Frequent Anti-V1V2 Responses Induced by HIV-DNA Followed by HIV-MVA with or without CN54rgp140/GLA-AF in Healthy African Volunteers
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Linear Peptide Microarray
2.3. Assessment of Binding Antibodies to the V1V2 Region of HIV-1 Env
2.3.1. Binding IgG Antibodies
2.3.2. Binding IgG1 and IgG3 Antibodies
2.4. Depletion of IgG from Serum Samples
2.5. Assessment of Env-Specific IgA Antibodies
2.6. Assessment of ADCC-Mediating Antibodies
2.7. Statistical Analysis
2.8. Ethical Considerations
3. Results
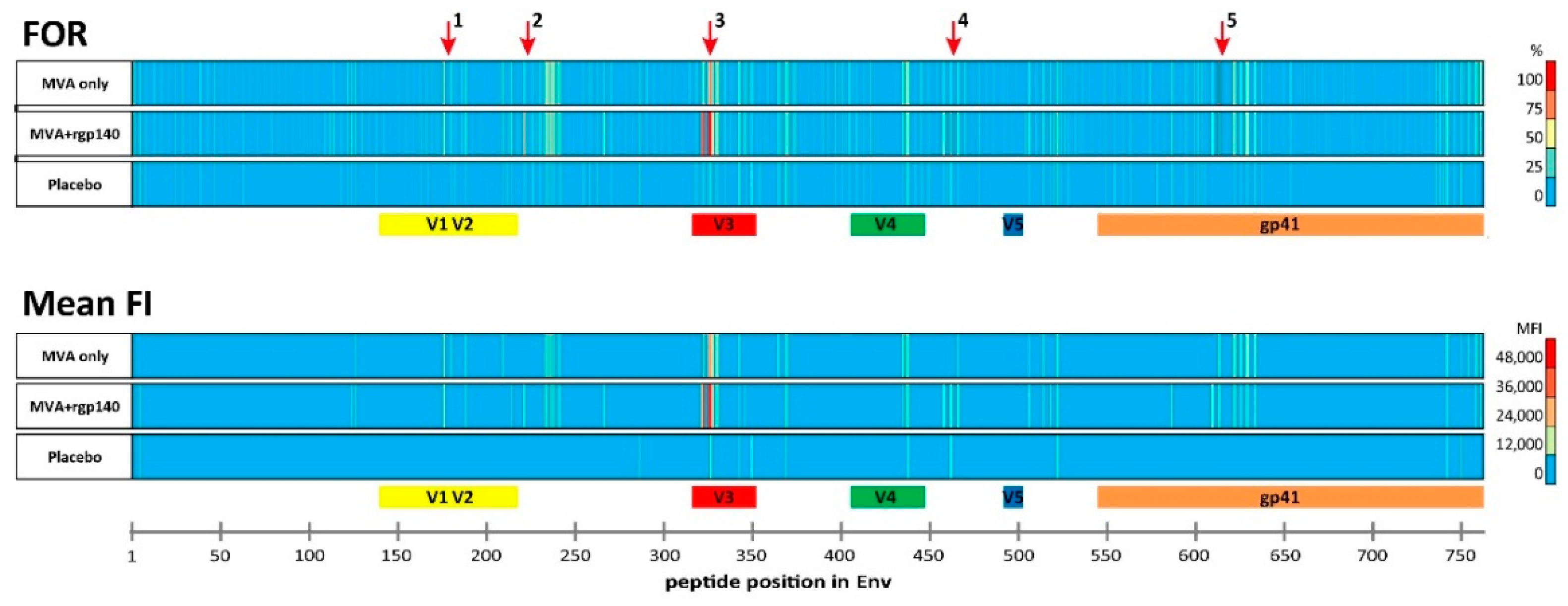
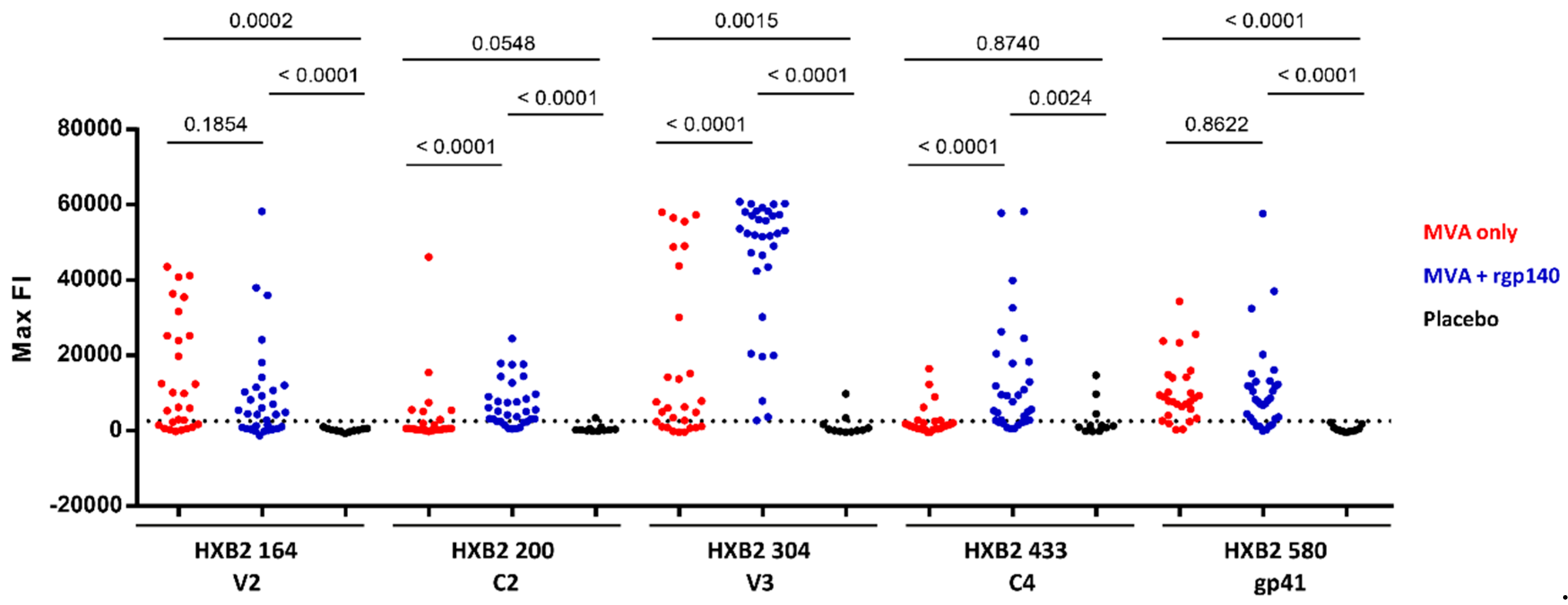
3.1. The Vaccination Regimen Induced IgG Antibodies Recognizing Linear Antigenic Epitopes in the V1V2, C2, V3, C4, and gp41 Env Regions
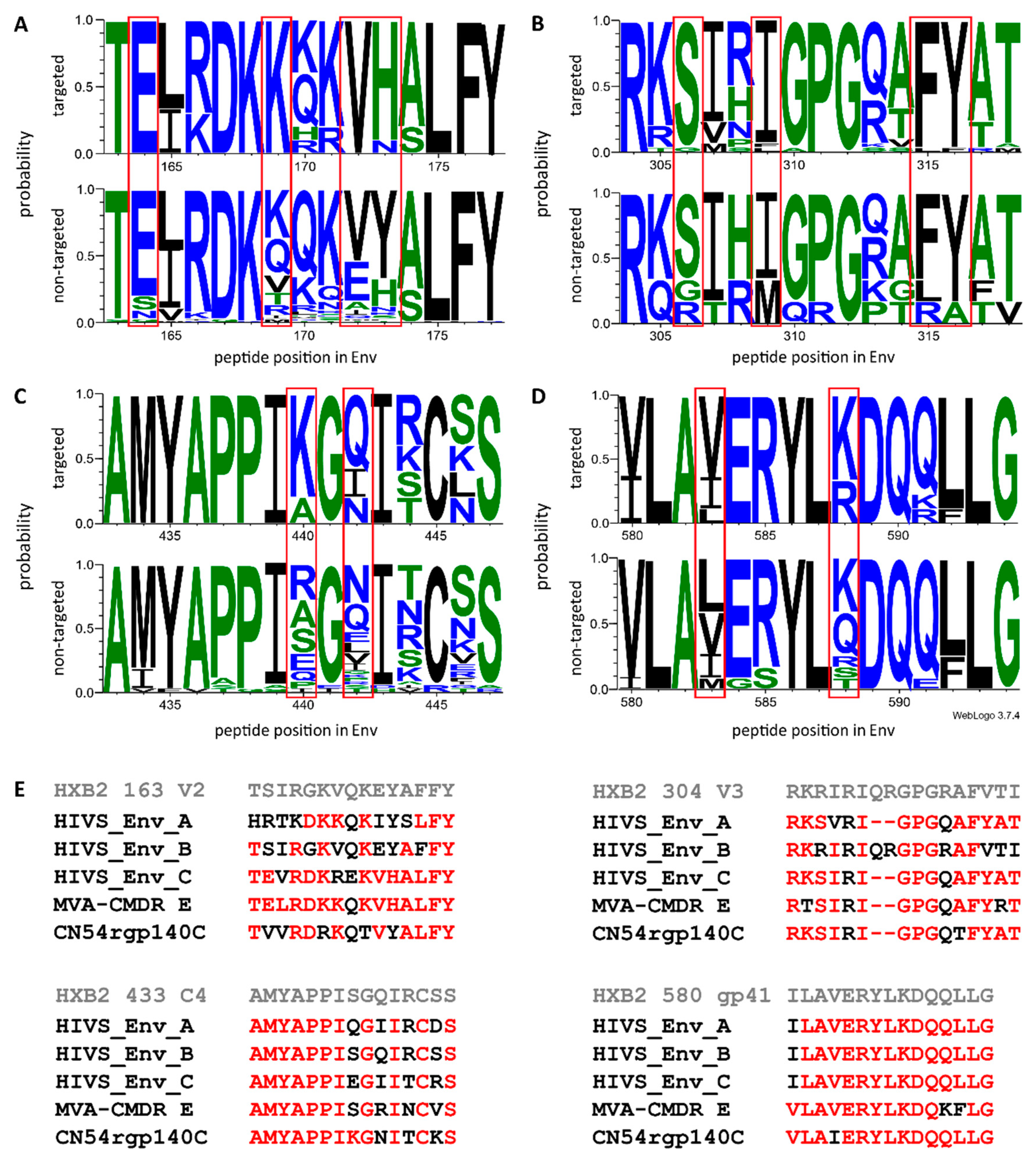
3.2. Comparison of Antigen Variant Recognition within the Five Immunodominant Regions
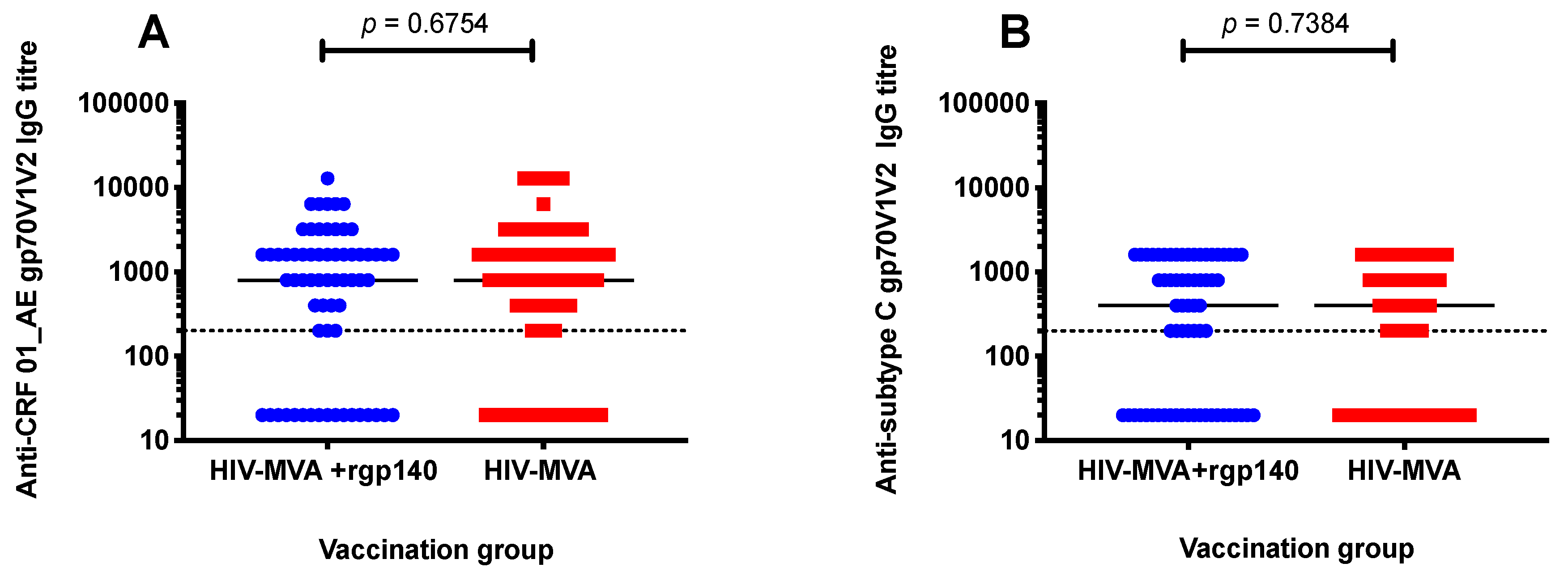
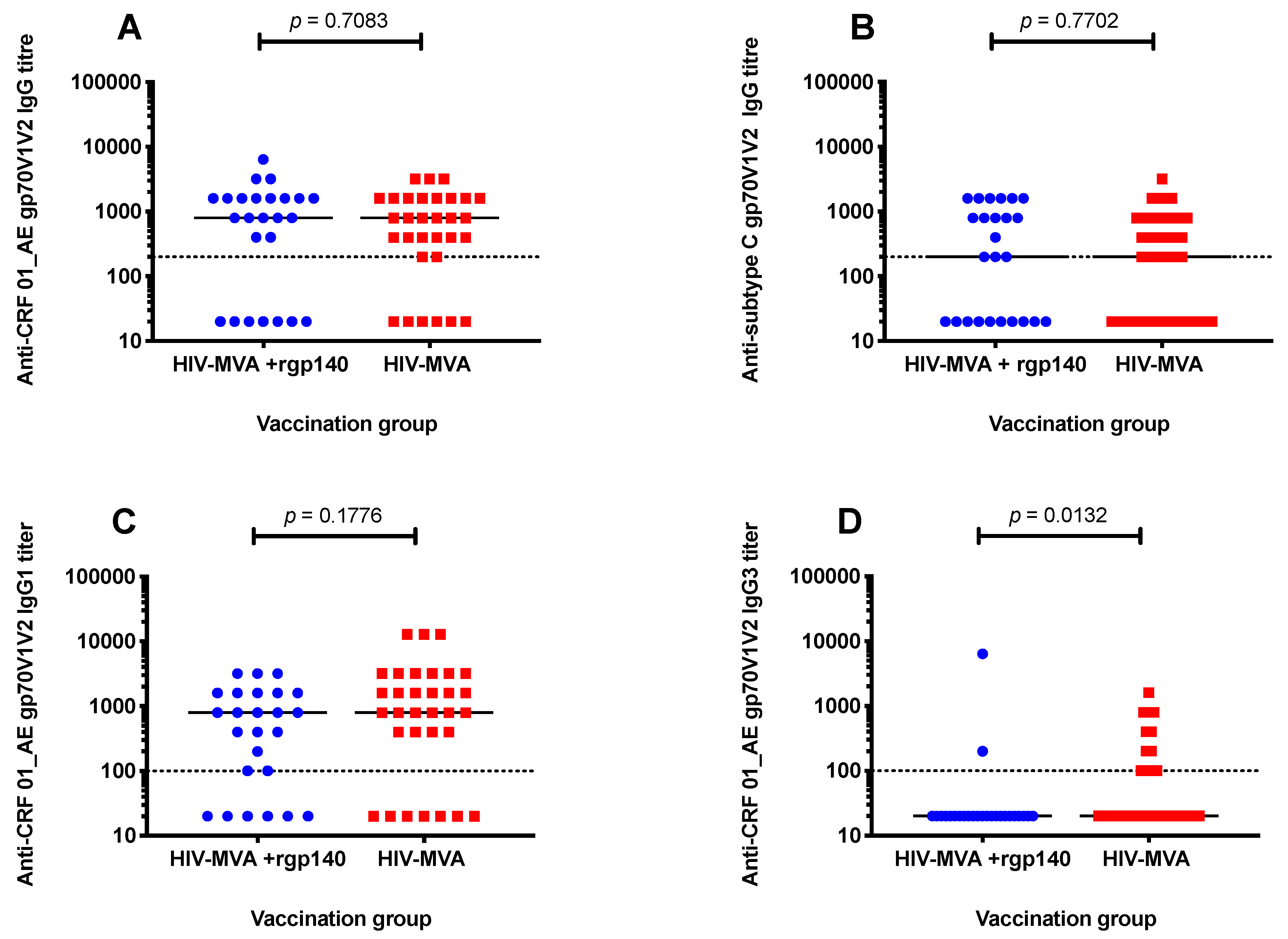
3.3. V1V2 Binding IgG Antibodies
3.4. Anti-V1V2 IgG1 and IgG3 Responses against CRF01_AE
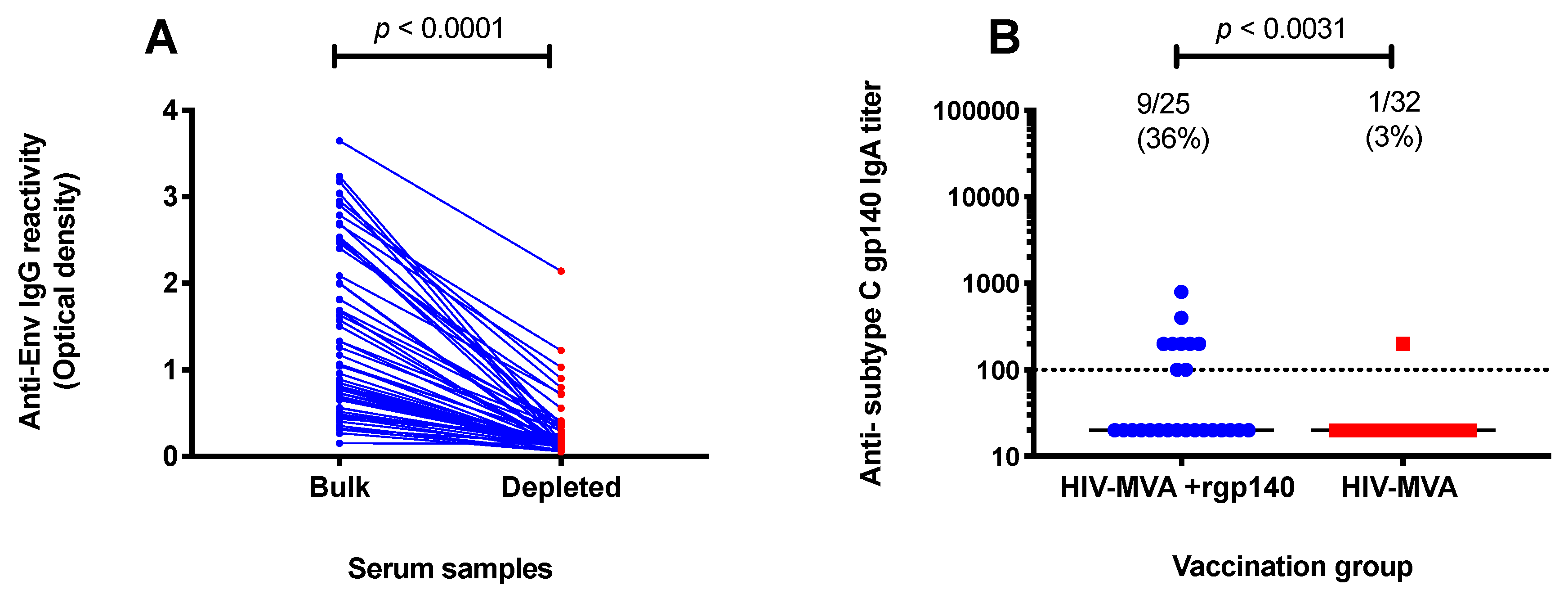
3.5. Subtype C gp140-Specific IgA Binding Antibodies
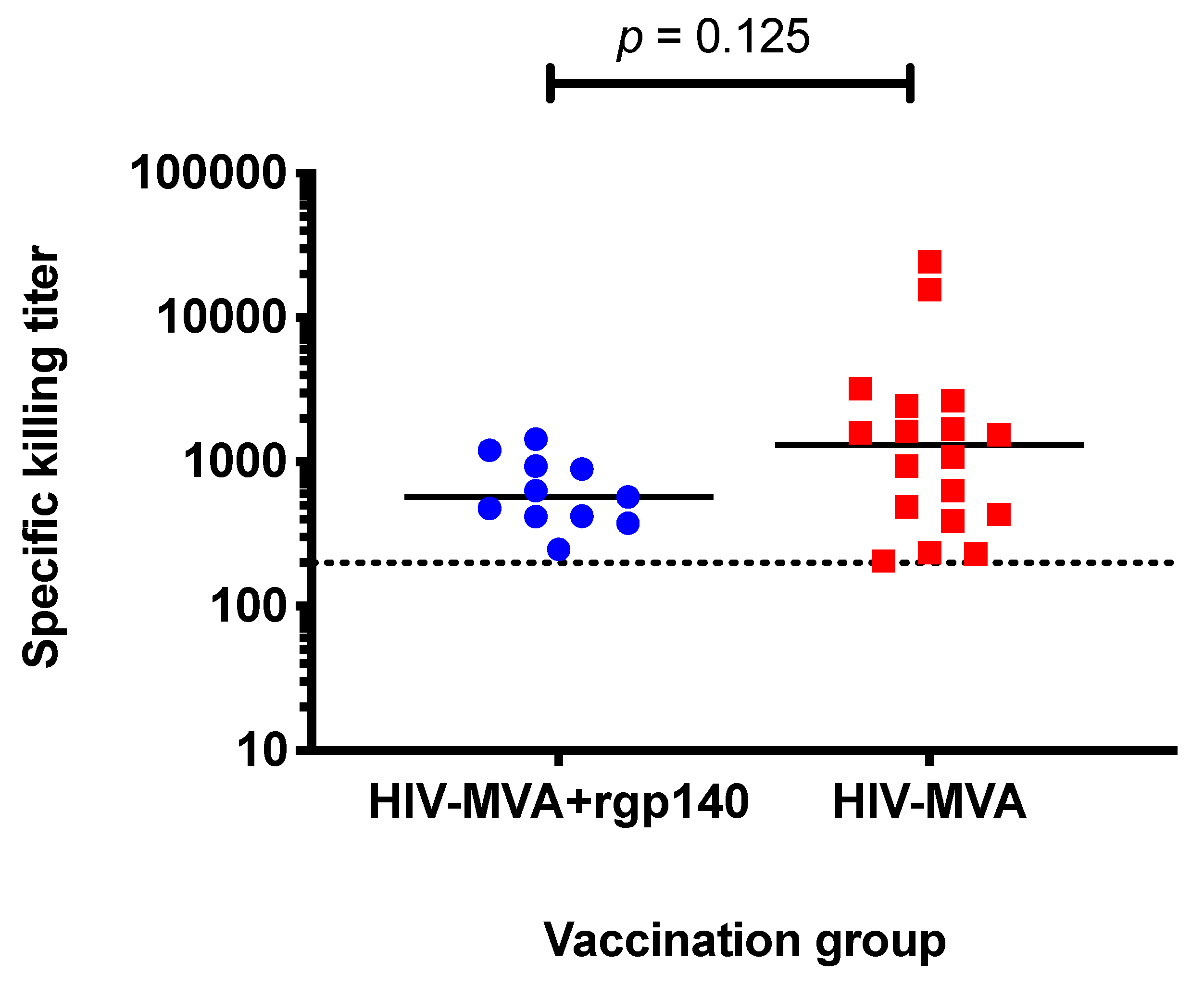
3.6. ADCC Activity against CM235 CRF01_AE Infected Target Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Fauci, A.S.; Folkers, G.K.; Marston, H.D. Ending the Global HIV/AIDS Pandemic: The Critical Role of an HIV Vaccine. Clin. Infect. Dis. 2014, 59, S80–S84. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S.; Marston, H.D. Ending AIDS—Is an HIV Vaccine Necessary? N. Engl. J. Med. 2014, 370, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Medlock, J.; Pandey, A.; Parpia, A.S.; Tang, A.; Skrip, L.A.; Galvani, A.P. Effectiveness of UNAIDS targets and HIV vaccination across 127 countries. Proc. Natl. Acad. Sci. USA 2017, 114, 4017–4022. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S. An HIV Vaccine Is Essential for Ending the HIV/AIDS Pandemic. J. Am. Med. Assoc. 2017, 318, 1535–1536. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.J.; Kim, J.H.; Corey, L.; Michael, N.L. Human Immunodeficiency Virus Vaccine Trials. Cold Spring Harb. Perspect. Med. 2012, 2, a007351. [Google Scholar] [CrossRef]
