Abstract
Potash tailing piles caused by fertilizer production shape their surroundings because of the associated salt impact. A previous study in these environments addressed the functional community “biocrust” comprising various micro- and macro-organisms inhabiting the soil surface. In that previous study, biocrust microalgae and cyanobacteria were isolated and morphologically identified amongst an ecological discussion. However, morphological species identification maybe is difficult because of phenotypic plasticity, which might lead to misidentifications. The present study revisited the earlier species list using an integrative approach, including molecular methods. Seventy-six strains were sequenced using the markers small subunit (SSU) rRNA gene and internal transcribed spacer (ITS). Phylogenetic analyses confirmed some morphologically identified species. However, several other strains could only be identified at the genus level. This indicates a high proportion of possibly unknown taxa, underlined by the low congruence of the previous morphological identifications to our results. In general, the integrative approach resulted in more precise species identifications and should be considered as an extension of the previous morphological species list. The majority of taxa found were common in saline habitats, whereas some were more likely to occur in nonsaline environments. Consequently, biocrusts in saline environments of potash tailing piles contain unique microalgae and cyanobacteria that will possibly reveal several new taxa in more detailed future studies and, hence, provide new data on the biodiversity, as well as new candidates for applied research.
1. Introduction
Biological soil crusts (biocrusts) are multidimensional communities consisting of various micro- and macro-organisms, which inhabit the first millimeters of the soil [1]. They include microalgae and cyanobacteria, as well as protists, heterotrophic bacteria, and fungi, and in later successional stages, also lichens and mosses might grow [1]. As ecosystem engineers, biocrusts increase the soil stability; alter hydrological cycles; and provide nutrients via primary production, aerial N-fixation, dust trapping, and bio-weathering [2,3]. Biocrusts occur in all climate zones worldwide; they have their greatest impacts where vascular plant growth is limited due to extreme abiotic stressors [2]. In drylands, biocrusts cover large areas, thus creating biodiversity hot spots in these hostile environments [2], and can reach a stable climax state [4]. However, biocrusts are vulnerable against disturbances, such as trampling [5,6,7,8], and endangered by climate change [9]. This stresses the relevance of biocrust research, along with the great interest on their multifunctional ecosystem services in the growing field of applied biocrust research [10,11,12,13,14].
Microalgae and cyanobacteria in biocrusts are well-adapted to their harsh, mainly arid environments [15]. Cyanobacteria and filamentous algae are key organisms in biocrusts and excrete sticky extracellular polysaccharides (EPS). EPS are important in promoting the cohesion of biocrust inhabitants and the connection to soil particles and act as water-holding biomolecules in drought tolerance [16]. Overall, desiccation stress in biocrusts has been intensively studied, with the focus on a variety of organisms such as green algae [17,18] and cyanobacteria [19,20] to address the large-scale ecological aspects [21,22], as well as the (eco-)physiological processes [23,24].
Similarly, the salt tolerance of cyanobacteria [25,26], as well as of algae, in marine habitats [27,28] is well-studied. Cells under saline conditions are exposed to water deprivation analogous to drought conditions but, also, show differences in some physiological processes [25,28]. In contrast to the total absence of water, salt-stressed cells can still be in contact with liquid water yet be subjected to a diminished water potential. The loss of intracellular water due to the high osmotic potential outside the cell can be hindered by inorganic and organic osmolytes. These compatible solutes increase the osmotic potential within the cell and thereby prevent water loss. In addition, the toxic Na+ ions are exchanged for the nutrient K+ to maintain the cellular ion balance. Due to their increased production of some commercially relevant secondary metabolites under salt stress, certain microalgae such as Dunaliella salina are of interest for large-scale biomass production [29,30]. Hypersaline wastewater from the potash industry can be used for this purpose [31].
Halophilic microbial communities have been addressed in a variety of studies, such as microbial mats in hypersaline lakes [32,33,34] and lagoonal mats [35,36], as well as benthic communities in solar saltern plants [37,38,39]. In these hypersaline aquatic habitats, photosynthetic organisms were found in salinities even higher than 30%, such as the green alga Dunaliella [40,41] and the cyanobacteria Geitlerinema, Phormidium and Komvophoron [42]. Further, several studies have described the biodiversity of algae and cyanobacteria in saline terrestrial habitats [35,42,43,44,45,46,47,48,49,50,51,52,53]. Most of them focused on natural saline habitats.
Anthropogenic salinization, however, is increasingly affecting aquatic [54], as well as terrestrial [55,56], ecosystems on a global scale. In Europe, one of the main drivers of anthropogenic salinization is the potash industry for fertilizer production [57]. The valuable components (KCl and MgSO4) of the mined potash salt are separated from the residue NaCl. Potash tailing piles, as a consequence, consist of highly saline overburden (mostly of NaCl). In central Germany, in particular, potash tailing piles, reaching a height of 200 m, shape the landscape. Rainfall dissolves the deposited salt and washes out highly saline pile wastewaters, which, in one potash tailing pile in Germany, contained dissolved concentrations up to 320 g/L (very close to saturation; predominantly Na+ and Cl−) [58]. Without a base seal, this process leads to a salinization of the surrounding ecosystems.
As a result, unique salt-affected ecosystems emerge, inhabited by adapted costal flora and fauna, despite a distance of about 400 km inland from the coast. Halophyte plant communities, with members such as the sea aster (Tripolium pannonicum) and glasswort (Salicornia sp.), have been well-studied close to these tailing piles [59,60,61]. However, higher plants occur only in the surroundings of potash tailing piles and do not inhabit the pile body itself, due to the extreme salinity of the overburden. Little is known about biocrusts in the surroundings of such hypersaline potash tailing piles. Eilmus et al. [62] found a rich prokaryotic community directly on the bare material of a German potash tailing pile, whereas our previous study described diverse communities of both cyanobacteria and eukaryotic microalgae in biological soil crusts between the less-saline vegetation line and the hyper-saline bare pile bodies of several potash tailing piles across Germany [63]. In this previous study, we found species numbers comparable to those in less-saline regions, which underlined the significance of research on those specific terrestrial habitats. However, that study was based on morphological observations only. Investigations on biocrust algae in saline terrestrial inland habitats using modern molecular phylogenetic approaches are still rare.
Morphological methods based on light microscopy have a long tradition and continue to be essential for microalgal and cyanobacteria species identification. However, the phenotypic plasticity of unicellular algae due to culture conditions, age, and reproductive stages complicate identifications. In particular, salinity has an impact on the morphology of algal cells [64]. In addition, microalgal and cyanobacteria cells often lack distinct morphological traits, which limits the use of light microscopy for accurate species identification. This limitation often results in a high proportion of identifications only to the family or genus level [65]. Sommer et al. [63], for example, identified only 32% of their microalgal strains from saline potash tailing piles to the species level.
To accurately identify algal strains, an integrative approach should be considered. This approach combines classical morphological methods with modern molecular phylogenetic analyses. It was introduced by Komárek [66] for cyanobacteria, followed by Pröschold and Leliaert [67] for green algae, and has been increasingly used in microalgal [65,68,69,70,71,72,73], as well as cyanobacterial [35,74,75,76,77], research. Some recent reviews stressed the advantages and challenges of this approach [78,79,80,81]. Still, some limitations on microalgal and cyanobacteria identification remain, such as a lack of identified sequences in databases; nevertheless, the combination of the mentioned methods highly improves the quality of the species lists, thereby providing precise phylogenetic, and not biodiversity, information.
Revisiting morphologically identified strains with molecular methods can lead to the finding of new species or rare taxa that would have been overlooked by using only light microscopy [65,72,82]. Particularly in extreme habitats, the probability of finding yet undiscovered species is high [83] due to the unique ecological niches inhabited by adapted organisms. In the extreme habitats of potash tailing piles, there might well be a high potential of discovering previously unknown, highly specialized cyanobacteria and microalgae.
The aim of our study was to complement the information on biocrust microalgal and cyanobacterial cultures from the surroundings of saline potash tailing piles as a continuation of the previous morphological observations [63]. Therefore, common conserved markers for the small subunit ribosomal ribonucleic acid (SSU rRNA) gene, as well as the variable internal transcribed spacer (ITS), were used to elucidate the identity and phylogenetic classification of these green algae and cyanobacteria. Using the integrative approach, we assumed that we would be able to generate more detailed phylogenetic information compared to the morphological observations alone.
2. Materials and Methods
2.1. Algal and Cyanobacteria Isolates and Their Maintenance
In total, 76 original strains were analyzed in this study. A total of 66 strains, including 52 green algae and 14 cyanobacteria strains isolated from the surroundings of potash tailing piles in Germany, were published earlier. This paper included the respective site descriptions, samplings, and isolation procedures [63]. In addition, ten unpublished green algae strains isolated from two of the sampling sites (TT and NN) of the previous publication [63], as well as from another location at the Teutschenthal tailing piles (TTF, Figure 1), were included in this study. The latter site was situated in the nearest environment to the tailing piles in an abandoned lignite mine (Figure 1). Rainwater dissolves some salt from the tailing pile. These strongly saline brines frequently flood the former mining area. Thin biocrusts were found between the brine channels and the bordering vegetation (Figure 1c). The sampling procedure and isolation of the unialgal strains were performed as described previously [63].
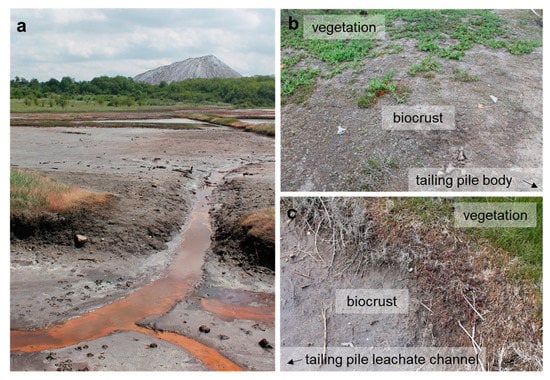
Figure 1.
The potash tailing pile in Teutschenthal, Germany. (a) The plain in front of the tailing pile represents an abandoned lignite mine, which is regularly flooded by highly saline tailing pile leachate. (b) Biocrusts close to the Teutschenthal tailing pile between the vegetation and tailing pile body and (c) between the vegetation and tailing pile leachate water channels.
Unialgal cultures were maintained on solidified 3N-BBM+V [84] and cyanobacteria cultures on BG-11 [85], both media enriched with 3% NaCl. Cultures were kept at 20 °C with a light/dark cycle of 16:8 h with 30-µm photons m−2 s−1 (Osram Lumilux Cool White lamps L36W/840, Munich, Germany). Isolates were morphologically evaluated using an Olympus BX51 light microscope with Nomarski DIC optics (Olympus Ltd., Hamburg, Germany), and photomicrographs were taken with the digital camera Olympus UC30 attached to the microscope and processed with the cellSense Entry imaging software (v. 2.1, Olympus Soft Imaging Solutions, Münster, Germany). The morphological species list of green algae and cyanobacteria isolates from potash tailing pile areas was published by Sommer et al. [63].
2.2. DNA Extraction, PCR, and Sequencing
The SSU region, as well as the ITS1-5.8S-ITS2 region (hereafter, ITS-1,2 regions), and in particular groups, the ITS-2 region for green algae and the SSU-LSU rRNA (large subunit rRNA) intergenic spacer for cyanobacteria were used as molecular markers. The cells of green algae and cyanobacteria were disrupted by shaking with 1.25–1.55-mm glass beads in combination with a threefold freezing and thawing cycle (liquid N, heating block 65 °C). The DNA was extracted with the NucleoSpin Plant II mini kit (Macherey Nagel, Düren, Germany) following the instructions. PCR was performed as described by Mikhailyuk et al. [86] using the primers EAF3 and ITS055R for green algae [87,88] and SSU-4-forw and ptLSU C-D-rev for cyanobacteria [89]. Sanger sequencing was conducted by GATC Sequencing Services (Eurofins Genomics Germany, Ebersberg, Germany) using the primers EAF3 and 1400R [87], N920R [88], 536R [90], 920F and 1400F [91], and GF and GR [92] for green algae and SSU-4-forw, Wil 6, Wil 12, Wil 14, Wil 5, Wil 9, Wil 16, and ptLSU C-D-rev [89,93] for cyanobacteria.
2.3. Phylogenetic Analyses
Bioinformatics steps were performed using the software Geneious 8.1.9 (Biomatters Ltd., v. 8.1.9, Auckland, New Zealand), if not stated differently. The sequences of the isolates were compared to identified taxa in the Gene Bank database using BLASTn [94]. Top BLAST hits, as well as some additional sequences, were downloaded (Supplementary Table S1) and aligned with the original isolates. The MUSCLE algorithm [95] was used with a maximum of eight iterations. The alignments were cut and checked for errors manually. Bayesian phylogenic trees were calculated with the MrBayes [96] add-in by using the GTR+G+I evolutionary model run for 5,000,000 generations and taking the trees every 500 generations. Of the four runs of the Markov chain Monte Carlo, two were run separately, with split frequencies at the end of the runs below 0.01. Maximum-likelihood (ML) analyses were performed to verify the Bayesian results, using GARLI [97] with 1000 bootstrap replicates. Phylogenetic consensus trees were edited in the MEGA-X 10.1 software [98] and finalized with PowerPoint (Microsoft Office, Standard 2013, Redmond, WA, USA).
