Comparative Genomics Analysis of Vibrio anguillarum Isolated from Lumpfish (Cyclopterus lumpus) in Newfoundland Reveal Novel Chromosomal Organizations
Abstract
1. Introduction
2. Material and Methods
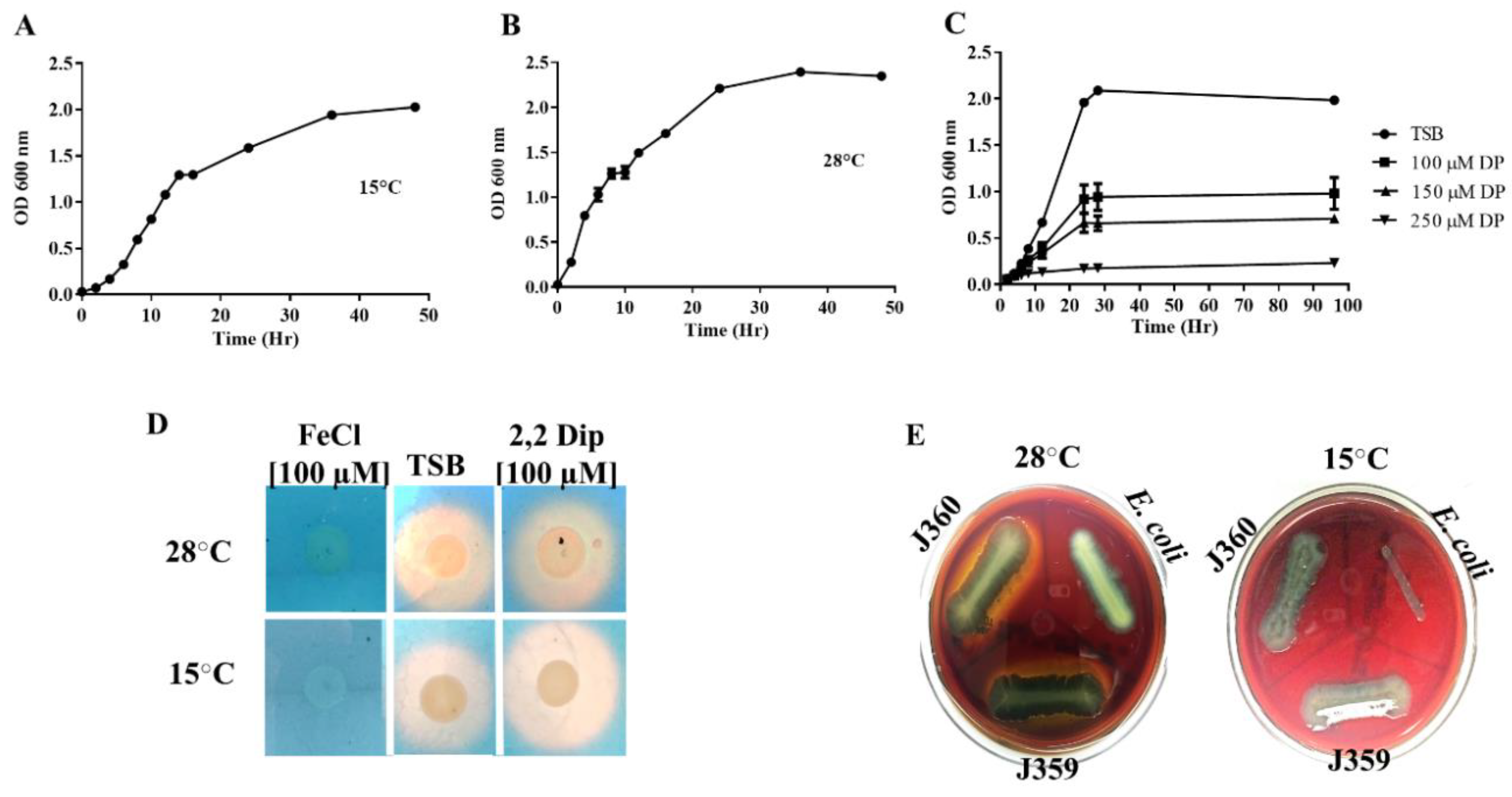
2.1. Bacterial Culture Conditions
2.2. V. anguillarum Isolation
2.3. Matrix-Assisted Laser Desorption/Ionization—Time-of-Flight (MALDI-TOF) Mass Spectrometry and Serotypification Analysis
2.4. Biochemical, Enzymatic, and Physiological Characterization
2.5. Siderophores Synthesis
2.6. Fish Holding
2.7. Infection Assay in Lumpfish
2.8. DNA Extraction and Sequencing
2.9. Genome Assembly, Annotation, and Mapping
2.10. Whole Genome Comparison and Phylogeny Analysis
2.11. Multi-Locus Sequence Analysis of V. anguillarum Housekeeping Genes
2.12. Genomic Islands Analysis
2.13. Comparative Analysis of V. aguillarum J360 Large Plasmid
2.14. Statistical Analysis
2.15. Ethics Statement
3. Results
3.1. Phenotypic, Biochemical, and Enzymatic Characterization
3.2. MALDI-TOF and Agglutination Analysis
3.3. Infection Assay in Specific Pathogen-Free Lumpfish
3.4. V. anguillarum J360 Genome Sequencing and Annotation
3.5. Whole Genome Alignment, Phylogeny, and Synteny
3.6. Multi-Locus Sequence Analysis (MLSA) and Phylogeny
3.7. Distribution of Genes Associated with Pathogenesis and Environmental Adaptation in V. anguillarum J360
3.8. Genomic Islands (GIs)
3.9. V. anguillarum Large Plasmids Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pruzzo, C.; Huq, A.; Colwell, R.; Donelli, G. Pathogenic Vibrio Species in the Marine and Estuarine Environment; Springe: Boston, MA, USA, 2005; pp. 217–252. [Google Scholar]
- Baker-Austin, C.; Oliver, J.; Alam, M.; Ali, A.; Waldor, M.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nat. Rev. Dis. Primers. 2018, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Stockley, L.; Rangdale, R.; Martinez-Urtaza, J. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: A European perspective. Environ. Microbiol. Rep. 2010, 2, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Gram, L.; Melchiorsen, J.; Spanggaard, B.; Huber, I.; Nielsen, T. Inhibition of Vibrio anguillarum by Pseudomonas fluorescens AH2, a possible probiotic treatment of fish. Appl. Environ. Microbiol. 1999, 65, 969–973. [Google Scholar] [CrossRef] [PubMed]
