Single Mathematical Parameter for Evaluation of the Microorganisms’ Growth as the Objective Function in the Optimization by the DOE Techniques
Abstract
:1. Introduction
2. Theoretical Background
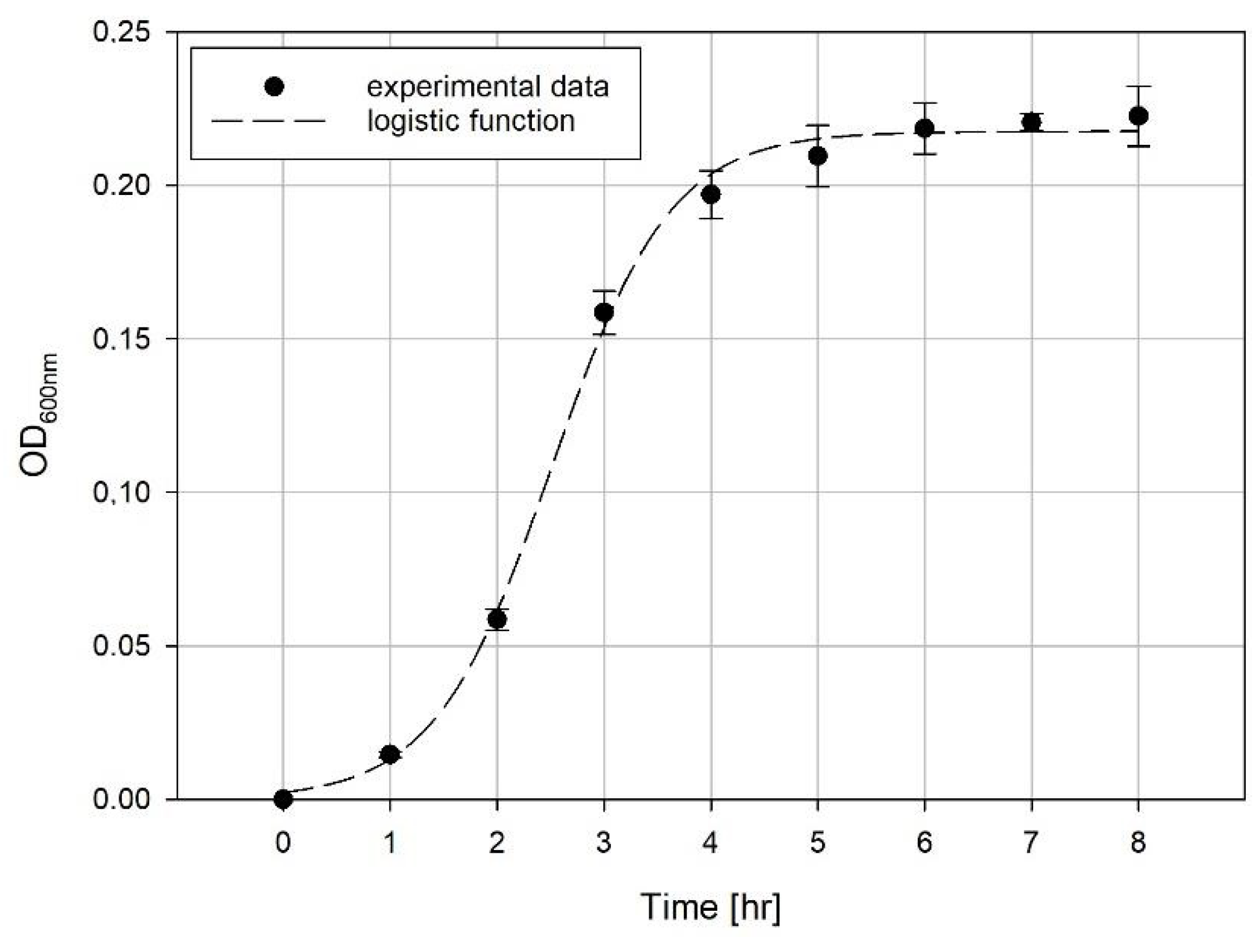
Evaluation of Growth Curves
3. Materials and Methods
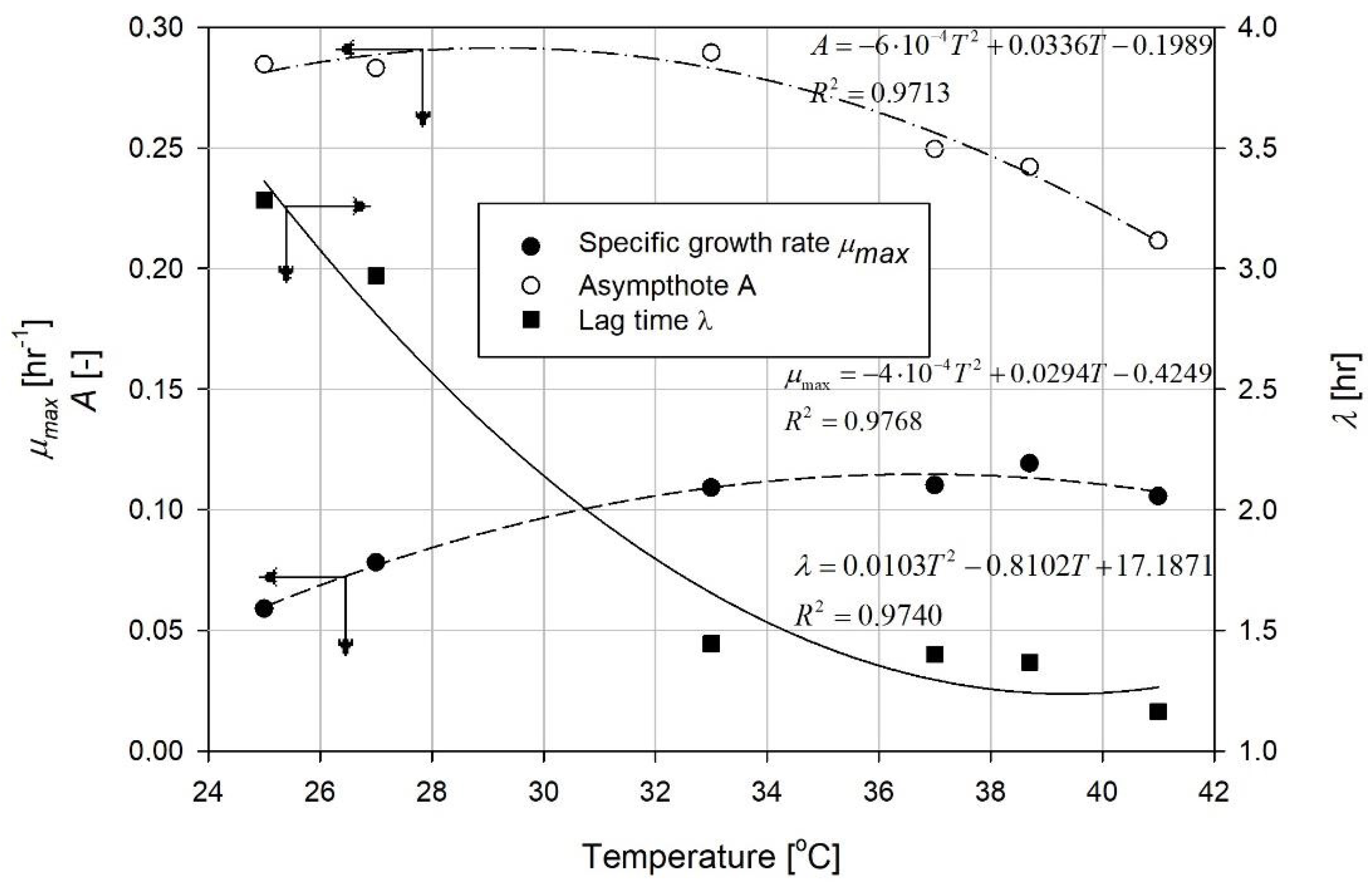
4. Results and Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cardoso, V.M.; Campani, G.; Santos, M.P.; Silva, G.G.; Pires, M.C.; Gonçalves, V.M.; Giordano, R.D.C.; Sargo, C.R.; Horta, A.C.; Zangirolami, T.C. Cost Analysis Based on Bioreactor Cultivation Conditions: Production of a Soluble Recombinant Protein Using Escherichia coli BL21(DE3). Biotechnol. Rep. 2020, 26, e00441. [Google Scholar] [CrossRef]
- Cha, J.W.; Jang, S.H.; Kim, Y.J.; Chang, Y.K.; Jeong, K.J. Engineering of Klebsiella Oxytoca for Production of 2,3-Butanediol Using Mixed Sugars Derived from Lignocellulosic Hydrolysates. GCB Bioenergy 2020, 12, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Tashiro, T.; Yoshimura, F. A Neo-Logistic Model for the Growth of Bacteria. Phys. A Stat. Mech. Appl. 2019, 525, 199–215. [Google Scholar] [CrossRef] [Green Version]
- Pereira, S.; Otero, A. Haematococcus Pluvialis Bioprocess Optimization: Effect of Light Quality, Temperature and Irradiance on Growth, Pigment Content and Photosynthetic Response. Algal Res. 2020, 51, 102027. [Google Scholar] [CrossRef]
- Derakhshandeh, M.; Un, U.T. Optimization of Microalgae Scenedesmus SP. Growth Rate Using a Central Composite Design Statistical Approach. Biomass Bioenergy 2019, 122, 211–220. [Google Scholar] [CrossRef]
- Gelain, L.; Van Der Wielen, L.; Van Gulik, W.M.; Pradella, J.G.D.C.; Da Costa, A.C. Mathematical Modelling for the Optimization of Cellulase Production Using Glycerol for Cell Growth and Cellulose as the Inducer Substrate. Chem. Eng. Sci. 2020, 116048. [Google Scholar] [CrossRef]