Taxa were named after current taxonomically accepted species or genus names stated in AlgaeBase [99] using the respective references (Table 1). Subsequently, the original habitats of all strains were classified as follows: aquatic (frequent presence of water, e.g., marine, freshwater, brackish water, bog water; both planktonic and benthic, or in microbial mats); terrestrial (in the absence of permanent water, e.g., on soil, bark, leaves, stones, facades, in biocrusts, or other nonaquatic biofilms); and endophytic/phycobiontic (e.g., phycobionts in lichens or endophytes in macrophytes, hereafter endophytic). The symbols are colored according to the salinity of the habitats (blue = nonsaline, e.g., freshwater and soil; orange = saline, e.g., marine, brackish and saline soil; black = other extreme conditions, e.g., acidity, heat, and radioactivity; and grey: unknown).

Table 1.
Integrative species identifications compared to morphological identifications in a previous study [63]. Possibly undescribed taxa are marked (•). Additional strains that were not published and morphologically identified previously are placed in parentheses.
3. Results
3.1. SSU rRNA Gene Phylogeny
The 76 original strains were distributed among the Chlorophyta (62) and cyanobacteria (14). All green-algal strains belonged to the phylum Chlorophyta; the phylum Streptophyta was absent. Of the Chlorophyta, 32% (20 isolates) fell into the Chlorophyceae. The majority of these original strains (11 isolates) were placed in the order Chlamydomonadales (Figure 2). We found lineages in the Moewusinia (genus Alvikia and two separate lineages close to the genera Spongiococcum/Actinochloris and Axilosphaera/Eubrownia), Chlorogonia (separate lineage close to Chlorogonium), and Chloromonadinia (Chloromonas), as well as an independent lineage formed by the genus Borodinellopsis. In the second order present, the Sphaeropleales (nine isolates, 14%; Figure 3), the strains clustered in the lineages formed by the genera Bracteacoccus and Tetradesmus.
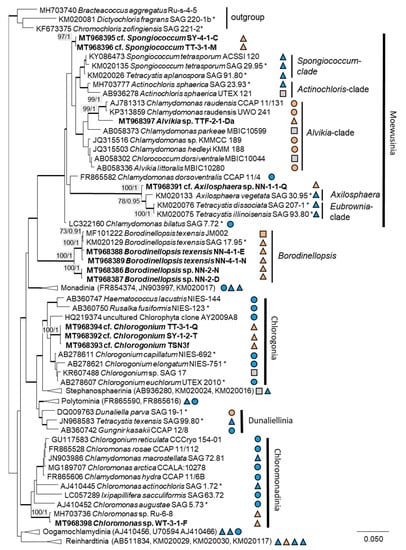
Figure 2.
Molecular phylogeny of the Chlamydomonadales based on small subunit ribosomal ribonucleic acid (SSU rRNA) gene sequencing. The phylogenetic tree was calculated by the Bayesian method, including posterior probabilities (PP) with additional maximum likelihood bootstrap values (BP); branches are supported by both methods in bold (BP > 60% and PP > 0.9). Authentic strains were marked with an asterisk, original strains from this study in bold. Habitats of the strains were classified according to habitat type (Δ terrestrial = absence of permanent water, o aquatic = presence of water, and □ unknown) and saline ( ); nonsaline (
); nonsaline ( ); and unknown (
); and unknown ( ) habitats.
) habitats.
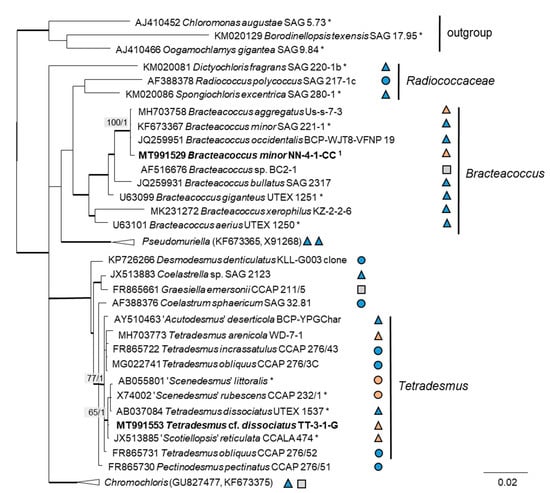
Figure 3.
Molecular phylogeny of the Sphaeropleales based on SSU rRNA gene sequencing. The phylogenetic tree was calculated by the Bayesian method, including posterior probabilities (PP) with additional maximum likelihood bootstrap values (BP); branches are supported by both methods in bold (BP > 60% and PP > 0.9). Authentic strains were marked with an asterisk, original strains from this study in bold. Habitats of the strains were classified according to habitat type (Δ terrestrial = absence of permanent water, o aquatic = presence of water, and □ unknown) and saline ( ); nonsaline (
); nonsaline ( ); and unknown (
); and unknown ( ) habitats. 1 Identical to Bracteacoccus minor NN-4-1-CC (MT991529), NN-4-1-H (MT991530), NN-4-1-D2 (MT968390), TTF-1-1-M (MT991533), TTF-2-1-A (MT991534), and TTF-2-1-J (MT991535).
) habitats. 1 Identical to Bracteacoccus minor NN-4-1-CC (MT991529), NN-4-1-H (MT991530), NN-4-1-D2 (MT968390), TTF-1-1-M (MT991533), TTF-2-1-A (MT991534), and TTF-2-1-J (MT991535).
Twenty-two percent (14 isolates) of the original strains belonging to Chlorophyta were distributed among the Ulvophyceae (Figure 4). They clustered in the order Ulvales, Desmochloris-clade (genera Desmochloris and Halochlorococcum), as well as in the order Ulotrichales, Planophila-clade (genus Planophila) and Acrosiphonia-clade (a separate lineage close to the clade with “Ulothrix” and Chlorothrix species). The majority of strains (45% of the Chlorophyta strains) fell in the Trebouxiophyceae (28 isolates, Figure 5) and were distributed among the genera Chloroidium, Watanabea, Diplosphaera, Pseudostichococcus, Pseudochlorella, Chlorella, and Nannochloris. Fourteen percent (two isolates) of the cyanobacteria isolates (Figure 6) clustered in the Nostocales (genus Cyanocohniella), and 86% (12 isolates) fell in the Synechococcales (genus Nodosilinea and an unresolved clade labeled “Phormidesmis”).
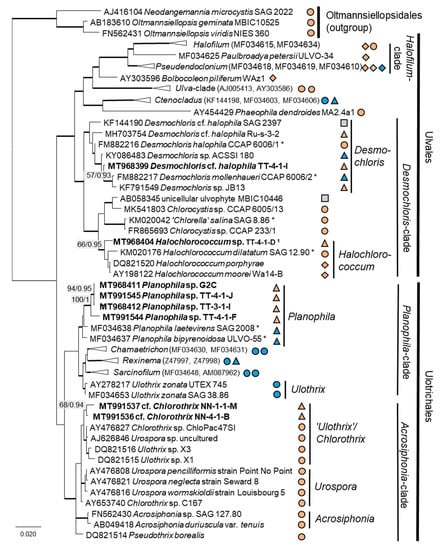
Figure 4.
Molecular phylogeny of the Ulvophyceae based on SSU rRNA gene sequencing. The phylogenetic tree was calculated by the Bayesian method, including posterior probabilities (PP) with additional maximum likelihood bootstrap values (BP); branches are supported by both methods in bold (BP > 60% and PP > 0.9). Authentic strains were marked with an asterisk, original strains from this study in bold. Habitats of the strains were classified according to habitat type (Δ terrestrial = absence of permanent water, o aquatic = presence of water, ◊ phycobiontic or endophytic, and □ unknown) and saline ( ); nonsaline (
); nonsaline ( ), and unknown (
), and unknown ( ) habitats. 1 Identical to Halochlorococcum sp. TT-4-1-M (MT968405), TT-4-1-O (MT968406), NN-4-1-Q (MT968401), NN-4-1-S (MT968402), and NN-4-1-T (MT968403).
) habitats. 1 Identical to Halochlorococcum sp. TT-4-1-M (MT968405), TT-4-1-O (MT968406), NN-4-1-Q (MT968401), NN-4-1-S (MT968402), and NN-4-1-T (MT968403).

Figure 5.
Molecular phylogeny of the Trebouxiophyceae based on SSU rRNA gene sequencing. The phylogenetic tree was calculated by the Bayesian method, including posterior probabilities (PP) with additional maximum likelihood bootstrap values (BP); branches are supported by both methods in bold (BP > 60% and PP > 0.9). Authentic strains were marked with an asterisk, original strains from this study in bold. Habitats of the strains were classified according to habitat type (Δ terrestrial = absence of permanent water, o aquatic = presence of water, ◊ phycobiontic or endophytic, and □ unknown) and saline ( ); nonsaline (
); nonsaline ( ); other extremes (
); other extremes ( e.g., acidity); and unknown (
e.g., acidity); and unknown ( ) habitats. 1 Identical to Watanabea sp. NN-1-2-X1B (MT968415) and 2 identical to Pseudostichococcus monallantoides SY-1-2-P (MT991549), WT-3-1-A (MW074294), WT-3-1-H (MT991551), and WT-3-1-P1 (MT991552).
) habitats. 1 Identical to Watanabea sp. NN-1-2-X1B (MT968415) and 2 identical to Pseudostichococcus monallantoides SY-1-2-P (MT991549), WT-3-1-A (MW074294), WT-3-1-H (MT991551), and WT-3-1-P1 (MT991552).
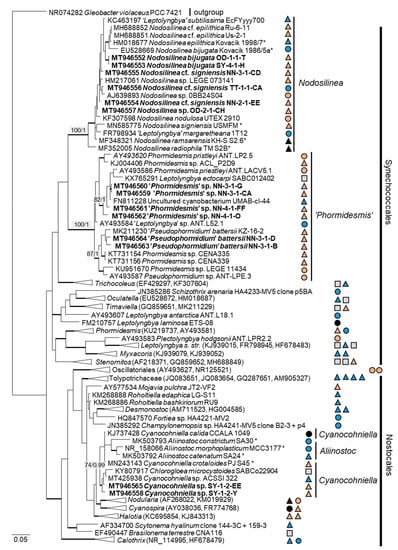
Figure 6.
Molecular phylogeny of the cyanobacteria based on SSU rRNA gene sequencing. The phylogenetic tree was calculated by the Bayesian method, including posterior probabilities (PP) with additional maximum likelihood bootstrap values (BP); branches are supported by both methods in bold (BP > 60% and PP > 0.9). Authentic strains were marked with an asterisk, original strains from this study in bold. Habitats of the strains were classified according to habitat type (Δ terrestrial = absence of permanent water, o aquatic = presence of water, ◊ phycobiontic or endophytic, and □ unknown) and saline ( ); nonsaline (
); nonsaline ( ); other extremes (
); other extremes ( e.g., radioactivity); and unknown (
e.g., radioactivity); and unknown ( ) habitats.
) habitats.
3.2. ITS Phylogeny
In order to define the original strains more precisely (to the species level), ITS-1,2 phylogenies of several Chlorophyta and cyanobacteria genera were calculated. Some original strains of the Sphaeropleales, Chlorophyceae, and Trebouxiophyceae clearly clustered in clades formed by known species: Bracteacoccus minor (Figure 7a), Pseudostichococcus monallantoides, Diplosphaera chodatii (Figure 8a), Chloroidium saccharophilum (Figure 9a), and Pseudochlorella signiensis (Figure 9c). A Tetradesmus strain clustered within the strongly supported group formed by several species perhaps representing the same taxon (“Scenedesmus” rubescens, “Scotiellopsis” reticulata, and Tetradesmus dissociatus (Figure 7b).
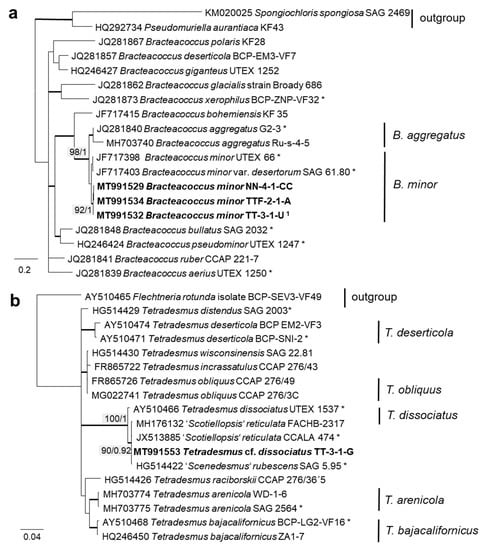
Figure 7.
Molecular phylogeny of Sphaeropleales genera based on ITS-1,2 sequencing. (a) Bracteacoccus (only ITS-2) and (b) Tetradesmus. The phylogenetic tree was calculated by the Bayesian method, including posterior probabilities (PP) with additional maximum likelihood bootstrap values (BP); branches are supported by both methods in bold (BP > 60% and PP > 0.9). Authentic strains marked with an asterisk, original strains from this study in bold. 1 Identical to Bracteacoccus minor TTF-1-1-M (MT991533), TTF-2-1-J (MT991535), TT-3-1-J (MT991531), NN-4-1-D2 (MT968390), and NN-4-1-H (MT991530).