- Kim, J.H.; Excler, J.L.; Michael, N.L. Lessons from the RV144 Thai Phase III HIV-1 Vaccine Trial and the Search for Correlates of Protection. Annu. Rev. Med. 2015, 66, 423–437. [Google Scholar] [CrossRef]
- Flynn, N.M.; Forthal, D.N.; Harro, C.D.; Judson, F.N.; Mayer, K.H.; Para, M.F. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 2005, 191, 654–665. [Google Scholar] [CrossRef]
- Pitisuttithum, P.; Gilbert, P.; Gurwith, M.; Heyward, W.; Martin, M.; van Griensven, F.; Hu, D.; Tappero, J.W.; Bangkok Vaccine Evaluation Group. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J. Infect. Dis. 2006, 194, 1661–1671. [Google Scholar] [CrossRef]
- Buchbinder, S.P.; Mehrotra, D.V.; Duerr, A.; Fitzgerald, D.W.; Mogg, R.; Li, D.; Gilbert, P.B.; Lama, J.R.; Marmor, M.; Del Rio, C.; et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): A double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 2008, 372, 1881–1893. [Google Scholar] [CrossRef]
- Gray, G.E.; Allen, M.A.; Moodie, Z.; Churchyard, G.; Bekker, L.-G.; Nchabeleng, M.; Mlisana, K.; Metch, B.; De Bruyn, G.; Latka, M.H.; et al. Safety and efficacy of the HVTN 503/Phambili Study of a clade-B-based HIV-1 vaccine in South Africa: A double-blind, randomised, placebo-controlled test-of-concept phase 2b study. Lancet Infect. Dis. 2011, 11, 507–515. [Google Scholar] [CrossRef]
- Hammer, S.M.; Sobieszczyk, M.E.; Janes, H.; Karuna, S.T.; Mulligan, M.J.; Grove, D.; Koblin, B.A.; Buchbinder, S.P.; Keefer, M.C.; Tomaras, G.D.; et al. Efficacy Trial of a DNA/rAd5 HIV-1 Preventive Vaccine. N. Engl. J. Med. 2013, 369, 2083–2092. [Google Scholar] [CrossRef] [PubMed]
- Rerks-Ngarm, S.; Pitisuttithum, P.; Nitayaphan, S.; Kaewkungwal, J.; Chiu, J.; Paris, R.; Premsri, N.; Namwat, C.; De Souza, M.; Adams, E.; et al. Vaccination with ALVAC and AIDSVAX to Prevent HIV-1 Infection in Thailand. N. Engl. J. Med. 2009, 361, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Pitisuttithum, P.; Marovich, M.A. Prophylactic HIV vaccine: Vaccine regimens in clinical trials and potential challenges. Expert Rev. Vaccines 2020, 19, 133–142. [Google Scholar] [CrossRef] [PubMed]
- UNAIDS. HVTN 702 Clinical Trial of an HIV Vaccine Stopped; Press Statement; UNAIDS: Geneva, Switzerland, 2020. [Google Scholar]
- ClinicalTrials.gov. Available online: https://www.clinicaltrials.gov/ct2/show/NCT03060629 (accessed on 29 July 2020).