- Medina-Cabrera, E.V.; Rühmann, B.; Schmid, J.; Sieber, V. Optimization of Growth and EPS Production in Two Porphyridum Strains. Bioresour. Technol. Rep. 2020, 11, 100486. [Google Scholar] [CrossRef]
- Lechowska, J.; Kordas, M.; Konopacki, M.; Fijałkowski, K.; Drozd, R.; Rakoczy, R. Hydrodynamic Studies in Magnetically Assisted External-Loop Airlift Reactor. Chem. Eng. J. 2019, 362, 298–309. [Google Scholar] [CrossRef]
- Rakoczy, R.; Konopacki, M.; Lechowska, J.; Bubnowska, M.; Hürter, A.; Kordas, M.; Fijałkowski, K. Gas to Liquid Mass Transfer in Mixing System with Application of Rotating Magnetic Field. Chem. Eng. Process. Process Intensif. 2018, 130, 11–18. [Google Scholar] [CrossRef]
- Konopacka, A.; Rakoczy, R.; Konopacki, M. The Effect of Rotating Magnetic Field on Bioethanol Production by Yeast Strain Modified by Ferrimagnetic Nanoparticles. J. Magn. Magn. Mater. 2019, 473, 176–183. [Google Scholar] [CrossRef]
- Yang, J.; Ma, C.; Tao, J.; Li, J.; Du, K.; Wei, Z.; Chen, C.; Wang, Z.; Zhao, C.; Ma, M. Optimization of Polyvinylamine-Modified Nanocellulose for Chlorpyrifos Adsorption by Central Composite Design. Carbohydr. Polym. 2020, 245, 116542. [Google Scholar] [CrossRef] [PubMed]
- Leili, M.; Khorram, N.S.; Godini, K.; Azarian, G.; Moussavi, R.; Peykhoshian, A. Application of Central Composite Design (CCD) for Optimization of Cephalexin Antibiotic Removal Using Electro-Oxidation Process. J. Mol. Liq. 2020, 313, 113556. [Google Scholar] [CrossRef]
- Kenawy, I.M.; Eldefrawy, M.M.; Eltabey, R.M.; Zaki, E. Melamine Grafted Chitosan-Montmorillonite Nanocomposite for Ferric Ions Adsorption: Central Composite Design Optimization Study. J. Clean. Prod. 2019, 241, 118189. [Google Scholar] [CrossRef]