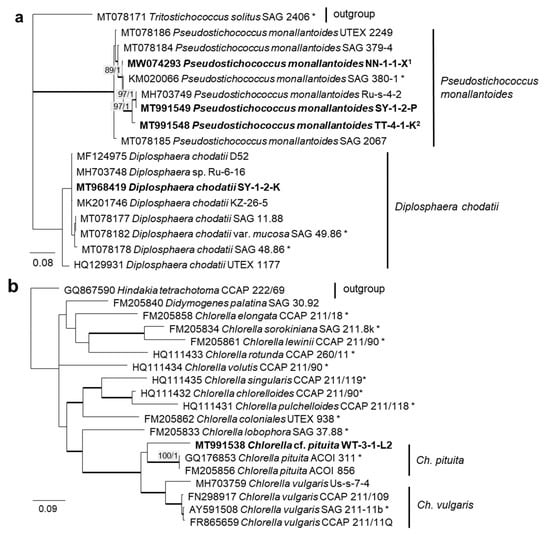
Figure 8.
Molecular phylogeny of Trebouxiophyceae genera based on ITS-1,2 sequencing. (a) Pseudostichococcus and Diplosphaera and (b) Chlorella. The phylogenetic tree was calculated by the Bayesian method, including posterior probabilities (PP) with additional maximum likelihood bootstrap values (BP); branches are supported by both methods in bold (BP >60% and PP > 0.9). Authentic strains were marked with an asterisk, original strains from this study in bold. 1 Identical to Pseudostichococcus monallantoides WT-3-1-H (MT991551) and WT-3-1-P1 (MT991552). 2 Identical to Pseudostichococcus monallantoides TTF-2-1-BB (MT991550).
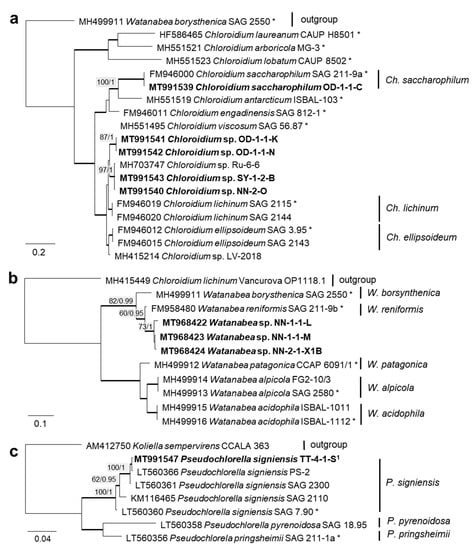
Figure 9.
Molecular phylogeny of the Trebouxiophyceae genera based on ITS-1,2 sequencing (2). (a) Chloroidium, (b) Watanabea, and (c) Pseudochlorella. The phylogenetic tree was calculated by the Bayesian method, including posterior probabilities (PP) with additional maximum likelihood bootstrap values (BP); branches are supported by both methods in bold (BP > 60% and PP > 0.9). Authentic strains were marked with an asterisk, original strains from this study in bold. 1 Identical to Pseudochlorella signiensis TSN1 (MT968421) and TSN3 (MT991546).
Some original strains clustered with strains of known species but formed distant sister lineages to the authentic strains possibly representing other taxa: Chlorella cf. pituita (Figure 8b) and Desmochloris cf. halophila (Figure 10b). Many other original strains were placed in separate lineages from the reference strains of known species and may represent still unknown species. This was the case in the Trebouxiophyceae: Chloroidium (the closest species was C. lichinum, Figure 9a) and Watanabea (closest species W. reniformis, Figure 9b), as well as the Ulvophyceae: Planophila (the closest species was P. laetevirens, Figure 10a). In the Acrosiphonia-clade of the Ulvophyceae, two original strains (cf. Chlorothrix) formed a strongly distinct separate branch without closely related reference strains, which may represent a new genus (Figure 10c).
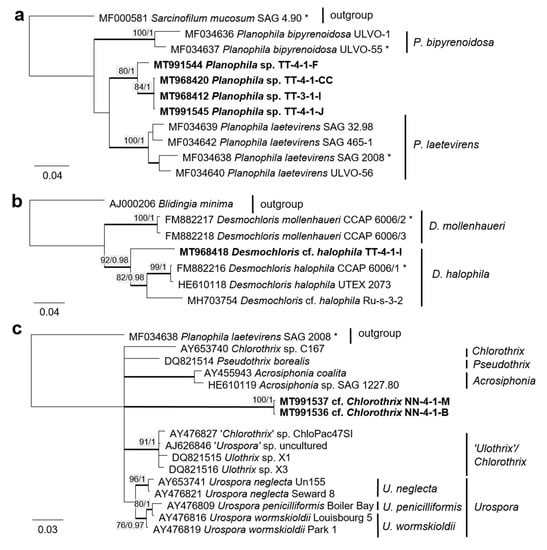
Figure 10.
Molecular phylogeny of selected genera of the Ulvophyceae based on ITS-1,2 sequencing. (a) Planophila, (b) Desmochloris, and (c) Acrosiphonia-clade. The phylogenetic tree was calculated by the Bayesian method, including posterior probabilities (PP) with additional maximum likelihood bootstrap values (BP); branches are supported by both methods in bold (BP > 60% and PP > 0.9). Authentic strains marked with an asterisk, original strains from this study in bold.
The combined SSU rRNA and SSU-LSU ITS phylogeny of the genus Nodosilinea (Synechococcales, cyanobacteria, Figure 11b) showed that two original strains clearly clustered with the species N. bijugata; the other four strains formed lineages close to the species Nodosilinea signiensis and the unidentified strain Nodosilinea KIOST-1. The SSU-LSU ITS phylogeny of a Nostocales isolate showed closeness to the Cyanocohniella and Aliinostoc species, rendering a paraphyletic position of Cyanocohniella (Figure 11a).
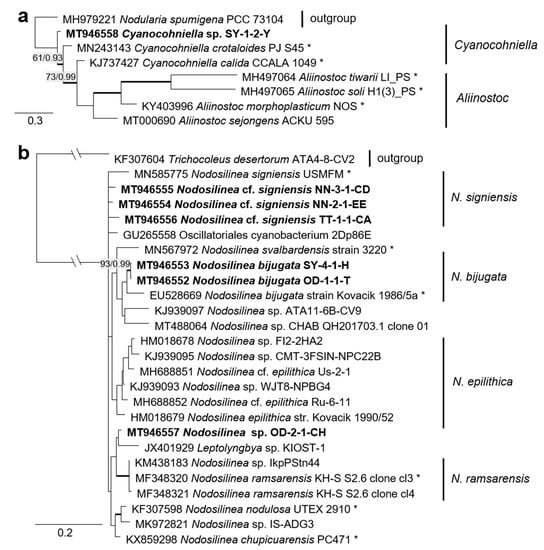
Figure 11.
Molecular phylogeny of selected genera of the cyanobacteria. (a) Cyanocohniella based on SSU-LSU ITS sequencing and (b) Nodosilinea based on combined SSU and SSU-LSU ITS sequencing. The phylogenetic tree was calculated by the Bayesian method, including posterior probabilities (PP) with additional maximum likelihood bootstrap values (BP); branches are supported by both methods in bold (BP > 60% and PP > 0.9). Authentic strains marked with an asterisk, original strains of this study in bold.
3.3. Morphological Observations
The morphology of the original strains was examined and is presented on Figure 12, Figure 13, Figure 14, Figure 15, Figure 16, Figure 17 and Figure 18. Forty strains were unambiguously identified to the species level using morphological characters solely. Five strains were tentatively identified because of unclear morphological characters or differences from morphological diagnoses of some species. Thirty-one strains were identified only to the genus level or higher because of morphological inconsistencies with known species or the absence of clear morphological characters. The list of these morphological species is included in Table 1 and has been published previously [63].
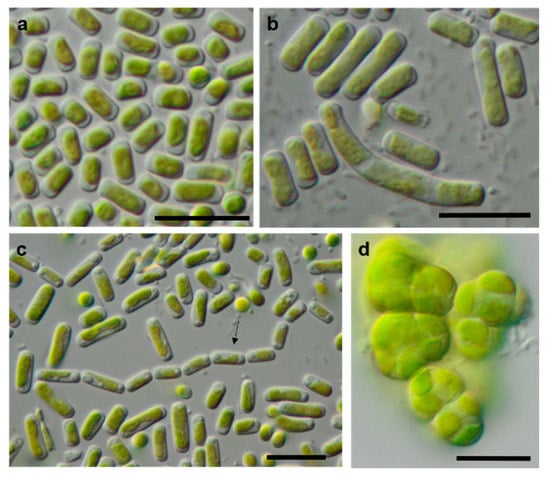
Figure 12.
Photomicrographs of Trebouxiophyceae isolates. (a,c): Pseudostichococcus monallantoides WT-3-1-A (a) young cells and (c) mature cells forming a filament (arrow)), (b) Pseudostichococcus monallantoides SY-1-2-X, and (d) Diplosphaera chodatii SY-1-2-K with cubic cell packages. Scale bars: 10 µm.
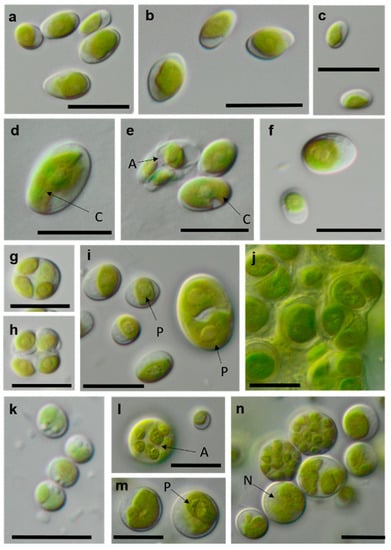
Figure 13.
Photomicrographs of Trebouxiophyceae isolates (2). (a–c) Chloroidium saccharophilum OD-1-1-C with a smooth chloroplast; (d,e) Chloroidium sp. OD-1-1-K with lobed chloroplast (d) mature cell, (e) autospore, and mature cells; (f–j) Pseudochlorella signiensis TSN-1 (f) young cells, (g) autosporangium, (h) autospores, and (i) cells with sharp pyrenoids; (j) mature cells in cell aggregate; (k) Nannochloris sp. TT-3-1-F; and (l–n) Watanabea sp. NN-1-1-E (l) autosporangium, (m) mature cells with parietal chloroplast, and prominent pyrenoid. Arrow labels: A = autospore, N = nucleus, and P = pyrenoid. Scale bars: 10 µm.
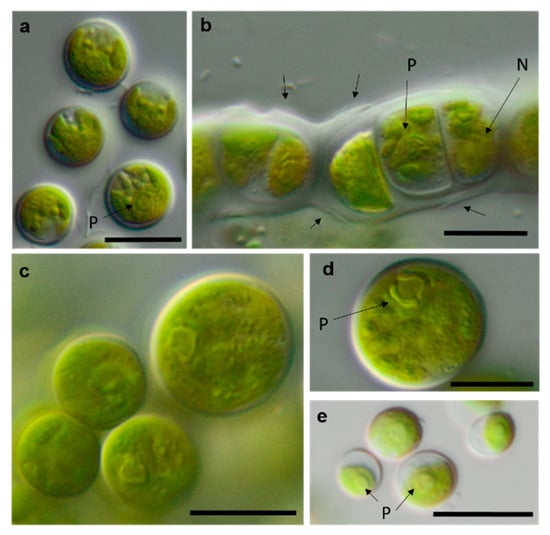
Figure 14.
Photomicrographs of Ulvophyceae isolates. (a) Desmochloris cf. halophila TT-4-1-I, (b) cf. Chlorothrix NN-1-1-M a corrugated cell wall, (c,d) Halochlorococcum sp. TT-4-1-O (d) single cell with lateral pyrenoid surrounded by two starch grains, and (e): Planophila sp. G2C with an unlobed chloroplast and one pyrenoid. Arrow labels: N = nucleus and P = pyrenoid. Scale bars: 10 µm.
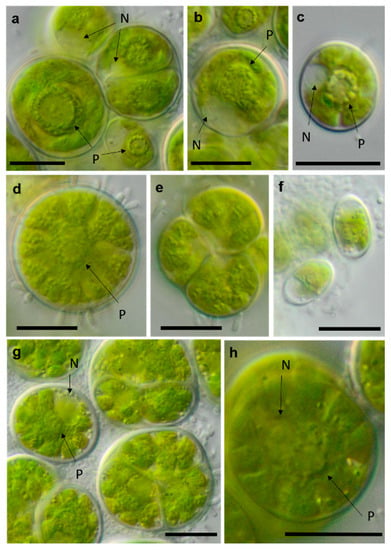
Figure 15.
Photomicrographs of Chlamydomonadales isolates. (a,b) cf. Spongiococcum TT-3-1-M; (c–e) Borodinellopsis sp. NN-2-D (c) juvenile cell and (d) mature cell with lobed chloroplast; and (f–h) Borodinellopsis texensis NN-4-1-E (f) zoospores, (h) single cell with prominent nucleus, and one pyrenoid surrounded by several fine starch grains. Arrow labels: N = nucleus and P = pyrenoid. Scale bars: 10 µm.
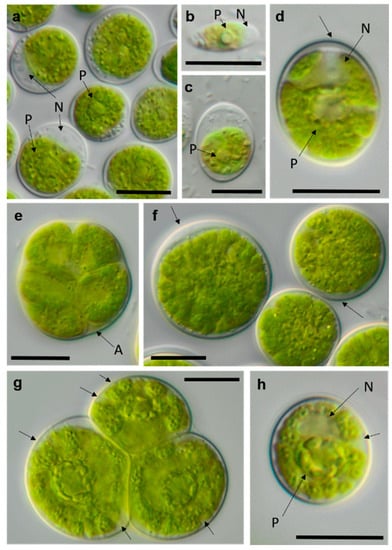
Figure 16.