- Frans, I.; Michiels, C.; Bossier, P.; Willems, K.; Lievens, B.; Rediers, H. Vibrio anguillarum as a fish pathogen: Virulence factors, diagnosis and prevention. J. Fish Dis. 2011, 34, 643–661. [Google Scholar] [CrossRef]
- Holm, K.; Nilsson, K.; Hjerde, E.; Willassen, N.; Milton, D. Complete genome sequence of Vibrio anguillarum strain NB10, a virulent isolate from the Gulf of Bothnia. Stand. Genom. Sci. 2015, 10, 60. [Google Scholar] [CrossRef]
- Li, G.; Mo, Z.; Li, J.; Xiao, P.; Hao, B. Complete Genome Sequence of Vibrio anguillarum M3, a Serotype O1 Strain Isolated from Japanese Flounder in China. Genome Announc. 2013, 1, e00769-13. [Google Scholar] [CrossRef]
- Naka, H.; Dias, G.; Thompson, C.; Dubay, C.; Thompson, F.; Crosa, J. Complete genome sequence of the marine fish pathogen Vibrio anguillarum harboring the pJM1 virulence plasmid and genomic comparison with other virulent strains of V. anguillarum and V. ordalii. Infect. Immun. 2011, 79, 2889–2900. [Google Scholar] [CrossRef][Green Version]
- Brooker, A.; Papadopoulou, A.; Gutierrez, C.; Rey, S.; Davie, A.; Migaud, H. Sustainable production and use of cleaner fish for the biological control of sea lice: Recent advances and current challenges. Vet. Rec. 2018, 183, 383. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.; Treasurer, C.; Pooley, A.; Keay, R.; Imsland, A.; Garcia de Leaniz, C. Use of lumpfish for sea-lice control in salmon farming:challenges and opportunities. Rev. Aqua. 2017, 10, 1–20. [Google Scholar] [CrossRef]
- Imsland, A.; Reynolds, P.; Eliassen, G.; Hangstad, A.; Foss, A.; Vikingstad, E.; Elvegård, T. The use of lumpfish (Cyclopterus lumpus L.) to control sea lice (Lepeophtheirus salmonis Krøyer) infestations in intensively farmed Atlantic salmon (Salmo salar L.). Aquaculture 2014, 424, 18–23. [Google Scholar] [CrossRef]
