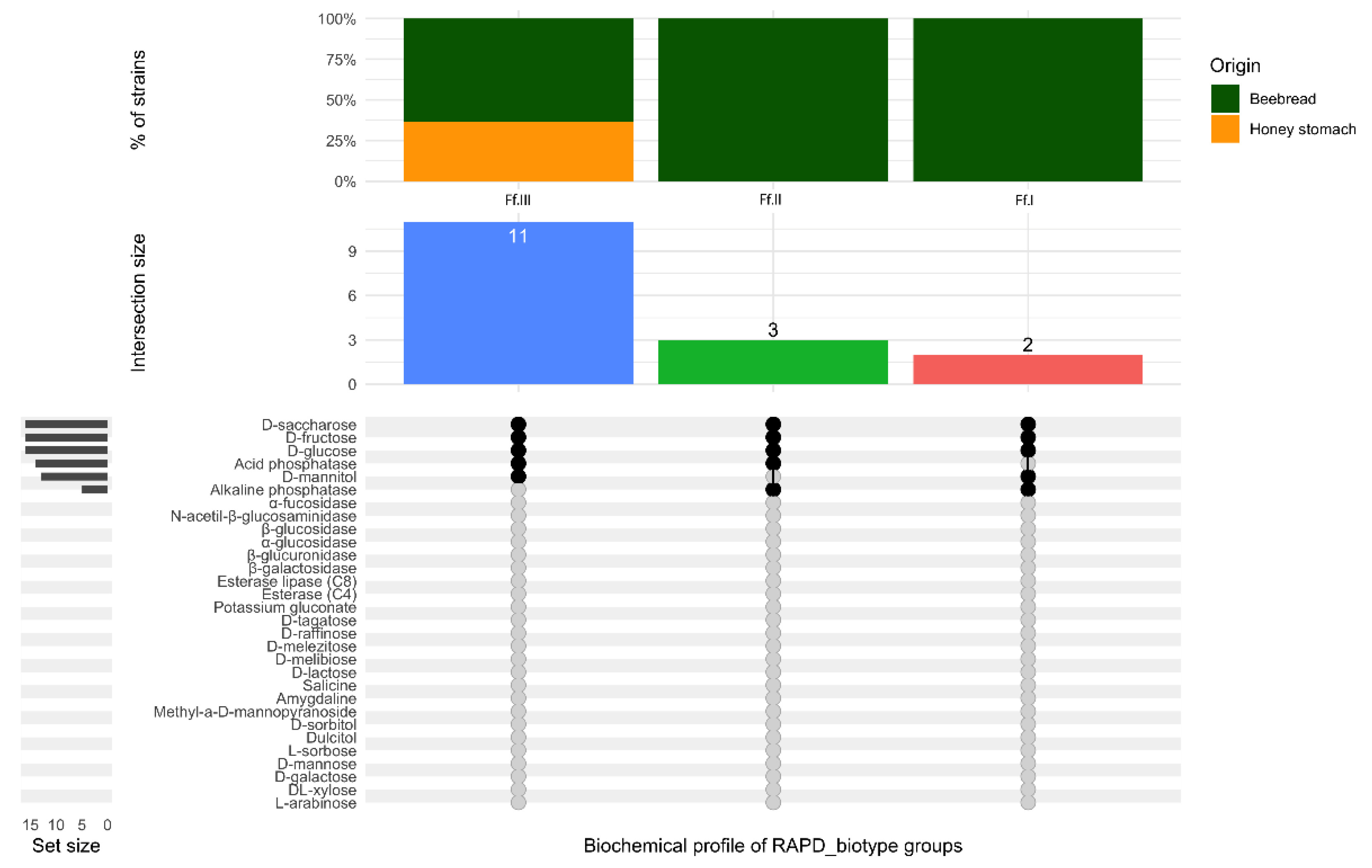
3.2. Biochemical Characterization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kwong, W.K.; Engel, P.; Koch, H.; Moran, N.A. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc. Natl. Acad. Sci. USA 2014, 111, 11509–11514. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Moran, N.A. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of acquisition of the gut microbiota of Apis mellifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387. [Google Scholar] [CrossRef]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.W.; Soh, E.J.Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic microbiome evolution in social bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Steele, M.I.; Leonard, S.P.; Motta, E.V.S.; Moran, N.A. Honey bees as models for gut microbiota research. Lab. Anim. (N. Y.) 2018, 47, 317–325. [Google Scholar] [CrossRef]
- Evans, J.D.; Lopez, D.L. Bacterial probiotics induce an immune response in the honey bee (Hymenoptera: Apidae). J. Econ. Entomol. 2004, 97, 752–756. [Google Scholar] [CrossRef]
- Cox-Foster, D.L.; Conlan, S.; Holmes, E.C.; Palacios, G.; Evans, J.D.; Moran, N.A.; Quan, P.-L.; Briese, T.; Hornig, M.; Geiser, D.M.; et al. A Metagenomic Survey of Microbes in Honey Bee Colony Collapse Disorder. Science 2007, 318, 283–287. [Google Scholar] [CrossRef]
- Evans, J.D.; Schwarz, R.S. Bees brought to their knees: Microbes affecting honey bee health. Trends Microbiol. 2011, 19, 614–620. [Google Scholar] [CrossRef]
- Cornman, R.S.; Tarpy, D.R.; Chen, Y.; Jeffreys, L.; Lopez, D.; Pettis, J.S.; van Engelsdorp, D.; Evans, J.D. Pathogen Webs in Collapsing Honey Bee Colonies. PLoS ONE 2012, 7, e43562. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Martinson, V.G.; Moran, N.A. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. USA 2012, 109, 11002–11007. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, R.S.; Moran, N.A.; Evans, J.D. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc. Natl. Acad. Sci. USA 2016, 113, 9345–9350. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef]
- Vasquez, A.; Forsgren, E.; Fries, I.; Paxton, R.J.; Flaberg, E.; Szekely, L.; Olofsson, T.C. Symbionts as major modulators of insect health: Lactic acid bacteria and honeybees. PLoS ONE 2012, 7, e33188. [Google Scholar] [CrossRef]
- Rokop, Z.P.; Horton, M.A.; Newton, I.L.G. Interactions between cooccurring lactic acid bacteria in honey bee hives. Appl. Environ. Microbiol. 2015, 81, 7261–7270. [Google Scholar] [CrossRef]
- Olofsson, T.C.; Butler, E.; Markowicz, P.; Lindholm, C.; Larsson, L.; Vasquez, A. Lactic acid bacterial symbionts in honey-bees–an unknown key to honey’s antimicrobial and therapeutic activities. Int. Wound J. 2016, 13, 668–679. [Google Scholar] [CrossRef]
- Andersen, J.M.; Barrangou, R.; Hachem, M.A.; Lahtinen, S.; Goh, Y.J.; Svensson, B.; Klaenhammer, T.R. Transcriptional and functional analysis of galactooligosaccharide uptake by lacS in Lactobacillus acidophilus. Proc. Natl. Acad. Sci. USA 2011, 108, 17785–17790. [Google Scholar] [CrossRef]