Photomicrographs of Chlamydomonadales isolates (2). (a–c) cf. Chlorogonium SY-1-2-T (a) mature cells and (b) zoospore; (d–f) Chloromonas sp. WT-3-1-F (d) young cell, (e) zoosporangium, and (f) mature cell with apical cell wall bulge (arrow); and (g,h) cf. Axilosphaera NN-1-1-Q, arrows indicating slightly open spaces between chloroplast lobes ((g) adult cells in tetrad and (h) young cell. Arrow labels: N = nucleus and P = pyrenoid. Scale bars: 10 µm.
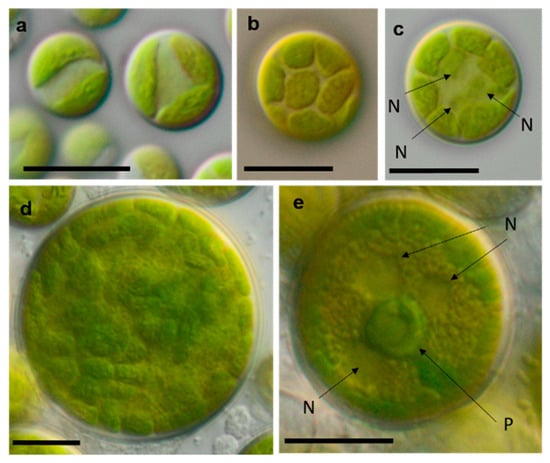
Figure 17.
Photomicrographs of Sphaeropleales isolates. (a–d) Bracteacoccus minor (a) young cells, TTF-2-1-M and (b,c) TTF-2-1-A (b) surface view of disc-shaped chloroplasts; (c) several nuclei are visible in the cell; (d) mature cell with multiple chloroplasts, NN-4-1-CC, (e) and Tetradesmus cf. dissocuatus TT-3-1-G, a large mature cell before division containing several nuclei and a central pyrenoid. Arrow labels: N = nucleus and P = pyrenoid. Scale bars: 10 µm.
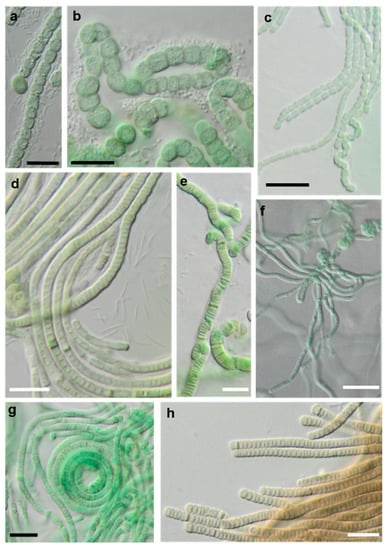
Figure 18.
Photomicrographs of cyanobacteria isolates: (a,b) Nostocales and (c–h) Synecchococcales. (a,b) Cyanocohniella sp. SY-1-2-Y, (c) Nodosilinea cf. signiensis NN-3-1-CD, (d) “Phormidesmis” sp. NN-3-1-CA, (e) “Pseudophormidium” battersii NN-3-1-D, (f) Nodosilinea cf. signiensis NN-2-1-EE, (g) Nodosilinea bijugata OD-1-1-T, and (h) “Pseudophormidium” battersii NN-3-1-B. Black scale bars: 10 µm and white scale bars: 20 µm.
4. Discussion
4.1. Morphological vs. Molecular Species Determination
Table 1 shows a comparison of the results from our study to the previously published morphological species list [63]. Several of the original isolates were previously correctly identified to the species level based solely on morphology. Usually, these taxa have distinct morphological features that can be recognized quickly. As an example, Borodinellopsis texensis is characterized by a prominent asteroid chloroplast, central pyrenoid, and the formation of cell packages (Figure 15f–h), and Diplosphaera chodatii is identified by the characteristic cubic cell packages, cell diameter, and the absence of coated pyrenoids (Figure 12d) [100].
Some strains were correctly identified to the genus level or morphological species complexes. For example, several strains of the morphologically prominent genus Chloroidium, initially identified as Ch. ellipsoideum, were redefined as the morphologically close species Ch. saccharophilum and Chloroidium sp. The cells of this genus can be easily recognized by their ellipsoid shapes and characteristic chloroplast structures (Figure 13a–e) [68]. Previous identifications of the small-celled Nannochloris sp. were also confirmed by the integrative approach. However, the group of Nannochloris-like algae is problematic for identification in general and requires taxonomic revision, including strains from ecologically different environments [101,102].
Several strains with Tetracystis/Chlorococcum-like morphology finally fell in different but related lineages in the Moewusinia (Chlorophyceae; Axilosphaera, Spongiococcum, Alvikia, and Chlorogonium), confirming the general polyphyly of this morphotype [103]. The strain that was morphologically identified as Radiosphaera negevensis (Moewusinia) due to the presence of almost spherical mature cells and typical asteroid chloroplasts was redefined as the Chloromonas sp. (Chloromonadinia). This disagreement in identification may occur due to the high level of morphological parallelism between nonrelated lineages of Chlorophyceae, previously discussed by Mikhailyuk et al. [65]. Possibly, the same explanation can be applied to the strains cf. Chlorothrix (Acrosiphonia-clade, Ulotrichales, and Ulvophyceae), which were originally identified as Ulothrix species (Figure 4, Planophila-clade).
Further, the identification of some cryptic taxa characterized by simple morphology was corrected based on the integrative approach. Several strains that were initially identified as morphologically different Stichococcus species were unambiguously identified as a single species in the newly revised genus Pseudostichococcus [119] (Figure 8a), namely P. monallantoides. Likewise, some strains of Bracteacoccus minor were initially morphologically identified as several small-celled Bracteacoccus (including B. minor) and Pseudomuriella species (Figure 7a).
The Parietochloris strains that were morphologically identified due to their specific structure of the pyrenoid envelope, chloroplast morphology, and general appearance of young and mature cells [100], however, were redefined as the recently revised genera Pseudochlorella and Watanabea [70,121]. Members of the same class (Trebouxiophyceae), these genera are related and, consequently, have similar morphology.
The strains morphologically identified as Spongiochloris excentrica (Chlorophyceae) mostly clustered in a completely different class (Ulvophyceae) in clades corresponding to the genera Desmochloris and Halochlorococcum. This may have occurred due to morphological similarities between the two classes: a high similarity of the spongiomorph chloroplast and the pyrenoid morphology, as well as the multinucleosis of vegetative cells in some stages of their life cycles. These morphological parallelisms have been discussed in a recent publication [65]. Halochlorococcum is poorly investigated [114] and absent from widely used identification books and guides [100], which underlines the difficulty of identifying this genus morphologically.
The strain TT-3-1-G was also identified as Spongiochloris excentrica on the basis of morphological features [100]; however, following the phylogenetic analysis, it clustered in another chlorophycean genus. This genus, Tetradesmus, is morphologically completely different from Spongiochloris. Our strain formed large multinucleate cells (see Figure 17e) that actually resemble Spongiochloris. These cells may represent a specific stage in the life cycle of Tetradesmus (before division and autospore formation) rather than a general phenotype.
The strains morphologically identified as Nostoc sensu lato and Leptolyngbya sensu lato, to give an example for cyanobacteria, were redetermined to be species of the newly described genera Cyanocohniella [76] and Nodosilinea [122], which still belong to the same orders (Nostocales and Synechococcales, respectively). Altogether, the molecular approach resulted in more precise identification and should be considered as an extension of the previous morphological species list.
As explained above, some of these misidentifications resulted in the splitting of one taxon into several morphologically identified taxa. This can be explained by two factors: age of cultures and phenotypic plasticity. Cultures of different ages may have shown differences in cell morphology. Young cells may be smaller and, for example, in the case of Halochlorococcum sp., are more likely to resemble Chlorella. Older cells may have been larger and potentially showed more morphological traits that were attributed to another taxon. Likewise, young cells of Planophila sp. did not form characteristic cell packets but showed a Chlorella-like morphology (Figure 14e), leading to the mismatch between the morphological approach and the molecular results.
Differences in morphology also appear with different culture conditions, such as temperature, light, and salinity. This effect is termed phenotypic plasticity. Some recent studies have revealed that phenotypic plasticity of several terrestrial green algae can be caused by different levels of salinity [119,124].
Some morphologically identified species from the potash tailing pile sites [63] could not be addressed with the methods used here. One example is the genus Pseudendoclonium, a filamentous branched green alga in the Ulvophyceae, which was frequently found in biocrusts of the tailing pile environments [63]. This genus is a widely distributed halotolerant alga and has also been identified as Dilabifilum in some studies [43,51,125]. In the previous study, the genus Planophila was represented by five morphologically identified species [63], e.g., characterized by dense cell aggregates. However, the present study identified only one species, which was single-celled (Figure 14e). Sequencing other morphotypes of this genus was not successful. Thus, our approach was not feasible for these isolates. For these reasons, the taxa identified in this study do not comprise a complete species list.
In sum, the integrative approach confirmed some of the morphological taxa, redefined others more precisely, and excluded a few strains due to methodological issues. Consequently, the results should be considered as an extension and revision of the species list published previously [63], underlining the general advantages of an integrative approach.
4.2. Taxa with Unclear Phylogenetic Positions
In the Chlamydomonadales, the genus Borodinellopsis was isolated quite often from our samples (four isolates, Figure 15c–h). Two original strains were clearly identified as Borodinellopsis texensis. Another two original strains formed a sister branch to this species (Figure 2). This lineage may represent another species of Borodinellopsis, either B. oleifera, which is morphologically characterized by the presence of oil drops [126], or a new taxon. However, no reference sequence of B. oleifera was available. Thus, more detailed morphological observation is necessary to reach a definitive conclusion.
Several strains were found among the Tetracystis/Chlorococcum-like isolates in the Moewusinia-clade, identified as cf. Axilosphaera, cf. Spongiococcus, and Alvikia sp. The isolate NN-1-1-Q (cf. Axilosphaera, Figure 16g,h) clustered with Eubrownia and Axilosphaera vegetata. Even though the latter represents the closest relative, the isolate analyzed in this study differed from these reference strains regarding the SSU rRNA gene sequence, indicating a possibly new taxon. Ettl and Gärtner [100] described Axilosphaera vegetata as morphologically similar to Borodinellopsis but distinguished by the absence of the larger cell complexes in Axilosphaera, which, in turn, are common for Borodinellopsis. We observed the isolate NN-1-1-Q in dyads or tetrads only (Figure 16g), which showed parietal cup-shaped and richly dissected chloroplasts (Figure 16g,h). The chloroplast of Axilosphaera is described as parietal cup-shaped and dissected, becoming almost asteroid in mature cells, which is similar to the cup-shaped and dissected chloroplasts of the sister lineage Eubrownia [105]. Two other Moewusinia isolates fell in new lineages (Figure 2). The isolates cf. Spongiococcum (SY-4-1-C and TT-3-1-M), which showed a Tetracystis-like morphology and a spongiomorph chloroplast with a prominent pyrenoid (Figure 15a,b), as well as the isolate Alvikia sp. (TTF-2-1-Da), which showed a Chlorococcum-like morphology, possibly representing new taxa. For final clarification, more thorough investigations are required.
The original isolates TT-3-1-Q, SY-1-2-T and TSN3f, placed in the Chlorogonia, were unique, since they were clearly separated from the closest Chlorogonium species. The genus Chlorogonium is flagellated, spindle-shaped, or ovoid and characterized by the presence of two or more pyrenoids [103]. Young cells of our isolates had Chlorogonium-like morphology (Figure 16b) and had one pyrenoid and a posterior nucleus; however, mature cells became ovoid to nearly spherical, never a spindle shape (Figure 16a,c). Consequently, our isolates may represent a new taxon.
Several strains belonged to the Ulvophyceae. In the order Ulvales, the isolate T-4-I-1 fell in the genus Desmochloris (Figure 4 and Figure 14a), which includes two species: D. mollenhauerii and D. halophila [112]. The original strain of this study was clearly separated from D. mollenhauerii and was placed in an intermediate position to the type of species of D. halophila. Thus, we identified our strains as Desmochloris cf. halophila, which may represent a new species for the genus.
Several other isolates within the Desmochloris-clade were placed in the interesting and insufficiently described genus Halochlorococcum (according to the SSU rRNA gene phylogeny, Figure 4). Seven species are known in this genus [114]. Even though the genus was recently re-established after having an invalid status for a long time [114], only the SSU rRNA gene sequences of three species (H. dilatatum, H. porphyrae, and H. moorei) were available in the database, which did not allow specification of the original isolates to the species level. This stresses the need for a general revision of the genus Halochlorococcum.
Four original strains of another ulvophyte fell in the genus Planophila, forming a distinct separate lineage between the two known species P. bipyrenoidosa and P. laetevirens (according to the ITS-1,2 phylogeny, Figure 10a). This result strongly indicates that these strains represent a new species.
A unique case consists of two original strains from the Ulotrichales. According to the ITS-1,2 phylogeny, these strains were placed in the Acrosiphonia-clade (Figure 4), forming a significant new branch close to the genera Pseudothrix, Chlorothrix, Urospora, and “Ulothrix” (Figure 10c). Urospora is a filamentous, uniseriate marine algae and has a multinucleate gametophyte [116], whereas the gametophyte thallus of Pseudothrix is initially uniseriate and develops into multiseriate filaments [117]. Mature cells of Ulothrix and Chlorothrix are uniseriate and uninucleate [127]. Thus, our isolates showing uniseriate and uninucleate filaments (Figure 14b) were morphologically most similar to Ulothrix and Clorothrix. The genus Ulothrix, however, is placed in the Planophila-clade of the Ulotrichales [69] (Figure 4), represented by strains identified as Ulothrix zonata (SAG 38.86, UTEX 745). Thus, we tentatively named our isolates (NN-4-1-M and NN-4-1-B) cf. Chlorothrix.