- McEwan, G.; Groner, M.; Cohen, A.; Imsland, A.; Revie, C. Modelling sea lice control by lumpfish on Atlantic salmon farms: Interactions with mate limitation, temperature and treatment rules. Dis. Aqua. Org. 2019, 133, 69–82. [Google Scholar] [CrossRef]
- Sayer, M.; Davenport, J. Hypometabolism in torpid goldsinny wrasse subjected to rapid reductions in seawater temperature. J. Fish Biol. 1996, 49, 64–75. [Google Scholar] [CrossRef]
- Costa, I.; Driedzic, W.; Gamperl, A. Metabolic and cardiac responses of cunner Tautogolabrus adspersus to seasonal and acute changes in temperature. Phy. Biochem. Zool. 2013, 86, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Deady, S.; Varian, S.; Fives, J. The use of cleaner-fish to control sea lice on two Irish salmon (Salmo salar) farms with particular reference to wrasse behaviour in salmon cages. Aquaculture 1995, 131, 73–90. [Google Scholar] [CrossRef]
- Blanco Gonzalez, E.; de Boer, F. The development of the Norwegian wrasse fishery and the use of wrasses as cleaner fish in the salmon aquaculture industry. Fish. Sci. 2017, 83, 661–670. [Google Scholar] [CrossRef]
- Sørensen, U.; Larsen, J. Serotyping of Vibrio anguillarum. Appl. Environ. Microbiol. 1986, 51, 593–597. [Google Scholar] [CrossRef]
- Pedersen, K.; Grisez, L.; van Houdt, R.; Tiainen, T.; Ollevier, F.; Larsen, J. Extended serotyping scheme for Vibrio anguillarum with the definition and characterization of seven provisional O-serogroups. Curr. Microbiol. 1999, 38, 183–189. [Google Scholar] [CrossRef]
- Aquaculture, Department of Aquaculture (Ed.) Fisheries and Land Resources: Newfoundland and Labrador Aquatic Animal Reportable and Notifiable Disease; Fisheries and Aquaculture: Saint John, NL, Canada, 2020; p. 2. Available online: https://www.gov.nl.ca/ffa/files/Newfoundland-and-Labrador-Aquatic-Animal-Reportable-and-Notifiable-Diseases-September-2020.pdf (accessed on 26 October 2020).