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT03964415 (accessed on 29 July 2020).
- Robb, M.L.; Rerks-Ngarm, S.; Nitayaphan, S.; Pitisuttithum, P.; Kaewkungwal, J.; Kunasol, P.; Khamboonruang, C.; Thongcharoen, P.; Morgan, P.; Benenson, M.; et al. Risk behaviour and time as covariates for efficacy of the HIV vaccine regimen ALVAC-HIV (vCP1521) and AIDSVAX B/E: A post-hoc analysis of the Thai phase 3 efficacy trial RV 144. Lancet Infect. Dis. 2012, 12, 531–537. [Google Scholar] [CrossRef]
- Churchyard, G.; Morgan, C.; Adams, E.; Hural, J.; Graham, B.S.; Moodie, Z.; Grove, D.; Gray, G.; Bekker, L.-G.; McElrath, M.J.; et al. A Phase IIA Randomized Clinical Trial of a Multiclade HIV-1 DNA Prime Followed by a Multiclade rAd5 HIV-1 Vaccine Boost in Healthy Adults (HVTN204). PLoS ONE 2011, 6, e21225. [Google Scholar] [CrossRef]
- Excler, J.L.; Tomaras, G.D.; Russell, N.D. Novel directions in HIV-1 vaccines revealed from clinical trials. Curr. Opin. HIV AIDS 2013, 8, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Goepfert, P.A.; Elizaga, M.L.; Sato, A.; Qin, L.; Cardinali, M.; Hay, C.M.; Hural, J.; DeRosa, S.C.; Defawe, O.D.; Tomaras, G.D.; et al. Phase 1 Safety and Immunogenicity Testing of DNA and Recombinant Modified Vaccinia Ankara Vaccines Expressing HIV-1 Virus-like Particles. J. Infect. Dis. 2011, 203, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Harari, A.; Bart, P.A.; Stöhr, W.; Tapia, G.; Garcia, M.; Medjitna-Rais, E.; Burnet, S.; Cellerai, C.; Erlwein, O.; Barber, T.; et al. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J. Exp. Med. 2008, 205, 63–77. [Google Scholar] [CrossRef]
- Mehendale, S.; Thakar, M.; Sahay, S.; Kumar, M.; Shete, A.; Sathyamurthi, P.; Verma, A.; Kurle, S.; Shrotri, A.; Gilmour, J.; et al. Safety and Immunogenicity of DNA and MVA HIV-1 Subtype C Vaccine Prime-Boost Regimens: A Phase I Randomised Trial in HIV-Uninfected Indian Volunteers. PLoS ONE 2013, 8, e558311. [Google Scholar] [CrossRef]
- Bakari, M.; Aboud, S.; Nilsson, C.; Francis, J.M.; Buma, D.; Moshiro, C.; Aris, E.A.; Lyamuya, E.F.; Janabi, M.; Godoy-Ramirez, K.; et al. Broad and potent immune responses to a low dose intradermal HIV-1 DNA boosted with HIV-1 recombinant MVA among healthy adults in Tanzania. Vaccine 2011, 29, 8417–8428. [Google Scholar] [CrossRef]