- Chang, K.-H. Multiobjective Optimization and Advanced Topics. In Design Theory and Methods Using CAD/CAE; Elsevier BV: Amsterdam, The Netherlands, 2015; pp. 325–406. [Google Scholar]
- Zwietering, M.H.; Jongenburger, I.; Rombouts, F.M.; Riet, K.V.T. Modeling of the Bacterial Growth Curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwietering, M.H.; De Koos, J.T.; Hasenack, B.E.; De Witt, J.C.; Riet, K.V. Modeling of Bacterial Growth as a Function of Temperature. Appl. Environ. Microbiol. 1991, 57, 1094–1101. [Google Scholar] [CrossRef] [Green Version]
- Konopacki, M.; Rakoczy, R. The Analysis of Rotating Magnetic Field as a Trigger of Gram-Positive and Gram-Negative Bacteria Growth. Biochem. Eng. J. 2019, 141, 259–267. [Google Scholar] [CrossRef]
- Grimont, P.A.D.; Grimont, F. Klebsiella. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–26. [Google Scholar] [CrossRef]
- Mattila, S.; Ruotsalainen, P.; Jalasvuori, M. On-Demand Isolation of Bacteriophages against Drug-Resistant Bacteria for Personalized Phage Therapy. Front. Microbiol. 2015, 6, 1271. [Google Scholar] [CrossRef] [Green Version]
- Grygorcewicz, B.; Grudziński, M.; Wasak, A.; Augustyniak, A.; Pietruszka, A.; Nawrotek, P. Bacteriophage- Mediated Reduction of Salmonella Enteritidis in Swine Slurry. Appl. Soil Ecol. 2017, 119, 179–182. [Google Scholar] [CrossRef]
- Augustyniak, A.; Grygorcewicz, B.; Nawrotek, P. Isolation of Multidrug Resistant Coliforms and Their Bacteriophages from Swine Slurry. Turk. J. Vet. Anim. Sci. 2018, 42, 319–325. [Google Scholar] [CrossRef]
- Danis-Wlodarczyk, K.; Vandenheuvel, D.; Bin Jang, H.; Briers, Y.; Olszak, T.; Arabski, M.; Wasik, S.; Drabik, M.; Higgins, G.; Tyrrell, J.; et al. A Proposed Integrated Approach for the Preclinical Evaluation of Phage Therapy in Pseudomonas Infections. Sci. Rep. 2016, 6, 28115. [Google Scholar] [CrossRef] [Green Version]