- Endo, A.; Futagawa-Endo, Y.; Dicks, L.M. Isolation and characterization of fructophilic lactic acid bacteria from fructose-rich niches. Syst. Appl. Microbiol. 2009, 32, 593–600. [Google Scholar] [CrossRef]
- Filannino, P.; Di Cagno, R.; Tlais, A.Z.A.; Cantatore, V.; Gobbetti, M. Fructose-rich niches traced the evolution of lactic acid bacteria toward fructophilic species. Crit. Rev. Microbiol. 2019, 45, 65–81. [Google Scholar] [CrossRef]
- Vásquez, A.; Olofsson, T. The lactic acid bacteria involved in the production of bee pollen and bee bread. J. Apicult. Res. 2009, 48, 189–195. [Google Scholar] [CrossRef]
- Neveling, D.P.; Endo, A.; Dicks, L.M. Fructophilic Lactobacillus kunkeei and Lactobacillus brevis isolated from fresh flowers, bees and beehives. Curr. Microbiol. 2012, 65, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Salminen, S. Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Syst. Appl. Microbiol. 2013, 36, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Testa, B.; Lombardi, S.J.; Ganassi, S.; Ianiro, M.; Letizia, F.; Succi, M.; Tremonte, P.; Vergalito, F.; Cozzolino, A.; et al. Antimicrobial Activity against Paenibacillus larvae and Functional Properties of Lactiplantibacillus plantarum Strains: Potential Benefits for Honeybee Health. Antibiotics 2020, 9, 442. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Lombardi, S.J.; Ganassi, S.; Testa, B.; Ianiro, M.; Letizia, F.; Succi, M.; Tremonte, P.; Vergalito, F.; Cozzolino, A.; et al. Antagonistic Activity against Ascosphaera apis and Functional Properties of Lactobacillus kunkeei Strains. Antibiotics 2020, 9, 262. [Google Scholar] [CrossRef] [PubMed]
- Pattabhiramaiah, M.; Reddy, M.S.; Brueckner, D. Detection of novel probiotic bacterium Lactobacillus spp. in the workers of Indian honeybee, Apis cerana indica. Int. J. Environ. Sci. 2012, 2, 1134–1143. [Google Scholar]
- Audisio, M.C. Gram-positive bacteria with probiotic potential for the Apis mellifera L. honey bee: The experience in the northwest of Argentina. Probiotics Antimicrob. Proteins 2017, 9, 22–31. [Google Scholar] [CrossRef]
- Alberoni, D.; Baffoni, L.; Gaggìa, F.; Ryan, P.; Murphy, K.; Ross, P.R.; Stanton, C.; Di Gioia, D. Impact of beneficial bacteria supplementation on the gut microbiota, colony development and productivity of Apis mellifera L. Benef. Microbes 2018, 9, 269–278. [Google Scholar] [CrossRef]
- Fanciotti, M.N.; Tejerina, M.; Benítez-Ahrendts, M.R.; Audisio, M.C. Honey yield of different commercial apiaries treated with Lactobacillus salivarius A3iob, a new bee-probiotic strain. Benef. Microbes 2018, 9, 291–298. [Google Scholar] [CrossRef]
- Fontana, P.; Costa, C.; di Prisco, G.; Ruzzier, E.; Annoscia, D.; Battisti, A.; Caoduro, G.; Carpana, C.; Contessi, A.; Dal Lago, A.; et al. Appeal for biodiversity protection of native honey bee subspecies of Apis mellifera in Italy (San Michele all’Adige declaration). Bull. Insectol. 2018, 71, 257–271. [Google Scholar]