- Holm, K.; Baekkedal, C.; Soderberg, J.; Haugen, P. Complete genome sequences of seven Vibrio anguillarum strains as derived from PacBio sequencing. Genome Biol. Evol. 2018, 10, 1127–1131. [Google Scholar] [CrossRef]
- Louden, B.; Haarmann, D.; Lynne, A. Use of Blue Agar CAS Assay for Siderophore detection. J. Microbiol. Biol. Edu. 2011, 12, 51–53. [Google Scholar] [CrossRef]
- Wood, E. Molecular Cloning. A Laboratory Manual. Biochem. Educ. 1983, 11, 82. [Google Scholar] [CrossRef]
- Cameron, M.; Barkema, H.; De Buck, J.; De Vliegher, S.; Chaffer, M.; Lewis, J.; Keefe, G. Identification of bovine-associated coagulase-negative staphylococci by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry using a direct transfer protocol. J. Dairy Sci. 2017, 100, 2137–2147. [Google Scholar] [CrossRef] [PubMed]
- Connors, E.; Soto-Dávila, M.; Hossain, A.; Vasquez, I.; Gnanagobal, H.; Santander, J. Identification and validation of reliable Aeromonas salmonicida subspecies salmonicida reference genes for differential gene expression analyses. Infect. Genet. Evol. 2019, 73, 314–321. [Google Scholar] [CrossRef]
- Myhr, E.; Larsen, J.; Lillehaug, A.; Gudding, R.; Heum, M.; Håstein, T. Characterization of Vibrio anguillarum and closely related species isolated from farmed fish in Norway. Appl. Environ. Microbiol. 1991, 57, 2750–2757. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, P.; Gunasekaran, D. Assessment of antibiotic sensitivity and pathogenicity of Vibrio spp. and Aeromonas spp. from aquaculture enviroment. Moj Ecol. Environ. Sci. 2018, 3, 128–136. [Google Scholar] [CrossRef]
- Chakraborty, S.; Cao, T.; Hossain, A.; Gnanagobal, H.; Vasquez, I.; Boyce, D.; Santander, J. Vibrogen-2 vaccine trial in lumpfish (Cyclopterus lumpus) against Vibrio anguillarum. J. Fish Dis. 2019, 42, 1057–1064. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Press: New York, NY, USA, 2001. [Google Scholar]
- Aziz, R.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.; Formsma, K.; Gerdes, S.; Glass, E.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 26 October 2020).
- Carver, T.; Thomson, N.; Bleasby, A.; Berriman, M.; Parkhill, J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics 2009, 25, 119–120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Microbiol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Jukes, T.; Cantor, C. Evolution of Protein Molecules. In Mammalian Protein Metabolism; Munro, H., Ed.; Academic Press: Cambridge, MA, USA, 1969; pp. 21–132. [Google Scholar]
- Teru, Y.; Hikima, J.; Kono, T.; Sakai, M.; Takano, T.; Hawke, J.; Takeyama, H.; Aoki, T. Whole-genome sequence of Photobacterium damselae subsp. piscicida strain 91–197, isolated from hybrid striped Bass (Morone sp.) in the United States. Genome Announc. 2017, 5, e00600-17. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.; Mau, B.; Blattner, F.; Perna, N. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, B.; Grotkjær, T.; Alvise, P.; Yin, G.; Zhang, F.; Bunk, B.; Spröer, C.; Bentzon-Tilia, M.; Gram, L. Vibrio anguillarum is genetically and phenotypically unaffected by Long-Term continuous exposure to the antibacterial compound tropodithietic acid. Appl. Environ. Microbiol. 2016, 82, 4802. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Gil, B.; Soto-Rodríguez, S.; García-Gasca, A.; Roque, A.; Vazquez-Juarez, R.; Thompson, F.; Swings, J. Molecular identification of Vibrio harveyi-related isolates associated with diseased aquatic organisms. Microbiology 2004, 150, 1769–1777. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Shread, P.; Furniss, A.; Bryant, T. Taxonomy and description of Vibrio fluvialis sp. nov. (synonym group F vibrios, group EF6). J. Appl. Bacteriol. 1981, 50, 73–94. [Google Scholar] [CrossRef] [PubMed]