- Konopacki, M.; Grygorcewicz, B.; Dołęgowska, B.; Kordas, M.; Rakoczy, R. PhageScore: A Simple Method for Comparative Evaluation of Bacteriophages Lytic Activity. Biochem. Eng. J. 2020, 161, 107652. [Google Scholar] [CrossRef]
- Zagólski, O.; Stręk, P.; Kasprowicz, A.; Białecka, A. Effectiveness of Polyvalent Bacterial Lysate and Autovaccines Against Upper Respiratory Tract Bacterial Colonization by Potential Pathogens: A Randomized Study. Med. Sci. Monit. 2015, 21, 2997–3002. [Google Scholar] [CrossRef] [Green Version]
- Wei, D.; Gu, J.; Zhang, Z.; Wang, C.; Kim, C.H.; Jiang, B.; Shi, J.; Hao, J. Production of Chemicals by Klebsiella Pneumoniae Using Bamboo Hydrolysate as Feedstock. J. Vis. Exp. 2017, 1–7. [Google Scholar] [CrossRef]
- Tsuchiya, K.; Cao, Y.-Y.; Kurokawa, M.; Ashino, K.; Yomo, T.; Ying, B.-W. A Decay Effect of the Growth Rate Associated with Genome Reduction in Escherichia coli. BMC Microbiol. 2018, 18, 101. [Google Scholar] [CrossRef] [PubMed]
- Peleg, M.; Corradini, M.G. Microbial Growth Curves: What the Models Tell Us and What They Cannot. Crit. Rev. Food Sci. Nutr. 2011, 51, 917–945. [Google Scholar] [CrossRef] [PubMed]
- Todor, H.; Dulmage, K.; Gillum, N.; Bain, J.R.; Muehlbauer, M.J.; Schmid, A.K. A Transcription Factor Links Growth Rate and Metabolism in the Hypersaline Adapted Archaeon Halobacterium Salinarum. Mol. Microbiol. 2014, 93, 1172–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sikora, P.; Augustyniak, A.; Cendrowski, K.; Nawrotek, P.; Mijowska, E. Antimicrobial Activity of Al2O3, CuO, Fe3O4, and ZnO Nanoparticles in Scope of Their Further Application in Cement-Based Building Materials. Nanomaterials 2018, 8, 212. [Google Scholar] [CrossRef] [Green Version]
- Provost, J.; Wallert, M. MTT Proliferation Assay Protocol. Available online: http://web.mnstate.edu/provost/MTT Proliferation Assay Protocol 2012.pdf (accessed on 15 September 2020).