- Munseri, P.J.; Kroidl, A.; Nilsson, C.; Joachim, A.; Geldmacher, C.; Mann, P.; Moshiro, C.; Aboud, S.; Lyamuya, E.; Maboko, L.; et al. Priming with a Simplified Intradermal HIV-1 DNA Vaccine Regimen followed by Boosting with Recombinant HIV-1 MVA Vaccine Is Safe and Immunogenic: A Phase IIa Randomized Clinical Trial. PLoS ONE 2015, 10, e0119629. [Google Scholar] [CrossRef]
- Nilsson, C.; Hejdeman, B.; Godoy-Ramirez, K.; Tecleab, T.; Scarlatti, G.; Bråve, A.; Earl, P.L.; Stout, R.R.; Robb, M.L.; Shattock, R.J.; et al. HIV-DNA Given with or without Intradermal Electroporation Is Safe and Highly Immunogenic in Healthy Swedish HIV-1 DNA/MVA Vaccinees: A Phase I Randomized Trial. PLoS ONE 2015, 10, e0131748. [Google Scholar] [CrossRef]
- Viegas, E.O.; Tembe, N.; Nilsson, C.; Meggi, B.; Maueia, C.; Augusto, O.; Stout, R.; Scarlatti, G.; Ferrari, G.; Earl, P.L.; et al. Intradermal HIV-1 DNA Immunization Using Needle-Free Zetajet Injection Followed by HIV-Modified Vaccinia Virus Ankara Vaccination Is Safe and Immunogenic in Mozambican Young Adults: A Phase I Randomized Controlled Trial. AIDS Res. Hum. Retrovir. 2018, 34, 193–205. [Google Scholar] [CrossRef]
- Joachim, A.; Nilsson, C.; Aboud, S.; Bakari, M.; Lyamuya, E.F.; Robb, M.L.; Marovich, M.A.; Earl, P.; Moss, B.; Ochsenbauer, C.; et al. Potent Functional Antibody Responses Elicited by HIV-I DNA Priming and Boosting with Heterologous HIV-1 Recombinant MVA in Healthy Tanzanian Adults. PLoS ONE 2015, 10, e0118486. [Google Scholar] [CrossRef]
- Sandström, E.; Nilsson, C.; Hejdeman, B.; Bråve, A.; Bratt, G.; Robb, M.; Cox, J.H.; VanCott, T.; Marovich, M.; Stout, R.; et al. Broad Immunogenicity of a Multigene, Multiclade HIV-1 DNA Vaccine Boosted with Heterologous HIV-1 Recombinant Modified Vaccinia Virus Ankara. J. Infect. Dis. 2008, 198, 1482–1490. [Google Scholar] [CrossRef]
- Nilsson, C.; Godoy-Ramirez, K.; Hejdeman, B.; Bråve, A.; Gudmundsdotter, L.; Hallengärd, D.; Currier, J.R.; Wieczorek, L.; Hasselrot, K.; Earl, P.L.; et al. Broad and Potent Cellular and Humoral Immune Responses After a Second Late HIV-Modified Vaccinia Virus Ankara Vaccination in HIV-DNA-Primed and HIV-Modified Vaccinia Virus Ankara-Boosted Swedish Vaccinees. AIDS Res. Hum. Retrovir. 2014, 30, 299–311. [Google Scholar] [CrossRef]