- Fujino, T.; Miwatani, T.; Yasuda, J.; Kondo, M.; Takeda, Y.; Akita, Y.; Kotera, K.; Okada, M.; Nishimune, H.; Shimizu, Y.; et al. Taxonomic studies on the bacterial strains isolated from cases of “shirasu” food-poisoning (Pasteurella parahaemolytica) and related microorganisms. Biken J. 1965, 8, 63–71. [Google Scholar] [PubMed]
- Le Roux, F.; Zouine, M.; Chakroun, N.; Binesse, J.; Saulnier, D.; Bouchier, C.; Zidane, N.; Ma, L.; Rusniok, C.; Lajus, A.; et al. Genome sequence of Vibrio splendidus: An abundant planctonic marine species with a large genotypic diversity. Environ. Microbiol. 2009, 11, 1959–1970. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Bertelli, C.; Hoad, G.; Winsor, G.; Brinkman, F.; Laird, M.; Lau, B.; Williams, K. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Villarreal-Chiu, J.; Quinn, J.; McGrath, J. The genes and enzymes of phosphonate metabolism by bacteria, and their distribution in the marine environment. Front. Microbiol. 2012, 3, 19. [Google Scholar] [CrossRef]
- Favrot, L.; Blanchard, J.; Vergnolle, O. Bacterial GCN5-Related N-Acetyltransferases: From resistance to regulation. Biochemistry 2016, 55, 989–1002. [Google Scholar] [CrossRef]
- Marcos-Lopez, M.; Donald, K.; Stagg, H.; McCarthy, U. Clinical Vibrio anguillarum infection in lumpsucker Cyclopterus lumpus in Scotland. Vet. Rec. 2013, 173, 319. [Google Scholar] [CrossRef] [PubMed]
- Tagliavia, M.; Salamone, M.; Bennici, C.; Quatrini, P.; Cuttitta, A. A modified culture medium for improved isolation of marine vibrios. Microbiol. Open. 2019, 8, e00835. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Mou, X.; Nelson, D. HlyU Is a Positive Regulator of hemolysin expression in Vibrio anguillarum. J. Bacteriol. 2011, 193, 4779. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boyce, D.; Ang, K.; Prickett, R. Cunner and lumpfish as cleaner fish species in Canada. In Cleaner fish biology and Aquaculture Applications; Treasurers, J.W., Ed.; 5m Publishing: Sheffield, UK, 2018. [Google Scholar]
- Castillo, D.; Alvise, P.; Xu, R.; Zhang, F.; Middelboe, M.; Gram, L. Comparative genome analyses of Vibrio anguillarum strains reveal a link with pathogenicity traits. mSystems 2017, 2, e00001-17. [Google Scholar] [CrossRef]
- Steinum, T.; Karatas, S.; Martinussen, N.; Meirelles, P.; Thompson, F.; Colquhoun, D. Multilocus sequence analysis of close relatives Vibrio anguillarum and Vibrio ordalii. Appl. Environ. Microbiol. 2016, 82, 5496–5504. [Google Scholar] [CrossRef]
- Glaeser, S.; Kämpfer, P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 2015, 38, 237–245. [Google Scholar] [CrossRef]
- Whittaker, B.; Consuegra, S.; Garcia de Leaniz, C. Genetic and phenotypic differentiation of lumpfish (Cyclopterus lumpus) across the North Atlantic: Implications for conservation and aquaculture. PeerJ 2018, 6, e5974. [Google Scholar] [CrossRef]