- Querol, A.; Barrio, E.; Ramon, D. A comparative study of different methods of yeast strains characterization. Syst. Appl. Microbiol. 1992, 15, 439–446. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory: New York, NY, USA, 1989. [Google Scholar]
- Cocolin, L.; Manzano, M.; Cantoni, C.; Comi, G. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 2001, 67, 5113–5121. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Urso, R.; Iacumin, L.; Cantoni, P.; Cattaneo, G.; Comi, G.; Cocolin, L. Culture dependent and–independent methods to investigate the microbial ecology of Italian fermented sausage. Appl. Environ. Microbiol. 2005, 71, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Sheffield, V.C.; Cox, D.R.; Lerman, L.S.; Myers, R.M. Attachment of a 40-base pairs G + C rich sequence (GC clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc. Natl. Acad. Sci. USA 1989, 86, 297–303. [Google Scholar] [CrossRef]
- Testa, B.; Lombardi, S.J.; Tremonte, P.; Succi, M.; Tipaldi, L.; Pannella, G.; Sorrentino, E.; Iorizzo, M.; Coppola, R. Biodiversity of Lactobacillus plantarum from traditional Italian wines. World J. Microbiol. Biotechnol. 2014, 30, 2299–2305. [Google Scholar] [CrossRef]
- Klijn, N.; Weerkamp, A.H.; de Vos, W.M. Identification of mesophilic lactic acid bacteria by using polymerase chain reaction amplified variable regions of 16S rRNA and specific DNA probes. Appl. Environ. Microbiol. 1991, 57, 3390–3393. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Shaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic. Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Huey, B.; Hall, J. Hypervariable DNA fingerprinting in Escherichia coli. Minisatellite probe from bacteriophage M13. J. Bacteriol. 1989, 171, 2528–2532. [Google Scholar] [CrossRef]
- Akopyanz, N.; Bukanov, N.O.; Westblom, T.U.; Kresovich, S.; Berg, D.E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR based RAPD fingerprinting. Nucleic. Acids Res. 1992, 20, 5137–5142. [Google Scholar] [CrossRef]
- Dong, Z.X.; Li, H.Y.; Chen, Y.F.; Wang, F.; Deng, X.Y.; Lin, L.B.; Zhang, Q.L.; Li, J.L.; Guo, J. Colonization of the gut microbiota of honey bee (Apis mellifera) workers at different developmental stages. Microbiol. Res. 2020, 231, 126370. [Google Scholar] [CrossRef]
- Feizabadi, F.; Sharifan, A.; Tajabadi, N. Isolation and identification of lactic acid bacteria from stored Apis mellifera honey. J. Apic. Res. 2020, 1–6. [Google Scholar] [CrossRef]
- De Leonardis, A.; Testa, B.; Macciola, V.; Lombardi, S.J.; Iorizzo, M. Exploring enzyme and microbial technology for the preparation of green table olives. Eur. Food Res. Technol. 2016, 242, 363–370. [Google Scholar] [CrossRef]
- Iorizzo, M.; Testa, B.; Lombardi, S.J.; García-Ruiz, A.; Muñoz-González, C.; Bartolomé, B.; Moreno-Arribas, M.V. Selection and technological potential of Lactobacillus plantarum bacteria suitable for wine malolactic fermentation and grape aroma release. LWT-Food Sci. Technol. 2016, 73, 557–566. [Google Scholar] [CrossRef]