- Joachim, A.; Munseri, P.J.; Nilsson, C.; Bakari, M.; Aboud, S.; Lyamuya, E.F.; Tecleab, T.; Liakina, V.; Scarlatti, G.; Robb, M.L.; et al. Three-Year Durability of Immune Responses Induced by HIV-DNA and HIV-Modified Vaccinia Virus Ankara and Effect of a Late HIV-Modified Vaccinia Virus Ankara Boost in Tanzanian Volunteers. AIDS Res. Hum. Retrovir. 2017, 33, 880–888. [Google Scholar] [CrossRef]
- Viegas, E.O.; Kroidl, A.; Munseri, P.J.; Missanga, M.; Nilsson, C.; Tembe, N.; Bauer, A.; Joachim, A.; Joseph, S.; Mann, P.; et al. Optimizing the immunogenicity of HIV prime-boost DNA-MVA-rgp140/GLA vaccines in a phase II randomized factorial trial design. PLoS ONE 2018, 13, e0206838. [Google Scholar] [CrossRef]
- Rao, M.; Peachman, K.K.; Kim, J.; Gao, G.; Alving, C.R.; Michael, N.L.; Rao, V.B. HIV-1 variable loop 2 and its importance in HIV-1 infection and vaccine development. Curr. HIV Res. 2013, 11, 427–438. [Google Scholar]
- O’Connell, R.J.; Kim, J.H.; Excler, J.-L. The HIV-1 gp120 V1V2 loop: Structure, function and importance for vaccine development. Expert Rev. Vaccines 2014, 13, 1489–1500. [Google Scholar] [CrossRef]
- Sullivan, N.; Thali, M.; Furman, C.; Ho, D.D.; Sodroski, J. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J. Virol. 1993, 67, 3674–3679. [Google Scholar] [CrossRef] [PubMed]
- Kwong, P.D.; Wyatt, R.; Robinson, J.; Sweet, R.W.; Sodroski, J.; Hendrickson, W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 1998, 393, 648–659. [Google Scholar] [CrossRef] [PubMed]
- Arthos, J.; Cicala, C.; Martinelli, E.; MacLeod, K.; Van Ryk, D.; Wei, D.; Xiao, Z.; Veenstra, T.D.; Conrad, T.P.; Lempicki, R.A.; et al. HIV-1 envelope protein binds to and signals through integrin α4β7, the gut mucosal homing receptor for peripheral T cells. Nat. Immunol. 2008, 9, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Haynes, B.F.; Gilbert, P.B.; McElrath, M.J.; Zolla-Pazner, S.; Tomaras, G.D.; Alam, S.M.; Evans, D.T.; Montefiori, D.C.; Karnasuta, C.; Sutthent, R.; et al. Immune-Correlates Analysis of an HIV-1 Vaccine Efficacy Trial. N. Engl. J. Med. 2012, 366, 1275–1286. [Google Scholar] [CrossRef]
- Karasavvas, N.; Billings, E.; Rao, M.; Williams, C.; Zolla-Pazner, S.; Bailer, R.T.; Koup, R.A.; Madnote, S.; Arworn, D.; Shen, X.; et al. The Thai Phase III HIV Type 1 Vaccine Trial (RV144) Regimen Induces Antibodies That Target Conserved Regions Within the V2 Loop of gp120. AIDS Res. Hum. Retrovir. 2012, 28, 1444–1457. [Google Scholar] [CrossRef]