Several different cultures were found in the Trebouxiophyceae. Five original isolates clustered within the genus Chloroidium (Figure 5). One of them (OD-1-1-C) was clearly identified as Ch. saccharophilum, since its ITS-1,2 sequence was identical to the authentic strain of this species (SAG 211-9a, Figure 9a). The other isolates formed two separate lineages together with the unidentified strain, originating from sand dunes of the Baltic Sea coast (Ru-6-6). These lineages are close to Ch.lichinum, as well as to Ch. ellipsoideum, and may represent new species.
Three other trebouxiophycean isolates clustered with Watanabea and formed a separate lineage closely related to the type strain of W. reniformis (SAG 211-9b). The chloroplast of W. reniformis does not show a pyrenoid [70], whereas the isolates of this study clearly did (Figure 13m). Another species in this clade, W. borsynthenica, in turn, has a pyrenoid [70]. However, the latter is clearly different from our isolates regarding the ITS-1,2 phylogeny, which leads to an unclear status of the original isolates.
In the Sphaeropleales, the original strain TT-3-1-G clustered in the genus Tetradesmus. It was close to the authentic strains of four species: “Scenedesmus” rubescens, “Scotiellopsis” reticulata, Tetradesmus dissociatus, and “Scenedesmus” littoralis, forming a highly supported cluster in the SSU rRNA gene and ITS-1,2 phylogenies (Figure 3 and Figure 7b). Indeed, the SSU rRNA gene and the ITS sequences were very similar (differing in only one nucleotide) and, also, in their ITS-2 region, which is used for species delimitation within the genus [128]. Thus, it is obvious that this cluster, including our original strain, most likely represents one distinct species. The findings of another study confirm the genetic similarity here [111]. However, there are some nomenclatural uncertainties, since the status of the oldest species name in that clade (Halochlorella rubescens) was recently declared as invalid [129]. Despite the genetic identity and a generally similar morphology of these four strains, they showed some minor, though characteristic, morphological differences: Scenedesmus rubescens forms cell packets [130], and Tetradesmus dissociatus shows characteristic thread-like remnants of the sporangial walls. We also observed some specific multinucleate stages in our original strain TT-3-1-G (Figure 17e), which may have caused its previous misidentification (see above). Therefore, further investigations are required for the final decision concerning this clade. Accordingly, we named the original isolate as Tetradesmus dissociatus, since it is the only valid species name available in this group.
The two cyanobacteria isolates in the Nostocales clustered with the rare and recently described and revised genus Cyanocohniella [75,76]. However, both the SSU and SSU-LSU intergenic spacer phylogeny showed a paraphyly of Cyanocohniella and the genetically close genus Aliinostoc (Figure 6 and Figure 11a). Therefore, the identification remains unclear. Moreover, both genera may require further revision.
Several original isolates from the Synechococcales with unclear Leptolyngbya-like morphology (Figure 18c,f,g) clustered within the genus Nodosilinea, which was defined recently [122], followed by the description of several new Nodosilinea species [123,131,132]. Some of our isolates fell in clades formed by known species (Figure 11b) and identified as N. bijugata and N. cf. signiensis. The original isolate OD-2-1-H, however, was placed in a distinct clade with an unidentified reference strain (KIOST-1), indicating its unclear status.
Six other original isolates belonging to Synechococcales and characterized by prominent, deeply constricted green, pale-green, and brown trichomes, as well as narrow cells (Figure 18d–h), were ordered into another clade containing sequences labeled as Phormidesmis and Pseudophormidium. However, a recent review addressed the genus Phormidesmis [133], which clearly represents another lineage. Previous studies that found strains of the same clade termed it “Phormidesmis” [65,72], which stresses its unclear status and, as discussed above, may represent an undescribed genus [72]. Clearly, identification of our strains is not possible.
Overall, our study showed a high proportion of unclear and possibly new taxa. For 24% of the taxa found in this study (16 isolates), identification even to genus level was doubtful or not possible from the integrative approach. This is twice as high as in a similar study on biocrusts in Baltic Sea coastal dunes, where the authors found unclear genera in 11% of their isolates [65]. Another recent study on microbial mats in hypersaline wind-tidal flats found that even 35% of the isolated cyanobacteria could not be assigned to the genus/family level [35]. This underlines the lack of knowledge in hypersaline environments, which is in congruence with our findings on biocrusts in the highly saline potash tailing pile environments.
The methods used in our study were not sufficient to formally describe the new taxa. Therefore, deeper analyses are needed. One crucial molecular method is the comparison of ITS secondary structures that clearly delimit the borders between species of both green algae and cyanobacteria. In addition, the morphology should be analyzed in detail. The original strains of this study were cultured on a solidified saline medium, which could result in changes in morphology. Phenotypic plasticity under saline conditions is known for several species [134]. To overcome this possible limitation, several growth media with or without added salt should be used, both solidified and liquid. In addition, the morphology of all reproductive stages, such as zoospores, gametes, or resting stages in green algae, as well as hormogonia or necridia in cyanobacteria, should be investigated in detail.
4.3. Congruence and Divergence of Phylogeny and Habitat Characteristics Regarding Salinity
Several species or genera identified in this study are already known to live in salt-affected habitats and, thus, are most probably adapted to saline conditions. It is not surprising that we identified many strains of Ulvophyceae, since this class mostly contains salt-tolerant marine species, such as the eponymous macrophyte genus Ulva. However, in terrestrial habitats, members of this class are mostly less common than the predominant Chlorophyceae and Trebouxiophyceae. Only a few studies have treated nonmarine Ulvophyceae, which were recently rearranged in the broad revision of Darienko and Pröschold [69].
The Ulvophyceae Desmochloris halophila was originally found in mixohaline water [113]. Other strains assigned to that genus/species also originated from salt-affected habitats [65,135]. Another Ulvophyceae, a close relative to the original strains of Halochlorococcum sp., was Halochlorococcum dilatatum (SAG 12.90), which was originally isolated from a rock pool in Helgoland, Germany. Rock pools have fluctuating salinities, which may strongly increase due to evaporation and reach hypersaline concentrations. The two other reference strains, H. porphyreae and H. moorei, were also found in marine environments [115,117]. Mettlig [136] classified the group as a genus, including soil algae; however, no record from a terrestrial environment was published thereafter. Thus, the findings of our study indicate the first record of Halochlorococcum in a biocrust.
The clade Tetradesmus, including “Scotiellopsis” reticulata, “Scenedesmus” rubescens, “S.” littoralis, and T. dissociates, is an interesting case in the Sphaeropleales concerning salinity. The reference strain “Scotiellopsis” reticulata (CCALA 474) was found in the sand on the coast of the Black Sea [137], and the type strain of “Scenedesmus” littoralis originated from coastal waters and tolerates up to 2% NaCl [138]. “Scenedesmus” rubescens was described based on a strain isolated from a culture of marine brown algae [130]. The reference strain Tetradesmus dissociatus is the only exception, since it was isolated from cornfield soil [139].
The order Chlamydomonadales also contains salt-tolerant species such as Borodinellopsis texensis, which was found in a sea salt farm (strain JM002, Figure 2) and in a hypersaline ecosystem [43]. The isolate Alvikia sp. (TTF-2-1-Da), interestingly, clustered with species found in saline environments [105] but not in terrestrial habitats. In contrast, the sister clade with Spongiococcum and Actinochloris included terrestrial isolates from nonsaline environments. Thus, our isolate may somehow represent a linkage between these clades as the first record from a salt-affected terrestrial site.
In the cyanobacteria, the closest reference to the two original strains, Cyanocohniella sp., was originally isolated from beach mats of the North Sea, The Netherlands [75]. In the sister genus Aliinostoc, some strains are known from other saline habitats, such as saline-alkaline lakes or mangroves [140]. In the “Phormidesmis” cluster, almost all of the respective reference strains were originally isolated from salt-affected habitats but of different types. These references originated from microbial mats in saline lakes [141,142] and from the coast of the brackish Sea of Azov [72]. Two strains of “Pseudophormidium” were isolated from the unusual habitat of salt-excreting leaves of the mangrove Avicennia schaueriana [143]. In sum, the original isolates belonging to the taxa discussed above match the saline habitat preferences of their closest phylogenetic relatives.
In contrast, several genera were identified that are more common in nonsaline environments. The Trebouxiophyceae comprises a wide variety of microalgae, including marine and freshwater, as well as numerous terrestrial taxa. In the genus Watanabea, most species were described from either aquatic, e.g., Watanabea reniformis (SAG 211-9b), or terrestrial habitats; there is no record from saline terrestrial environments yet known. Interestingly, several strains have been isolated from highly acidic environments [70,144]. Thus, the original isolates of this study underline the presence of extremophiles within the genus Watanabea and represent a unique finding in a saline biocrust.
A study focusing on Nannochloris-like algae described a new genus of marine/saline Nannochloris-like strains, namely Picochlorum, whereas freshwater Nannochloris-like strains formed a second cluster [101]. The respective strains of our study did not fall into the marine Picochlorum but, rather, in the freshwater Nannochloris.
The Trebouxiophyceae genus Chlorella is among one of the best-known genera of microalgae, but its phylogenetic status has long remained unresolved. A recent study addressed the question of the existence of true marine Chlorella species and, indeed, found three marine isolates belonging to this genus, namely Chlorella vulgaris [145]. The original isolate of this study, however, clustered with Chlorella pituita. The epitype strain of Chlorella pituita, though, was isolated from freshwater [97].
Consequently, we found several isolates that represent taxa that are typically found in saline habitats or that have some close relatives that inhabit such environments. Other taxa were more typical for nonsaline habitats. To withstand the harmful saline environment, salt-tolerant microalgae evolved protective mechanisms such as the excretion of extracellular polysaccharides (EPS) and the production of intracellular organic osmolytes. However, the biocrust as a multidimensional layer potentially provides shelter for less salt-tolerant organisms. Species adapted to salty conditions may form a barrier layer for sensitive species by EPS excretion. Thus, both tolerant as well as nontolerant taxa may appear in salt-affected biocrusts, although intolerant taxa may not survive outside the shelter of the biocrust community. Further, we cannot state whether the species lived under their optimum conditions or at the edge of their salt tolerance. Competitive pressure in less saline soils in these environments may lead to a migration of taxa to more saline soils on the edge of their tolerance. Future eco-physiological studies on the isolated microalgae and cyanobacteria strains should address their plasticity towards salinity by thoroughly identifying the tolerance range, as well as the optimum salinity.
Moreover, the mechanisms of salt tolerance are closely linked to drought tolerance, a trait that is generally necessary for microalgae outside the aquatic environment, such as in a biocrust. Though there are some differences in the effect, as well as the protective mechanisms of these two stressors, common terrestrial microalgae that are already adapted to water deprivation have a good potential to withstand saline conditions as well. This is in congruence with our findings of several common terrestrial algae and cyanobacteria, such as Pseudostichococcus [119], Bracteacoccus, Chloroidium [68], and Nodosilinea [122], in biocrusts of potash tailing pile environments.
5. Conclusions
Many taxa of already known salt-tolerant green algae and cyanobacteria were found in biocrusts of extremely saline potash tailing pile habitats using an integrative approach. Several strains were the first record of that taxon in a biocrust. However, there was a high proportion of original isolates with unclear taxonomic positions, indicating new species or genera. Thus, biocrusts in potash tailing pile areas revealed unique communities of microalgae and cyanobacteria. Studies in other saline terrestrial habitats were often based on morphology, and new species could have been overlooked in these cases. Comparing our results to the previous morphological observations on the original isolates, our findings should be considered as an extension and improvement of the morphologically based species list. However, more detailed research is needed for deeper analyses of the newly found lineages. Consequently, our findings illustrated the benefits of an integrative approach but also underlined the limits of the molecular methods used in this study for describing new species.
The original stains examined in this study reflected a highly diverse and unique community of biocrust algae and cyanobacteria from potash tailing pile environments, which may be specialized for these extreme habitats. Their phylogenetic identification is an important but very first step in the ongoing research and will be crucial for the interpretation of subsequent eco-physiological experiments. Assessing the salt tolerance of the isolates is a necessary next step in understanding the ecology of microalgae and cyanobacteria in these specialized biocrusts. In the future, they may be of interest for the young field of applied biocrust research. Since biocrusts are multifunctional communities that are involved in several ecological processes, such as soil formation and surface stabilization, they could potentially be used to cover potash tailing piles [146]. The microalgae and cyanobacteria identified in this study represent candidates for artificial biocrust formation due to their origin in unique habitats close to the tailing piles and the high proportion of possibly new taxa.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/11/1667/s1: Table S1: Gene Bank Accession Number and origin/habitat description of the reference strains used for phylogenetic tree construction.
Author Contributions
Conceptualization, V.S., K.G., and U.K.; methodology, V.S., T.M., and K.G.; software, V.S., T.M., and K.G.; validation, T.M. and K.G.; writing—original draft preparation, V.S.; writing—review and editing, K.G, T.M., and U.K.; and supervision, U.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a fellowship of Deutsche Bundesstiftung Umwelt (DBU) to VS, a Max-Buchner-Forschungsstipendium (MBFSt 3583) from the Gesellschaft für Chemische Technik und Biotechnologie (DECHEMA), as well as a DFG Grant GL 909/1-1, to KG, and a Georg-Forster research fellowship from the Alexander von Humboldt Foundation to TM. Further funding was provided by upi UmweltProjekt Ingenieurgesellschaft mbH and K+S Minerals and Agriculture GmbH.