- Succi, M.; Pannella, G.; Tremonte, P.; Tipaldi, L.; Coppola, R.; Iorizzo, M.; Lombardi, S.J.; Sorrentino, E. Sub-optimal pH preadaptation improves the survival of Lactobacillus plantarum strains and the malic acid consumption in wine-like medium. Front. Microbiol. 2017, 8, 470. [Google Scholar] [CrossRef]
- Tremonte, P.; Pannella, G.; Succi, M.; Tipaldi, L.; Sturchio, M.; Coppola, R.; Luongo, D.; Sorrentino, E. Antimicrobial activity of Lactobacillus plantarum strains isolated from different environments: A preliminary study. Int. Food Res. J. 2017, 21, 852–859. [Google Scholar]
- Pannella, G.; Lombardi, S.J.; Coppola, F.; Vergalito, F.; Iorizzo, M.; Succi, M.; Tremonte, P.; Iannini, C.; Sorrentino, E.; Coppola, R. Effect of Biofilm Formation by Lactobacillus plantarum on the Malolactic Fermentation in Model Wine. Foods 2020, 9, 797. [Google Scholar] [CrossRef]
- Di Cagno, R.; Filannino, P.; Cantatore, V.; Gobbetti, M. Novel solid-state fermentation of bee-collected pollen emulating the natural fermentation process of bee bread. Food Microbiol. 2019, 82, 218–230. [Google Scholar] [CrossRef]
- Aizenberg-Gershtein, Y.; Izhaki, I.; Halpern, M. Do honeybees shape the bacterial community composition in floral nectar? PLoS ONE 2013, 8, e67556. [Google Scholar] [CrossRef]
- Donkersley, P.; Rhodes, G.; Pickup, R.W.; Jones, K.C.; Wilson, K. Bacterial communities associated with honeybee food stores are correlated with land use. Ecol. Evol. 2018, 8, 4743–4756. [Google Scholar] [CrossRef]
- Czipa, N.; Phillips, C.J.; Kovács, B. Composition of acacia honeys following processing, storage and adulteration. J. Food Sci. Technol. 2019, 56, 1245–1255. [Google Scholar] [CrossRef]
- Thakur, M.; Nanda, V. Composition and functionality of bee pollen: A review. Trends Food Sci. Technol. 2020, 98, 82–106. [Google Scholar] [CrossRef]
- Anderson, K.E.; Carroll, M.J.; Sheehan, T.I.M.; Mott, B.M.; Maes, P.; Corby-Harris, V. Hive-stored pollen of honey bees: Many lines of evidence are consistent with pollen preservation, not nutrient conversion. Mol. Ecol. 2014, 23, 5904–5917. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Ricigliano, V.A.; Mott, B.M.; Copeland, D.C.; Floyd, A.S.; Maes, P. The queen’s gut refines with age: Longevity phenotypes in a social insect model. Microbiome 2018, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Trumbo, S.T.; Huang, Z.Y.; Robinson, G.E. Division of labor between undertaker specialists and other middle-aged workers in honey bee colonies. Behav. Ecol. Sociobiol. 1997, 41, 151–163. [Google Scholar] [CrossRef]