- Gottardo, R.; Bailer, R.T.; Korber, B.; Gnanakaran, S.; Phillips, J.; Shen, X.; Tomaras, G.D.; Turk, E.; Imholte, G.; Eckler, L.; et al. Plasma IgG to Linear Epitopes in the V2 and V3 Regions of HIV-1 gp120 Correlate with a Reduced Risk of Infection in the RV144 Vaccine Efficacy Trial. PLoS ONE 2013, 8, e75665. [Google Scholar] [CrossRef]
- Yates, N.L.; Liao, H.-X.; Fong, Y.; DeCamp, A.; Vandergrift, N.A.; Williams, W.T.; Alam, S.M.; Ferrari, G.; Yang, Z.-Y.; Seaton, K.E.; et al. Vaccine-Induced Env V1-V2 IgG3 Correlates with Lower HIV-1 Infection Risk and Declines Soon After Vaccination. Sci. Transl. Med. 2014, 6, 228ra39. [Google Scholar] [CrossRef]
- Canfield, S.M.; Morrison, S.L. The binding affinity of human IgG for its high affinity Fc receptor is determined by multiple amino acids in the CH2 domain and is modulated by the hinge region. J. Exp. Med. 1991, 173, 1483–1491. [Google Scholar] [CrossRef]
- Bruhns, P.; Iannascoli, B.; England, P.; Mancardi, D.A.; Fernandez, N.; Jorieux, S.; Daëron, M. Specificity and affinity of human Fcγ receptors and their polymorphic variants for human IgG subclasses. Blood 2009, 113, 3716–3725. [Google Scholar] [CrossRef]
- Vidarsson, G.; Dekkers, G.; Rispens, T. IgG Subclasses and Allotypes: From Structure to Effector Functions. Front. Immunol. 2014, 5, 520. [Google Scholar] [CrossRef]
- Chung, A.W.; Ghebremichael, M.; Robinson, H.; Brown, E.; Choi, I.; Lane, S.; Dugast, A.-S.; Schoen, M.K.; Rolland, M.; Suscovich, T.J.; et al. Polyfunctional Fc-Effector Profiles Mediated by IgG Subclass Selection Distinguish RV144 and VAX003 Vaccines. Sci. Transl. Med. 2014, 6, 228ra38. [Google Scholar] [CrossRef] [PubMed]
- Nadai, Y.; Held, K.; Joseph, S.; Ahmed, M.I.M.; Hoffmann, V.S.; Peterhoff, D.; Missanga, M.; Bauer, A.; Joachim, A.; Reimer, U.; et al. Envelope-Specific Recognition Patterns of HIV Vaccine-Induced IgG Antibodies Are Linked to Immunogen Structure and Sequence. Front. Immunol. 2019, 10, 717. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Sambor, A.; Garcia, A.; Berrong, M.; Pickeral, J.; Brown, S.; Rountree, W.; Sánchez, A.; Pollara, J.; Frahm, N.; Keinonen, S.; et al. Establishment and maintenance of a PBMC repository for functional cellular studies in support of clinical vaccine trials. J. Immunol. Methods 2014, 409, 107–116. [Google Scholar] [CrossRef]