- Cuzon, G.; Naas, T.; Truong, H.; Villegas, M.-V.; Wisell, K.T.; Carmeli, Y.; Gales, A.C.; Navon-Venezia, S.; Quinn, J.P.; Nordmann, P. Worldwide Diversity of Klebsiella Pneumoniae That Produce β-lactamase blaKPC-2Gene. Emerg. Infect. Dis. 2010, 16, 1349–1356. [Google Scholar] [CrossRef]
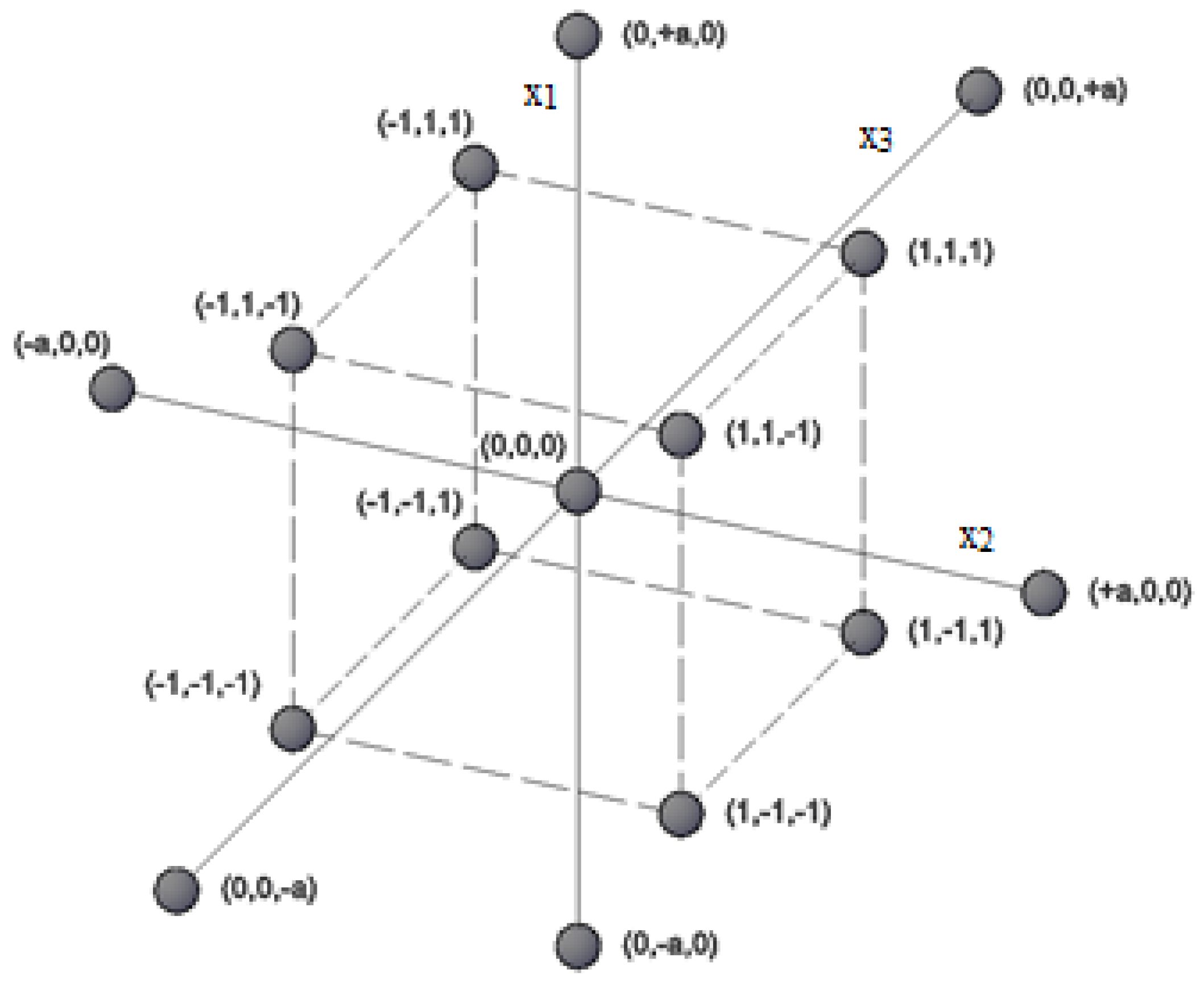



| Experiment | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| x1 | 1 | −1 | 1 | −1 | 1 | −1 | 1 | −1 | −a | +a | 0 | 0 | 0 | 0 | 0 |
| x2 | 1 | 1 | −1 | −1 | 1 | 1 | -1 | −1 | 0 | 0 | −a | +a | 0 | 0 | 0 |
| x3 | 1 | 1 | 1 | 1 | −1 | −1 | −1 | −1 | 0 | 0 | 0 | 0 | −a | +a | 0 |
| y | y1 | y2 | y3 | y4 | y5 | y6 | y7 | y8 | y9 | y10 | y11 | y12 | y13 | y14 | y15 |
| Temperature [°C] | Function Coefficients | R2 | ||
|---|---|---|---|---|
| a | b | c | ||
| 25 | 0.2846 | 4.7225 | 0.8289 | 0.9981 |
| 27.3 | 0.2831 | 5.2786 | 1.1033 | 0.9929 |
| 33 | 0.2895 | 4.1777 | 1.5073 | 0.9959 |
| 37 | 0.2495 | 1.7655 | 1.7655 | 0.9991 |
| 38.7 | 0.2421 | 4.6926 | 1.9688 | 0.9998 |
| 41 | 0.2116 | 4.3208 | 1.9952 | 0.9978 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konopacki, M.; Augustyniak, A.; Grygorcewicz, B.; Dołęgowska, B.; Kordas, M.; Rakoczy, R. Single