- Tan, K.; Guo, Y.H.; Nicolson, S.W.; Radloff, S.E.; Song, Q.S.; Hepburn, H.R. Honeybee (Apiscerana) Foraging Responses to the Toxic Honey of Tripterygium hypoglaucum (Celastraceae): Changing Threshold of Nectar Acceptability. J. Chem. Ecol. 2007, 33, 2209–2217. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.A.; Robertson, A.W.; Biggs, P.J.; Richards, K.K.; Jones, D.F.; Parkar, S.G. The effect of carbohydrate sources: Sucrose, invert sugar and components of mānuka honey, on core bacteria in the digestive tract of adult honey bees (Apis mellifera). PLoS ONE 2019, 14, e0225845. [Google Scholar] [CrossRef]
- Ricigliano, V.A.; Fitz, W.; Copeland, D.C.; Mott, B.M.; Maes, P.; Floyd, A.S.; Dockstader, A.; Anderson, K.E. The impact of pollen consumption on honey bee (Apis mellifera) digestive physiology and carbohydrate metabolism. Arch. Insect Biochem. Physiol. 2017, 96, e21406. [Google Scholar] [CrossRef]
- Zheng, H.; Perreau, J.; Powell, J.E.; Han, B.; Zhang, Z.; Kwong, W.K.; Tringe, S.G.; Moran, N.A. Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc. Natl. Acad. Sci. USA 2019, 116, 25909–25916. [Google Scholar] [CrossRef]
- Kaznowski, A.; Szymas, B.; Jazdzinska, E.; Kazimierczak, M.; Paetz, H.; Mokracka, J. The effects of probiotic supplementation on the content of intestinal microflora and chemical composition of worker honey bees (Apis mellifera). J. Apic. Res. 2005, 44, 10–14. [Google Scholar] [CrossRef]
- Michlmayr, H.; Kneifel, W. β-Glucosidase activities of lactic acid bacteria: Mechanisms, impact on fermented food and human health. FEMS Microbiol. Lett. 2014, 352, 1–10. [Google Scholar] [CrossRef]
- Johnson, R.M. Honey Bee Toxicology. Annu. Rev. Entomol. 2015, 60, 415–434. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Nishida, A.; Kwong, W.K.; Koch, H.; Engel, P.; Steele, M.I.; Moran, N.A. Metabolism of Toxic Sugars by Strains of the Bee Gut Symbiont Gilliamella apicola. mBio 2016, 7, e01326-e16. [Google Scholar] [CrossRef] [PubMed]
- Lee, F.J.; Rusch, D.B.; Stewart, F.J.; Mattila, H.R.; Newton, I.L.G. Saccharide break down and fermentation by the honey bee gut microbiome. Environ. Microbiol. 2015, 17, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Seeburger, V.C.; D’Alvise, P.; Shaaban, B.; Schweikert, K.; Lohaus, G.; Schroeder, A.; Hasselmann, M. The trisaccharide melezitose impacts honey bees and their intestinal microbiota. PLoS ONE 2020, 15, e0230871. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, T.C.; Vásquez, A.; Sammataro, D.; Macharia, J. A scientific note on the lactic acid bacterial flora within the honeybee subspecies Apis mellifera (Buckfast), A. m. scutellata, A. m. mellifera, and A. m. monticola. Apidologie 2011, 42, 696–699. [Google Scholar] [CrossRef]
- Janashia, I.; Carminati, D.; Rossetti, L.; Zago, M.; Fornasari, M.E.; Haertlé, T.; Chanishvili, N.; Giraffa, G. Characterization of fructophilic lactic microbiota of Apis mellifera from the Caucasus Mountains. Annal. Microbiol. 2016, 66, 1387–1395. [Google Scholar] [CrossRef]
- Pachla, A.; Wicha, M.; Ptaszyńska, A.A.; Borsuk, G.; Łaniewska–Trokenheim, L.; Małek, W. The molecular and phenotypic characterization of fructophilic lactic acid bacteria isolated from the guts of Apis mellifera L. derived from a Polish apiary. J. Appl. Genet. 2018, 59, 503–514. [Google Scholar] [CrossRef]