- Trkola, A.; Matthews, J.; Gordon, C.; Ketas, T.; Moore, J.P. A Cell Line-Based Neutralization Assay for Primary Human Immunodeficiency Virus Type 1 Isolates That Use either the CCR5 or the CXCR4 Coreceptor. J. Virol. 1999, 73, 8966–8974. [Google Scholar] [CrossRef]
- Adepoju, P. Moving on from the failed HIV vaccine clinical trial. Lancet HIV 2020, 7, e161. [Google Scholar] [CrossRef]
- Joachim, A.; Ahmed, M.I.M.; Pollakis, G.; Rogers, L.; Hoffmann, V.S.; Munseri, P.; Aboud, S.; Lyamuya, E.F.; Bakari, M.; Robb, M.L.; et al. Induction of Identical IgG HIV-1 Envelope Epitope Recognition Patterns After Initial HIVIS-DNA/MVA-CMDR Immunization and a Late MVA-CMDR Boost. Front. Immunol. 2020, 11, 719. [Google Scholar] [CrossRef]
- Rolland, M.; Edlefsen, P.T.; Larsen, B.B.; Tovanabutra, S.; Sanders-Buell, E.; Hertz, T.; DeCamp, A.C.; Carrico, C.; Menis, S.; Magaret, C.A.; et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature 2012, 490, 417–420. [Google Scholar] [CrossRef]
- Tomaras, G.D.; Haynes, B.F. HIV-1-specific antibody responses during acute and chronic HIV-1 infection. Curr. Opin. HIV AIDS 2009, 4, 373–379. [Google Scholar] [CrossRef]
- Karnasuta, C.; Akapirat, S.; Madnote, S.; Savadsuk, H.; Puangkaew, J.; Rittiroongrad, S.; Rerks-Ngarm, S.; Nitayaphan, S.; Pitisuttithum, P.; Kaewkungwal, J.; et al. Comparison of Antibody Responses Induced by RV144, VAX003, and VAX004 Vaccination Regimens. AIDS Res. Hum. Retrovir. 2017, 33, 410–423. [Google Scholar] [CrossRef]
- Tomaras, G.D.; Ferrari, G.; Shen, X.; Alam, S.M.; Liao, H.-X.; Pollara, J.; Bonsignori, M.; Moody, M.A.; Fong, Y.; Chen, X.; et al. Vaccine-induced plasma IgA specific for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG. Proc. Natl. Acad. Sci. USA 2013, 110, 9019–9024. [Google Scholar] [CrossRef] [PubMed]
- Easterhoff, D.; Pollara, J.; Luo, K.; Janus, B.; Gohain, N.; Williams, L.D.; Tay, M.Z.; Monroe, A.; Peachman, K.; Choe, M.; et al. HIV vaccine delayed boosting increases Env variable region 2-specific antibody effector functions. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT04066881 (accessed on 29 July 2020).