Acknowledgments
We thank the state environmental offices of the Peine and Lüchow-Dannenberg Districts, the Wietze community, the GESA (Gesellschaft zur Entwicklung und Sanierung von Altstandorten MBH), and Albrecht Palm, as well as the K+S Minerals and Agriculture GmbH, for fieldwork permits and/or access to the sites. Furthermore, we thank Alexandra Wölk, Ramona Kern, Matthias Kockx, and Christopher Lubs for their assistance during sampling. Special thanks are extended to Laura Fuchs for her strong support in the laboratory work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Belnap, J.; Büdel, B.; Weber, B. Biological soil crusts as an organizing principle in drylands. In Biological Soil Crusts: An Organizing Principle in Drylands; Weber, B., Büdel, B., Belnap, J., Eds.; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-30212-6. [Google Scholar]
- Weber, B.; Büdel, B.; Belnap, J. Biological Soil Crusts: An Organizing Principle in Drylands; Weber, B., Büdel, B., Belnap, J., Eds.; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-30212-6. [Google Scholar]
- Evans, R.D.; Johansen, J.R. Microbiotic Crusts and Ecosystem Processes. Crit. Rev. Plant Sci. 1999, 18, 183–225. [Google Scholar] [CrossRef]
- Chen, N.; Yu, K.; Jia, R.; Teng, J.; Zhao, C. Biocrust as one of multiple stable states in global drylands. Sci. Adv. 2020, 6, eaay3763. [Google Scholar] [CrossRef] [PubMed]
- Pietrasiak, N.; Johansen, J.R.; Ladoux, T.; Graham, R.C. Comparison of Disturbance Impacts to and Spatial Distribution of Biological Soil Crusts in the Little San Bernardino Mountains of Joshua Tree National Park, California. West. North Am. Nat. 2011, 71, 539–552. [Google Scholar] [CrossRef]
- Eldridge, D.J.; Woodhouse, J.N.; Curlevski, N.J.A.; Hayward, M.; Brown, M.V.; Neilan, B.A. Soil-foraging animals alter the composition and co-occurrence of microbial communities in a desert shrubland. ISME J. 2015, 9, 2671–2681. [Google Scholar] [CrossRef]
- Rao, B.; Liu, Y.; Lan, S.; Wu, P.; Wang, W.; Li, D. Effects of sand burial stress on the early developments of cyanobacterial crusts in the field. Eur. J. Soil Biol. 2012, 48, 48–55. [Google Scholar] [CrossRef]
- Cantón, Y.; Chamizo, S.; Rodriguez-Caballero, E.; Lázaro, R.; Roncero-Ramos, B.; Román, J.R.; Solé-Benet, A. Water Regulation in Cyanobacterial Biocrusts from Drylands: Negative Impacts of Anthropogenic Disturbance. Water 2020, 12, 720. [Google Scholar] [CrossRef]
- Rodriguez-Caballero, E.; Belnap, J.; Büdel, B.; Crutzen, P.J.; Andreae, M.O.; Pöschl, U.; Weber, B. Dryland photoautotrophic soil surface communities endangered by global change. Nat. Geosci. 2018, 11, 185–189. [Google Scholar] [CrossRef]
- Antoninka, A.; Faist, A.; Rodriguez-Caballero, E.; Young, K.E.; Chaudhary, V.B.; Condon, L.A.; Pyke, D.A. Biological soil crusts in ecological restoration: Emerging research and perspectives. Restor. Ecol. 2020, 28, S3–S8. [Google Scholar] [CrossRef]
- Antoninka, A.; Bowker, M.A.; Chuckran, P.; Barger, N.N.; Reed, S.; Belnap, J. Maximizing establishment and survivorship of field-collected and greenhouse-cultivated biocrusts in a semi-cold desert. Plant Soil 2017, 429, 213–225. [Google Scholar] [CrossRef]
- Muñoz-Rojas, M.; Chilton, A.; Liyanage, G.S.; Erickson, T.E.; Merritt, D.J.; Neilan, B.A.; Ooi, M.K.J. Effects of indigenous soil cyanobacteria on seed germination and seedling growth of arid species used in restoration. Plant Soil 2018, 429, 91–100. [Google Scholar] [CrossRef]
- Xiao, B.; Zhao, Y.; Wang, Q.; Li, C. Development of artificial moss-dominated biological soil crusts and their effects on runoff and soil water content in a semi-arid environment. J. Arid. Environ. 2015, 117, 75–83. [Google Scholar] [CrossRef]
- Chen, L.; Xie, Z.; Hu, C.; Li, D.; Wang, G.; Liu, Y. Man-made desert algal crusts as affected by environmental factors in Inner Mongolia, China. J. Arid. Environ. 2006, 67, 521–527. [Google Scholar] [CrossRef]
- Green, T.G.A.; Proctor, M.C.F. Physiology of Photosynthetic Organisms within Biological Soil Crusts: Their Adaptation, Flexibility, and Plasticity. In Biological Soil Crusts: An Organizing Principle in Drylands; Weber, B., Büdel, B., Belnap, J., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 347–381. [Google Scholar]
- Rossi, F.; Mugnai, G.; De Philippis, R. Complex role of the polymeric matrix in biological soil crusts. Plant Soil 2017, 429, 19–34. [Google Scholar] [CrossRef]
- Karsten, U.; Holzinger, A. Green algae in alpine biological soil crust communities: Acclimation strategies against ultraviolet radiation and dehydration. Biodivers. Conserv. 2014, 23, 1845–1858. [Google Scholar] [CrossRef] [PubMed]
- Pfaff, S.; Borchhardt, N.; Boy, J.; Karsten, U.; Gustavs, L. Desiccation tolerance and growth-temperature requirements of Coccomyxa (Trebouxiophyceae, Chlorophyta) strains from Antarctic biological soil crusts. Algological Stud. 2016, 151, 3–19. [Google Scholar] [CrossRef]
- Couradeau, E.; Felde, V.J.M.N.L.; Parkinson, D.; Uteau, D.; Rochet, A.; Cuellar, C.; Winegar, G.; Peth, S.; Northen, T.R.; Garcia-Pichel, F. In Situ X-ray Tomography Imaging of Soil Water and Cyanobacteria from Biological Soil Crusts Undergoing Desiccation. Front. Environ. Sci. 2018, 6, 65. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Polerecky, L.; Al-Habsi, A.; Oetjen, J.; Strous, M.; De Beer, D. Rapid Recovery of Cyanobacterial Pigments in Desiccated Biological Soil Crusts following Addition of Water. PLoS ONE 2014, 9, e112372. [Google Scholar] [CrossRef] [PubMed]
- Whitney, K.M.; Vivoni, E.R.; Duniway, M.C.; Bradford, J.B.; Reed, S.C.; Belnap, J. Ecohydrological role of biological soil crusts across a gradient in levels of development. Ecohydrology 2017, 10, e1875. [Google Scholar] [CrossRef]
- Lafuente, A.; Berdugo, M.; De Guevara, M.L.; Gozalo, B.; Maestre, F.T. Simulated climate change affects how biocrusts modulate water gains and desiccation dynamics after rainfall events. Ecohydrology 2018, 11, e1935. [Google Scholar] [CrossRef]
- Holzinger, A.; Lütz, C.; Karsten, U. Desiccation stress causes structural and ultrastructural alterations in the aeroterrestrial green alga Klebsormidium crenulatum (Klebsormidiophyceae, Streptophyta) isolated from an alpine soil crust. J. Phycol. 2011, 47, 591–602. [Google Scholar] [CrossRef]
- Karsten, U.; Herburger, K.; Holzinger, A. Dehydration, temperature, and light tolerance in members of the aeroterrestrial green algal genus Interfilum (Streptophyta) from biogeographically different temperate soils. J. Phycol. 2014, 50, 804–816. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 2011, 35, 87–123. [Google Scholar] [CrossRef] [PubMed]
- Pade, N.; Hagemann, M. Salt Acclimation of Cyanobacteria and Their Application in Biotechnology. Life 2014, 5, 25–49. [Google Scholar] [CrossRef]
- Kirst, G.O. Salinity Tolerance of Eukaryotic Marine Algae. Annu. Rev. Plant Biol. 1990, 41, 21–53. [Google Scholar] [CrossRef]
- Karsten, U. Seaweed acclimation to salinity and desiccation stress. In Seaweed Biology: Novel Insights into Ecophysiology, Ecology and Utilization. Ecological Studies 219; Wiencke, C., Bischof, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 87–107. ISBN 978-3-642-28451-9. [Google Scholar]
- Church, J.; Hwang, J.-H.; Kim, K.-T.; McLean, R.; Oh, Y.-K.; Nam, B.; Joo, J.C.; Lee, W.H. Effect of salt type and concentration on the growth and lipid content of Chlorella vulgaris in synthetic saline wastewater for biofuel production. Bioresour. Technol. 2017, 243, 147–153. [Google Scholar] [CrossRef]
- Ishika, T.; Bahri, P.A.; Laird, D.W.; Moheimani, N.R. The effect of gradual increase in salinity on the biomass productivity and biochemical composition of several marine, halotolerant, and halophilic microalgae. J. Appl. Phycol. 2018, 30, 1453–1464. [Google Scholar] [CrossRef]
- Menke, S.; Sennhenn, A.; Sachse, J.-H.; Majewski, E.; Huchzermeyer, B.; Rath, T. Screening of Microalgae for Feasible Mass Production in Industrial Hypersaline Wastewater Using Disposable Bioreactors. CLEAN Soil Air Water 2012, 40, 1401–1407. [Google Scholar] [CrossRef]
- Sabbe, K.; Hodgson, D.A.; Verleyen, E.; Taton, A.; Wilmotte, A.; Vanhoutte, K.; Vyverman, W. Salinity, depth and the structure and composition of microbial mats in continental Antarctic lakes. Freshw. Biol. 2004, 49, 296–319. [Google Scholar] [CrossRef]
- Krumbein, W.E.; Gorbushina, A.A.; Holtkamp-Tacken, E. Hypersaline Microbial Systems of Sabkhas: Examples of Life’s Survival in “Extreme” Conditions. Astrobiology 2004, 4, 450–459. [Google Scholar] [CrossRef]
- John, J.; Hay, M.; Paton, J. Cyanobacteria in benthic microbial communities in coastal salt lakes in Western Australia. Algological Stud. 2009, 130, 125–135. [Google Scholar] [CrossRef]
- Huang, I.-S.; Pinnell, L.J.; Turner, J.W.; Abdulla, H.; Boyd, L.; Linton, E.W.; Zimba, P.V. Preliminary Assessment of Microbial Community Structure of Wind-Tidal Flats in the Laguna Madre, Texas, USA. Biology 2020, 9, 183. [Google Scholar] [CrossRef]
- Ramos, V.M.C.; Castelo-Branco, R.; Leão, P.N.; Martins, J.; Carvalhal-Gomes, S.; Da Silva, F.S.; Filho, J.G.M.; Vasconcelos, V.M. Cyanobacterial Diversity in Microbial Mats from the Hypersaline Lagoon System of Araruama, Brazil: An in-depth Polyphasic Study. Front. Microbiol. 2017, 8, 1233. [Google Scholar] [CrossRef] [PubMed]
- Santhanakrishnan, T.; Lakshmanan, C.; Radhakrishnan, V. Mineralogical composition and microalgae communities of solar salt tumuli from Tuticorin, Southeast coast of India. Indian J. Geo-Marine Sci. 2018, 47, 498–502. [Google Scholar]
- Chatchawan, T.; Peerapornpisal, Y.; Komarek, J. Diversity of cyanobacteria in man-made solar saltern, Petchaburi Province, Thailand—A pilot study. Fottea 2011, 11, 203–214. [Google Scholar] [CrossRef]
- Sørensen, K.B.; Canfield, D.E.; Oren, A. Salinity Responses of Benthic Microbial Communities in a Solar Saltern (Eilat, Israel). Appl. Environ. Microbiol. 2004, 70, 1608–1616. [Google Scholar] [CrossRef][Green Version]
- Brock, T.D. Salinity and the Ecology of Dunaliella from Great Salt Lake. J. Gen. Microbiol. 1975, 89, 285–292. [Google Scholar] [CrossRef]
- Conner, A.J.; Benison, K.C. Acidophilic Halophilic Microorganisms in Fluid Inclusions in Halite from Lake Magic, Western Australia. Astrobiology 2013, 13, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Kirkwood, A.E.; Buchheim, J.A.; Buchheim, M.A.; Henley, W.J. Cyanobacterial Diversity and Halotolerance in a Variable Hypersaline Environment. Microb. Ecol. 2007, 55, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Vinogradova, O.N.; Darienko, T.M. Algae of Azovo-Syvashsky National Nature Park (Ukraine). Int. J. Algae 2008, 10, 163–178. [Google Scholar] [CrossRef]
- Canfora, L.; Vendramin, E.; Antisari, L.V.; Papa, G.L.; Dazzi, C.; Benedetti, A.; Iavazzo, P.; Adamo, P.; Jungblut, A.D.; Pinzari, F. Compartmentalization of gypsum and halite associated with cyanobacteria in saline soil crusts. FEMS Microbiol. Ecol. 2016, 92, 1–13. [Google Scholar] [CrossRef]