- Cilia, G.; Fratini, F.; Tafi, E.; Turchi, B.; Mancini, S.; Sagona, S.; Nanetti, A.; Cerri, D.; Felicioli, A. Microbial Profile of the Ventriculum of Honey Bee (Apis mellifera ligustica Spinola, 1806) Fed with Veterinary Drugs, Dietary Supplements and Non-Protein Amino Acids. Vet. Sci. 2020, 7, 76. [Google Scholar] [CrossRef]
- Ellegaard, K.M.; Engel, P. Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Ellegaard, K.M.; Engel, P. Beyond 16S rRNA community profiling: Intra-species diversity in the gut microbiota. Front. Microbiol. 2016, 7, 1475. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Hsu, T.; Sirota-Madi, A.; Shafquat, A.; Abu-Ali, G.; Morgan, X.C.; Huttenhower, C. Sequencing and beyond: Integrating molecular’omics’ for microbial community profiling. Nat. Rev. Microbiol. 2015, 13, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Sunagawa, S.; Mende, D.R.; Bork, P. Inter-individual differences in the gene content of human gut bacterial species. Genome Biol. 2015, 16, 82. [Google Scholar] [CrossRef] [PubMed]
- Ellegaard, K.M.; Suenami, S.; Miyazaki, R.; Engel, P. Vast differences in strain-level diversity in the gut microbiota of two closely related honey bee species. Curr. Biol. 2020, 30, 2520–2531.E7. [Google Scholar] [CrossRef] [PubMed]
- Sinotte, V.M.; Renelies-Hamilton, J.; Taylor, B.A.; Ellegaard, K.M.; Sapountzis, P.; Cognet, M.V.; Poulsen, M. Synergies between division of labor and gut microbiomes of social insects. Front. Ecol. Evol. 2020, 7. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iorizzo, M.; Pannella, G.; Lombardi, S.J.; Ganassi, S.; Testa, B.; Succi, M.; Sorrentino, E.; Petrarca, S.; De Cristofaro, A.; Coppola, R.; et al. Inter- and Intra-Species Diversity of Lactic Acid Bacteria in Apis mellifera ligustica Colonies. Microorganisms 2020, 8, 1578. https://doi.org/10.3390/microorganisms8101578
Iorizzo M, Pannella G, Lombardi SJ, Ganassi S, Testa B, Succi M, Sorrentino E, Petrarca S, De Cristofaro A, Coppola R, et al. Inter- and Intra-Species Diversity of Lactic Acid Bacteria in Apis mellifera ligustica Colonies. Microorganisms. 2020; 8(10):1578. https://doi.org/10.3390/microorganisms8101578
Chicago/Turabian StyleIorizzo, Massimo, Gianfranco Pannella, Silvia Jane Lombardi, Sonia Ganassi, Bruno Testa, Mariantonietta Succi, Elena Sorrentino, Sonia Petrarca, Antonio De Cristofaro, Raffaele Coppola, and et al. 2020. "Inter- and Intra-Species Diversity of Lactic Acid Bacteria in Apis mellifera ligustica Colonies" Microorganisms 8, no. 10: 1578. https://doi.org/10.3390/microorganisms8101578
APA StyleIorizzo, M., Pannella, G., Lombardi, S. J., Ganassi, S., Testa, B., Succi, M., Sorrentino, E., Petrarca, S., De Cristofaro, A., Coppola, R., & Tremonte, P. (2020). Inter- and Intra-Species Diversity of Lactic Acid Bacteria in Apis mellifera ligustica Colonies. Microorganisms, 8(10), 1578. https://doi.org/10.3390/microorganisms8101578