| First Randomization (Immunizations at Weeks 0, 4, 12) | ||||
| Group | A | B | C | |
| Immunization * | Two ID HIV-DNA injections (total 600 μg) | Two ID HIV-DNA injections (total 600 μg) with EP | One ID HIV-DNA injection (total 600 μg) with EP | |
| Second Randomization (Immunizations at Weeks 24, 40) | ||||
| Group | 1 | 2 | ||
| Immunization | 108 pfu HIV-MVA IM | HIV-MVA 108 pfu IM plus 100 μg CN54rgp140/GLA-AF | ||
| IDR | Peptide Position | HXB2 Position | Env Region | Representative Sequence | FOR (%) | Mean FI | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| MVA Only | MVA+rgp140 | Placebo | MVA Only | MVA+rgp140 | Placebo | |||||
| 1_V2 | 176 | 164 | V2 | ELRDKKQKVHALFYK | 68 | 63 | 0 | 20,587 | 14,486 | 0 |
| 2_C2 | 221 | 200 | C2 | AITQACPKVTFDPIP | 25 | 72 | 9 | 0 | 9188 | 0 |
| 3_V3 | 321 | 300 | V3 | GNNTRKSIRIGPGQT | 21 | 84 | 9 | 0 | 18,855 | 0 |
| 322 | 301 | V3 | NNTRKSIRIGPGQTF | 64 | 91 | 18 | 23,543 | 40,829 | 0 | |
| 325 | 304 | V3 | RKSIRIGPGSTFYAT | 68 | 100 | 18 | 25,586 | 45,585 | 0 | |
| 326 | 305 | V3 | KSVRIGPGQTFYATG | 82 | 97 | 27 | 28,859 | 43,633 | 12,656 | |
| 4_C4 | 461 | 433 | C4 | AMYAPPIAGNITCKS | 25 | 75 | 27 | 0 | 16,693 | 9640 |
| 5_gp41 | 612 | 580 | gp41 | VLAVERYLKDQKFLG | 86 | 78 | 0 | 11,773 | 13,694 | 0 |
| Antibody | Antigen (gp70V1V2) | HIV Subtype | Frequency of Responses (Positive/Tested, %) | ||
|---|---|---|---|---|---|
| HIV-MVA | HIV-MVA + CN54rgp140/GLA-AF | p value | |||
| IgG | A244 | CRF01_AE | 25/31 (81) | 18/25 (72) | 0.5318 |
| IgG | CN54 | C | 20/31 (65) | 15/25 (60) | 0.7859 |
| IgG1 | A244 | CRF01_AE | 25/32 (78) | 19/25 (76) | >0.9999 |
| IgG3 | A244 | CRF01_AE | 12/32 (38) | 2/25 (8) | 0.0132 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Msafiri, F.; Joachim, A.; Held, K.; Nadai, Y.; Chissumba, R.M.; Geldmacher, C.; Aboud, S.; Stöhr, W.; Viegas, E.; Kroidl, A.; et al. Frequent Anti-V1V2 Responses Induced by HIV-DNA Followed by HIV-MVA with or without CN54rgp140/GLA-AF in Healthy African Volunteers. Microorganisms 2020, 8, 1722. https://doi.org/10.3390/microorganisms8111722
Msafiri F, Joachim A, Held K, Nadai Y, Chissumba RM, Geldmacher C, Aboud S, Stöhr W, Viegas E, Kroidl A, et al. Frequent Anti-V1V2 Responses Induced by HIV-DNA Followed by HIV-MVA with or without CN54rgp140/GLA-AF in Healthy African Volunteers. Microorganisms. 2020; 8(11):1722. https://doi.org/10.3390/microorganisms8111722
Chicago/Turabian StyleMsafiri, Frank, Agricola Joachim, Kathrin Held, Yuka Nadai, Raquel Matavele Chissumba, Christof Geldmacher, Said Aboud, Wolfgang Stöhr, Edna Viegas, Arne Kroidl, and et al. 2020. "Frequent Anti-V1V2 Responses Induced by HIV-DNA Followed by HIV-MVA with or without CN54rgp140/GLA-AF in Healthy African Volunteers" Microorganisms 8, no. 11: 1722. https://doi.org/10.3390/microorganisms8111722
APA StyleMsafiri, F., Joachim, A., Held, K., Nadai, Y., Chissumba, R. M., Geldmacher, C., Aboud, S., Stöhr, W., Viegas, E., Kroidl, A., Bakari, M., Munseri, P. J., Wahren, B., Sandström, E., Robb, M. L., McCormack, S., Joseph, S., Jani, I., Ferrari, G., ... Nilsson, C. (2020). Frequent Anti-V1V2 Responses Induced by HIV-DNA Followed by HIV-MVA with or without CN54rgp140/GLA-AF in Healthy African Volunteers. Microorganisms, 8(11), 1722. https://doi.org/10.3390/microorganisms8111722