Mathematical Parameter for Evaluation of the Microorganisms’ Growth as the Objective Function in the Optimization by the DOE Techniques. Microorganisms 2020, 8, 1706. https://doi.org/10.3390/microorganisms8111706
Konopacki M, Augustyniak A, Grygorcewicz B, Dołęgowska B, Kordas M, Rakoczy R. Single Mathematical Parameter for Evaluation of the Microorganisms’ Growth as the Objective Function in the Optimization by the DOE Techniques. Microorganisms. 2020; 8(11):1706. https://doi.org/10.3390/microorganisms8111706
Chicago/Turabian StyleKonopacki, Maciej, Adrian Augustyniak, Bartłomiej Grygorcewicz, Barbara Dołęgowska, Marian Kordas, and Rafał Rakoczy. 2020. "Single Mathematical Parameter for Evaluation of the Microorganisms’ Growth as the Objective Function in the Optimization by the DOE Techniques" Microorganisms 8, no. 11: 1706. https://doi.org/10.3390/microorganisms8111706
APA StyleKonopacki, M., Augustyniak, A., Grygorcewicz, B., Dołęgowska, B., Kordas, M., & Rakoczy, R. (2020). Single Mathematical Parameter for Evaluation of the Microorganisms’ Growth as the Objective Function in the Optimization by the DOE Techniques. Microorganisms, 8(11), 1706. https://doi.org/10.3390/microorganisms8111706