- Tsujimura, S.; Nakahara, H.; Kosaki, T.; Ishida, N.; Iskakov, A.R. Distribution of soil algae in salinized irrigation land in the arid region of Central Asia. Soil Sci. Plant Nutr. 1998, 44, 67–76. [Google Scholar] [CrossRef]
- Kirkwood, A.E.; Henley, W.J. Algal community dynamics and halotolerance in a terrestrial, hypersaline environment. J. Phycol. 2006, 42, 537–547. [Google Scholar] [CrossRef]
- Buchheim, M.A.; Kirkwood, A.E.; Buchheim, J.A.; Verghese, B.; Henley, W.J. Hypersaline soil supports a diverse community of Dunaliella (Chlorophyceae). J. Phycol. 2010, 46, 1038–1047. [Google Scholar] [CrossRef]
- Arabadzhy-Tipenko, L.I.; Solonenko, A.N.; Bren, A.G. Cyanoprokaryota of the Salt Marshes at the Pryazov National Natural Park, Ukraine. Int. J. Algae 2019, 21, 299–310. [Google Scholar] [CrossRef]
- Novichkova-Ivanova, L.N. Soil Algae in Phytocenosis of the Sachara-Gobi Desert Region; Nauka: Leningrad, Russia, 1980. (In Russian) [Google Scholar]
- Prikhodkova, L.P. Blue-Green Algae of Soils of the Steppe Zone of Ukraine; Naukova Dumka Press: Kiev, Ukraine, 1992. (In Russian) [Google Scholar]
- Vinogradova, O.M.; Darienko, T.M. Terrestrial algae of hypersaline environments of the Central Syvash islands (Kherson Region, Ukraine). Biologia 2008, 63, 813–823. [Google Scholar] [CrossRef]
- Vinogradova, O.M. Cyanoprokaryota of Hypersaline Ecosystems of Ukraine; Alterpress: Kiev, Ukraine, 2012. (In Ukrainian) [Google Scholar]
- Patzelt, D.J.; Hodač, L.; Friedl, T.; Pietrasiak, N.; Johansen, J.R. Biodiversity of soil cyanobacteria in the hyper-arid Atacama Desert, Chile. J. Phycol. 2014, 50, 698–710. [Google Scholar] [CrossRef]
- Kaushal, S.S.; Likens, G.E.; Pace, M.L.; Utz, R.M.; Haq, S.; Gorman, J.; Grese, M. Freshwater salinization syndrome on a continental scale. Proc. Natl. Acad. Sci. USA 2018, 115, E574–E583. [Google Scholar] [CrossRef]
- Wang, H.; Jia, G. Satellite-based monitoring of decadal soil salinization and climate effects in a semi-arid region of China. Adv. Atmos. Sci. 2012, 29, 1089–1099. [Google Scholar] [CrossRef]
- Litalien, A.; Zeeb, B. Curing the earth: A review of anthropogenic soil salinization and plant-based strategies for sustainable mitigation. Sci. Total Environ. 2020, 698, 134235. [Google Scholar] [CrossRef]
- Cañedo-Argüelles, M.; Brucet, S.; Carrasco, S.; Flor-Arnau, N.; Ordeix, M.; Ponsá, S.; Coring, E. Effects of potash mining on river ecosystems: An experimental study. Environ. Pollut. 2017, 224, 759–770. [Google Scholar] [CrossRef]
- Siefert, B.; Büchel, G.; Lebküchner-Neugebauer, J. Potash mining waste pile Sollstedt (Thuringia): Investigations of the spreading of waste solutes in the Roethian Karst. Grundwasser 2006, 11, 99–110. [Google Scholar] [CrossRef]
- John, H. Zur Ausbreitung von Halophyten und salztoleranten Pflanzen in der Umgebung von Kali-Rückstandshalden am Beispiel des FND “Salzstelle bei Teutschenthal-Bahnhof“ (Saalkreis). Mitt. Florist. Kart. Sachsen-Anhalt 2000, 5, 175–197. [Google Scholar]
- Garve, E.; Garve, V. Halophyten an Kalihalden in Deutschland und Frankreich (Eisass). Tuexenia 2000, 20, 375–417. [Google Scholar]
- Guder, C. Kalihalden als Modellobjekte der kleinräumigen Florendynamik dargestellt an Untersuchungen im nördlichen Harzvorland. Braunschw. Nat. Schr. 1998, 5, 641–665. [Google Scholar]
- Eilmus, S.; Rösch, C.; Bothe, H. Prokaryotic life in a potash-polluted marsh with emphasis on N-metabolizing microorganisms. Environ. Pollut. 2007, 146, 478–491. [Google Scholar] [CrossRef]
- Sommer, V.; Karsten, U.; Glaser, K. Halophilic Algal Communities in Biological Soil Crusts Isolated from Potash Tailings Pile Areas. Front. Ecol. Evol. 2020, 8, 46. [Google Scholar] [CrossRef]
- Gustavs, L.; Eggert, A.; Michalik, D.; Karsten, U. Physiological and biochemical responses of green microalgae from different habitats to osmotic and matric stress. Protoplasma 2010, 243, 3–14. [Google Scholar] [CrossRef]
- Mikhailyuk, T.; Glaser, K.; Tsarenko, P.; Demchenko, E.; Karsten, U. Composition of biological soil crusts from sand dunes of the Baltic Sea coast in the context of an integrative approach to the taxonomy of microalgae and cyanobacteria. Eur. J. Phycol. 2019, 54, 263–290. [Google Scholar] [CrossRef]
- Komarek, J. Cyanobacterial Taxonomy: Current Problems and Prospects for the Integration of Traditional and Molecular Approaches. ALGAE 2006, 21, 349–375. [Google Scholar] [CrossRef]
- Pröschold, T.; Leliaert, F. Systematics of the green algae: Conflict of classic and modern approaches. In Unravelling the Algae: The Past, Present, and Future of Algal Systematics; Brodie, J., Lewis, J., Eds.; CRC Press: Boca Raton, FL, USA, 2007; pp. 123–154. [Google Scholar]
- Darienko, T.; Gustavs, L.; Mudimu, O.; Menendez, C.R.; Schumann, R.; Karsten, U.; Friedl, T.; Pröschold, T. Chloroidium, a common terrestrial coccoid green alga previously assigned to Chlorella (Trebouxiophyceae, Chlorophyta). Eur. J. Phycol. 2010, 45, 79–95. [Google Scholar] [CrossRef]
- Darienko, T.; Pröschold, T. Toward a monograph of non-marine Ulvophyceae using an integrative approach (Molecular phylogeny and systematics of terrestrial Ulvophyceae II.). Phytotaxa 2017, 324, 1–41. [Google Scholar] [CrossRef]
- Darienko, T.; Pröschold, T. Reevaluation and discovery of new species of the rare genus Watanabea and establishment of Massjukichlorella gen. nov. (Trebouxiophyceae, Chlorophyta) using an integrative approach. J. Phycol. 2019, 55, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Jung, P.; Briegel-Williams, L.; Schermer, M.; Büdel, B. Strong in combination: Polyphasic approach enhances arguments for cold-assigned cyanobacterial endemism. MicrobiolyOpen 2018, 8, e00729. [Google Scholar] [CrossRef]
- Mikhailyuk, T.I.; Vinogradova, O.M.; Glaser, K.; Demchenko, E.; Karsten, U. Diversity of Terrestrial Algae of Cape Kazantip (the Sea of Azov, Ukraine) and Some Remarks on their Phylogeny and Ecology. Int. J. Algae 2018, 20, 313–338. [Google Scholar] [CrossRef]
- Williams, L.; Loewen-Schneider, K.; Maier, S.; Büdel, B. Cyanobacterial diversity of western European biological soil crusts along a latitudinal gradient. FEMS Microbiol. Ecol. 2016, 92, 157. [Google Scholar] [CrossRef]
- Kabirnataj, S.; Nematzadeh, G.A.; Talebi, A.F.; Saraf, A.; Suradkar, A.; Tabatabaei, M.; Singh, P. Description of novel species of Aliinostoc, Desikacharya and Desmonostoc using a polyphasic approach. Int. J. Syst. Evol. Microbiol. 2020, 70, 3413–3426. [Google Scholar] [CrossRef] [PubMed]
- Jung, P.; Mikhailyuk, T.; Emrich, D.; Baumann, K.; Dultz, S.; Büdel, B. Shifting Boundaries: Ecological and Geographical Range extension Based on Three New Species in the Cyanobacterial Genera Cyanocohniella, Oculatella, and, Aliterella. J. Phycol. 2020, 56, 1216–1231. [Google Scholar] [CrossRef]
- Kaštovský, J.; Gomez, E.B.; Hladil, J.; Johansen, J.R. Cyanocohniella calida gen. et sp. nov. (Cyanobacteria: Aphanizomenonaceae) a new cyanobacterium from the thermal springs from Karlovy Vary, Czech Republic. Phytotaxa 2014, 181, 279–292. [Google Scholar] [CrossRef]
- Pietrasiak, N.; Osorio-Santos, K.; Shalygin, S.; Martin, M.P.; Johansen, J.R. When Is A Lineage A Species? A Case Study in Myxacorys gen. nov. (Synechococcales: Cyanobacteria) with the Description of Two New Species from the Americas. J. Phycol. 2019, 55, 976–996. [Google Scholar] [CrossRef]
- Komárek, J. A polyphasic approach for the taxonomy of cyanobacteria: Principles and applications. Eur. J. Phycol. 2016, 51, 346–353. [Google Scholar] [CrossRef]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Komarek, J. Quo vadis, taxonomy of cyanobacteria (2019). Fottea 2020, 20, 104–110. [Google Scholar] [CrossRef]
- Fawley, M.W.; Fawley, K.P. Identification of eukaryotic microalgal strains. J. Appl. Phycol. 2020, 32, 2699–2709. [Google Scholar] [CrossRef]
- Mikhailyuk, T.; Vinogradova, O.M.; Holzinger, A.; Glaser, K.; Samolov, E.; Karsten, U. New record of the rare genus Crinalium Crow (Oscillatoriales, Cyanobacteria) from sand dunes of the Baltic Sea, Germany: Epitypification and emendation of Crinalium magnum Fritsch et John based on an integrative approach. Phytotaxa 2019, 400, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Samolov, E.; Mikhailyuk, T.; Lukešová, A.; Glaser, K.; Büdel, B.; Karsten, U. Usual alga from unusual habitats: Biodiversity of Klebsormidium (Klebsormidiophyceae, Streptophyta) from the phylogenetic superclade G isolated from biological soil crusts. Mol. Phylogenetics Evol. 2019, 133, 236–255. [Google Scholar] [CrossRef] [PubMed]
- Starr, R.C.; Zeiskus, J.A. UTEX—The Culture Collection of Algae at the University of Texas at Austin. J. Phycol. 1993, 29, 1–106. [Google Scholar] [CrossRef]
- Stanier, R.Y.; Kunisawa, R.; Mandel, M.; Cohen-Bazire, G. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 1971, 35, 171–205. [Google Scholar] [CrossRef]
- Mikhailyuk, T.; Lukešová, A.; Glaser, K.; Holzinger, A.; Obwegeser, S.; Nyporko, S.; Friedl, T.; Karsten, U. New Taxa of Streptophyte Algae (Streptophyta) from Terrestrial Habitats Revealed Using an Integrative Approach. Protist 2018, 169, 406–431. [Google Scholar] [CrossRef]
- Marin, B.; Palm, A.; Klingberg, M.; Melkonian, M. Phylogeny and Taxonomic Revision of Plastid-Containing Euglenophytes based on SSU rDNA Sequence Comparisons and Synapomorphic Signatures in the SSU rRNA Secondary Structure. Protist 2003, 154, 99–145. [Google Scholar] [CrossRef]
- Marin, B.; Klingberg, M.; Melkonian, M. Phylogenetic Relationships among the Cryptophyta: Analyses of Nuclear-Encoded SSU rRNA Sequences Support the Monophyly of Extant Plastid-Containing Lineages. Protist 1998, 149, 265–276. [Google Scholar] [CrossRef]
- Wilmotte, A.; Van Der Auwera, G.; De Wachter, R. Structure of the 16 S ribosomal RNA of the thermophilic cyanobacterium chlorogloeopsis HTF (‘mastigocladus laminosus HTF’) strain PCC7518, and phylogenetic analysis. FEBS Lett. 1993, 317, 96–100. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: Hoboken, NJ, USA, 1991; pp. 115–175. [Google Scholar]
- Hoef-Emden, K.; Melkonian, M. Revision of the Genus Cryptomonas (Cryptophyceae): A Combination of Molecular Phylogeny and Morphology Provides Insights into a Long-Hidden Dimorphism. Protist 2003, 154, 371–409. [Google Scholar] [CrossRef]
- Goff, L.J.; Moon, D.A. PCR amplification of nuclear and plastid genes from algal herbarium specimens and algal spores. J. Phycol. 1993, 29, 381–384. [Google Scholar] [CrossRef]
- Marin, B.; Nowack, E.C.M.; Melkonian, M. A Plastid in the Making: Evidence for a Second Primary Endosymbiosis. Protist 2005, 156, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Bock, C.; Krienitz, L.; Pröschold, T. Taxonomic reassessment of the genus Chlorella (Trebouxiophyceae) using molecular signatures (barcodes), including description of seven new species. Fottea 2011, 11, 293–312. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase; World-Wide Electronic Publication. National University of Ireland: Galway, Ireland, 2020. Available online: https://www.algaebase.org (accessed on 4 September 2020).
- Ettl, H.; Gärtner, G. Syllabus der Boden- Luft- und Flechtenalgen, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2014; ISBN 978-3-642-39461-4. [Google Scholar]
- Henley, W.J.; Hironaka, J.L.; Guillou, L.; Buchheim, M.A.; Buchheim, J.A.; Fawley, M.W.; Fawley, K.P. Phylogenetic analysis of the ‘Nannochloris-like’ algae and diagnoses of Picochlorum oklahomensis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta). Phycologia 2004, 43, 641–652. [Google Scholar] [CrossRef]
- Hodač, L.; Hallmann, C.; Spitzer, K.; Elster, J.; Faßhauer, F.; Brinkmann, N.; Lepka, D.; Diwan, V.; Friedl, T. Widespread green algae Chlorella and Stichococcus exhibit polar-temperate and tropical-temperate biogeography. FEMS Microbiol. Ecol. 2016, 92, fiw122. [Google Scholar] [CrossRef]
- Nakada, T.; Nozaki, H.; Pröschold, T. Molecular phylogeny, ultrastructure, and taxonomic revision Ofchlorogonium (Chlorophyta): Emendation Ofchlorogonium and description of Gungnir gen. nov. and Rusalka gen. nov. J. Phycol. 2008, 44, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Dykstra, R.F. Borodinellopsis texensis: Gen. et sp. nov. A new alga from the Texas Gulf coast. In Contributions in Phycology; Parker, B., Brown, R.M.J., Eds.; Phycological Society of America: Lawrence, KS, USA, 1971; pp. 1–8. [Google Scholar]
- Watanabe, S.; Lewis, L.A. Phylogenetic interpretation of light and electron microscopic features of selected members of the phylogroup Moewusinia (Chlorophyceae), with new generic taxonomy. Phycologia 2017, 56, 329–353. [Google Scholar] [CrossRef]
- Matsuzaki, R.; Nakada, T.; Hara, Y.; Nozaki, H. Description of Chloromonas kasaiae sp. nov. (Volvocales, Chlorophyceae), based on comparative electron microscopy and molecular data. Phycologia 2013, 52, 239–245. [Google Scholar] [CrossRef]
- Matsuzaki, R.; Hara, Y.; Nozaki, H. A taxonomic revision of Chloromonas reticulata (Volvocales, Chlorophyceae), the type species of the genus Chloromonas, based on multigene phylogeny and comparative light and electron microscopy. Phycologia 2012, 51, 74–85. [Google Scholar] [CrossRef]
- Pröschold, T.; Marin, B.; Schlösser, U.G.; Melkonian, M. Molecular Phylogeny and Taxonomic Revision of (Chlorophyta). I. Emendation of Chlamydomonas ehrenberg and Chloromonas gobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist 2001, 152, 265–300. [Google Scholar] [CrossRef] [PubMed]
- Fuíková, K.; Flechtner, V.R.; Lewis, L.A. Revision of the genus Bracteacoccus Tereg (Chlorophyceae, Chlorophyta) based on a phylogenetic approach. Nova Hedwigia 2013, 96, 15–59. [Google Scholar] [CrossRef]
- Wynne, M.J.; Hallan, J.K. Reinstatement of Tetradesmus G. M. Smith (Sphaeropleales, Chlorophyta). Feddes Repert. 2015, 126, 83–86. [Google Scholar] [CrossRef]
- Kaufnerová, V.; Eliáš, M. The demise of the genus Scotiellopsis Vinatzer (Chlorophyta). Nova Hedwigia 2013, 97, 415–428. [Google Scholar] [CrossRef]
- Darienko, T.; Friedl, T.; Pröschold, T. Desmochloris mollenhaueri a new terrestrial ulvophycean alga from south-west African soils. (Molecular phylogeny and systematics of terrestrial Ulvophyceae I.). Algol. Stud. 2009, 129, 25–40. [Google Scholar] [CrossRef]
- Watanabe, S.; Kuroda, N.; Maiwa, F. Phylogenetic status of Helicodictyon planctonicum and Desmochloris halophila gen. et comb. nov. and the definition of the class Ulvophyceae (Chlorophyta). Phycologia 2001, 40, 421–434. [Google Scholar] [CrossRef]
- Dangeard, P.J.L.; Guiry, M.D. Validation of some invalid names for green algae introduced by P.J.L. Dangeard. Not. Algarum 2017, 42, 1–2. [Google Scholar]
- O’Kelly, C.J.; Wysor, B.; Bellows, W.K. Collinsiella (Ulvophyceae, Chlorophyta) and other ulotrichalean taxa with shell-boring sporophytes form a monophyletic clade. Phycologia 2004, 43, 41–49. [Google Scholar] [CrossRef]
- Lindstrom, S.C.; Hanic, L.A. The phylogeny of North American Urospora (Ulotrichales, Chlorophyta) based on sequence analysis of nuclear ribosomal genes, introns and spacers. Phycologia 2005, 44, 194–201. [Google Scholar] [CrossRef]
- Hanic, L.A.; Lindstrom, S.C. Life History and Systematic Studies of Pseudothrix borealis gen. et sp. nov. (=North Pacific Capsosiphon groenlandicus, Ulotrichaceae, Chlorophyta). ALGAE 2008, 23, 119–133. [Google Scholar] [CrossRef]
- Darienko, T.; Lukešová, A.; Pröschold, T. The polyphasic approach revealed new species of Chloroidium (Trebouxiophyceae, Chlorophyta). Phytotaxa 2018, 372, 51–66. [Google Scholar] [CrossRef]
- Pröschold, T.; Darienko, T. The green puzzle Stichococcus (Trebouxiophyceae, Chlorophyta): New generic and species concept among this widely distributed genus. Phytotaxa 2020, 441, 113–142. [Google Scholar] [CrossRef]
- Fawley, M.W.; Fawley, K.P.; Buchheim, M.A. Molecular Diversity among Communities of Freshwater Microchlorophytes. Microb. Ecol. 2004, 48, 489–499. [Google Scholar] [CrossRef]
- Darienko, T.; Gustavs, L.; Pröschold, T. Species concept and nomenclatural changes within the genera Elliptochloris and Pseudochlorella (Trebouxiophyceae) based on an integrative approach. J. Phycol. 2016, 52, 1125–1145. [Google Scholar] [CrossRef]
- Perkerson, R.B.I.; Johansen, J.R.; Kovácik, L.; Brand, J.; Kaštovský, J.; Casamatta, D.A. A unique pseudanabaenalean (Cyanobacteria) genus Nodosilinea gen. nov. based on morphological and molecular data. J. Phycol. 2011, 47, 1397–1412. [Google Scholar] [CrossRef]
- Radzi, R.; Muangmai, N.; Broady, P.; Omar, W.M.W.; Lavoue, S.; Convey, P.; Merican, F.M.M.S. Nodosilinea signiensis sp. nov. (Leptolyngbyaceae, Synechococcales), a new terrestrial cyanobacterium isolated from mats collected on Signy Island, South Orkney Islands, Antarctica. PLoS ONE 2019, 14, e0224395. [Google Scholar] [CrossRef]
- Darienko, T.; Gustavs, L.; Eggert, A.; Wolf, W.; Pröschold, T. Evaluating the Species Boundaries of Green Microalgae (Coccomyxa, Trebouxiophyceae, Chlorophyta) Using Integrative Taxonomy and DNA Barcoding with Further Implications for the Species Identification in Environmental Samples. PLoS ONE 2015, 10, e0127838. [Google Scholar] [CrossRef]
- Mikhailyuk, T.I. Terrestrial Algae from the Granite Outcrops of River Valleys of the Ukraine. Int. J. Algae 2013, 15, 311–330. [Google Scholar] [CrossRef]
- Schwarz, K. Neue Bodenalgen aus Dalmatien. Plant Syst. Evol. 1979, 131, 193–209. [Google Scholar] [CrossRef]
- Jónsson, S. The status of the Acrosiphoniales (Chlorophyta). Rit Fiskid. 1999, 16, 187–196. [Google Scholar]
- Sciuto, K.; Lewis, L.A.; Verleyen, E.; Moro, I.; La Rocca, N. Chodatodesmus australis sp. nov. (Scenedesmaceae, Chlorophyta) from Antarctica, with the emended description of the genus Chodatodesmus, and circumscription of Flechtneria rotunda gen. et sp. nov. J. Phycol. 2015, 51, 1172–1188. [Google Scholar] [CrossRef] [PubMed]
- Wynne, M.J.; Furnari, G. A census of J.P.L.Dangeard’s invalid taxa with proposals to resolve the nomenclatural problems of some of them. Nova Hedwigia 2014, 98, 515–527. [Google Scholar] [CrossRef]
- Dangeard, P. Sur quelques algues vertes marines nouvelles observées en culture. Botaniste 1966, 49, 5–45. [Google Scholar]
- Davydov, D.; Shalygin, S.; Vilnet, A.A. New cyanobacterium Nodosilinea svalbardensis sp. nov. (Prochlorotrichaceae, Synechococcales) isolated from alluvium in Mimer river valley of the Svalbard archipelago. Phytotaxa 2020, 442, 61–79. [Google Scholar] [CrossRef]
- Vázquez-Martínez, J.; Gutierrez-Villagomez, J.M.; Fonseca-García, C.; Ramirez-Chavez, E.; Mondragón-Sánchez, M.L.; Partida-Martínez, L.; Johansen, J.R.; Molina-Torres, J. Nodosilinea chupicuarensis sp. nov. (Leptolyngbyaceae, Synechococcales) a subaerial cyanobacterium isolated from a stone monument in central Mexico. Phytotaxa 2018, 334, 167–182. [Google Scholar] [CrossRef]
- Raabová, L.; Kovacik, L.; Elster, J.; Strunecký, O. Review of the genus Phormidesmis (Cyanobacteria) based on environmental, morphological, and molecular data with description of a new genus Leptodesmis. Phytotaxa 2019, 395, 1–16. [Google Scholar] [CrossRef]
- Pocock, T.; Vetterli, A.; Falk, S. Evidence for phenotypic plasticity in the Antarctic extremophile Chlamydomonas raudensis Ettl. UWO 241. J. Exp. Bot. 2011, 62, 1169–1177. [Google Scholar] [CrossRef]
- Qiao, K.; Takano, T.; Liu, S. Discovery of two novel highly tolerant NaHCO3 Trebouxiophytes: Identification and characterization of microalgae from extreme saline–alkali soil. Algal Res. 2015, 9, 245–253. [Google Scholar] [CrossRef]
- Metting, B. The systematics and ecology of soil algae. Bot. Rev. 1981, 47, 195–312. [Google Scholar] [CrossRef]
- Punčochářová, M.; Kalina, T. Taxonomy of the genus Scotiellopsis Vinatzer (Chlorococcales, Chlorophyta). Arch. Hydrobiol. 1981, 27, 119–147. [Google Scholar] [CrossRef]
- Hanagata, N. New species of Coelastrella and Scenedesmus (Chlorophyceae, Chlorophyta). J. Jpn. Bot. 2001, 76, 129–136. [Google Scholar]
- Verses, P.A.; Trainor, F.R. Dactylococcus dissociatus, a New Species from a Connecticut Cornfield Soil. Phycologia 1966, 6, 79–82. [Google Scholar] [CrossRef]
- Bagchi, S.N.; Dubey, N.; Singh, P. Phylogenetically distant clade of Nostoc-like taxa with the description of Aliinostoc gen. nov. and Aliinostoc morphoplasticum sp. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 3329–3338. [Google Scholar] [CrossRef] [PubMed]
- Taton, A.; Grubisic, S.; Ertz, D.; Hodgson, D.A.; Piccardi, R.; Biondi, N.; Tredici, M.R.; Mainini, M.; Losi, D.; Marinelli, F.; et al. Polyphasic study of antarctic cyanobacterial strains. J. Phycol. 2006, 42, 1257–1270. [Google Scholar] [CrossRef]
- Cole, J.K.; Hutchison, J.R.; Renslow, R.S.; Kim, Y.-M.; Chrisler, W.B.; Engelmann, H.E.; Dohnalkova, A.; Hu, D.; Metz, T.O.; Fredrickson, J.K.; et al. Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: Model systems for the study of autotroph-heterotroph interactions. Front. Microbiol. 2014, 5, 109. [Google Scholar] [CrossRef]
- Alvarenga, D.O.; Rigonato, J.; Branco, L.H.Z.; Melo, I.S.; Fiore, M.F. Phyllonema aviceniicola gen. nov., sp. nov. and Foliisarcina bertiogensis gen. nov., sp. nov., epiphyllic cyanobacteria associated with Avicennia schaueriana leaves. Int. J. Syst. Evol. Microbiol. 2016, 66, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Barcytė, D.; Hodač, L. Watanabea acidotolerans: A new trebouxiophyte lineage (Chlorophyta) inhabiting low pH environments from Europe to South America. Phycol. Res. 2018, 67, 120–127. [Google Scholar] [CrossRef]
- Darienko, T.; Rad-Menéndez, C.; Campbell, C.; Pröschold, T. Are there any true marine Chlorella species? Molecular phylogenetic assessment and ecology of marine Chlorella-like organisms, including a description of Droopiella gen. nov. Syst. Biodivers. 2019, 17, 811–829. [Google Scholar] [CrossRef]
- Sommer, V.; Kockx, M.; Wölk, A.; Glaser, K. Von Vogelkot zu grünen Teppichen. Biol. Unserer Zeit 2019, 49, 122–130. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).