- Marsh, J.; Taylor, R. Genetic and transcriptional analyses of the Vibrio cholerae mannose-sensitive hemagglutinin type 4 pilus gene locus. J. Bacteriol. 1999, 181, 1110–1117. [Google Scholar] [CrossRef]
- Weber, B.; Hasic, M.; Chen, C.; Wai, S.; Milton, D. Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ. Microbiol. 2009, 11, 3018–3028. [Google Scholar] [CrossRef]
- Hirono, I.; Masuda, T.; Aoki, T. Cloning and detection of the hemolysin gene of Vibrio anguillarum. Microb. Pathogens. 1996, 21, 173–182. [Google Scholar] [CrossRef]
- Rock, J.; Nelson, D. Identification and characterization of a hemolysin gene cluster in Vibrio anguillarum. Infect. Immun. 2006, 74, 2777–2786. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Naka, H.; Lopez, C.; Crosa, J. Role of the pJM1 plasmid-encoded transport proteins FatB, C and D in ferric anguibactin uptake in the fish pathogen Vibrio anguillarum. Environ. Microbiol. Rep. 2010, 2, 104–111. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Milton, D.; Hardman, A.; Camara, M.; Chhabra, S.; Bycroft, B.; Stewart, G.; Williams, P. Quorum sensing in Vibrio anguillarum: Characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-L-homoserine lactone. J. Bacteriol. 1997, 179, 3004–3012. [Google Scholar] [CrossRef] [PubMed]
- Busschaert, P.; Frans, I.; Crauwels, S.; Zhu, B.; Willems, K.; Bossier, P.; Michiels, C.; Verstrepen, K.; Lievens, B.; Rediers, H. Comparative genome sequencing to assess the genetic diversity and virulence attributes of 15 Vibrio anguillarum isolates. J. Fish Dis. 2015, 38, 795–807. [Google Scholar] [CrossRef]






| Species and Serotype | Geographic Origin, Host Species | Genome Size (bp) | Accession Numbers | References |
|---|---|---|---|---|
| V. anguillarum 775/O1 | USA, Pacific coast/Oncorhynchus kisutch | 4,117,056 | CP002284/5 | [8] |
| V. anguillarum M3/O1 | China, Shandong/Paralichthys olivaceus | 4,117,885 | CP006699/700 | [7] |
| V. anguillarum NB10/O1 | Sweden, Baltic Sea/Oncorhynchus mykiss | 4,373,835 | LK021130/29 | [6] |
| V. anguillarum VIB12/O2 | Greece/Dicentrarchus labrax | 4,897,690 | CP023310/11 | [20] |
| V. anguillarum VIB43/O1 | Scotland, UK/Dicentrarchus labrax | 4,407,865 | CP023054/5 | [20] |
| V. anguillarum CNEVA/O3 | France/Dicentrarchus labrax | 4,256,429 | CP022103/4 | [20] |
| V. anguillarum MHK3/O1 | China/Flounder | 4,015,925 | CP022468/9 | - |
| V. anguillarum 425 | China/Sea bass | 4,373,373 | CP020533/4 | - |
| V. anguillarum 87-9-116/O1 | Finland/Salmo salar | 4,338,125 | CP021980/1 | [20] |
| V. anguillarum JLL237/O1 | Denmark/Oncorhynchus mykiss | 4,286,989 | CP022101/2 | [20] |
| V. anguillarum ATCC-68554/O1 | USA, Pacific coast/Oncorhynchus kisutch | 4,141,906 | CP023209/8 | [20] |
| V. anguillarum 90-11-286 | Denmark/Farm-water sample | 4,342,224 | CP011460/1 | [37] |
| V. anguillarum S3/O1 | Denmark/Oncorhynchus mykiss | 4,272,973 | CP022099/100 | [20] |
| V. campbelli ATCC 25920 | USA, Hawaii/Seawater isolate | 5,178,103 | CP015863/4 | [38] |
| V. fluvialis ATCC 33809 | Bangladesh/Homo sapiens | 4,827,733 | CP014034/5 | [39] |
| V. parahaemolyticus ATCC 17802 | Japan/Seawater isolate | 5,152,461 | CP014046/7 | [40] |
| V. tasmaniensis LGP32 | France/Crassostrea gigas | 4,974,818 | FM954972/3 | [41] |
| Photobacterium damselae 91-197 | USA/Hybrid Striped Bass (Morone sp.) | 4,293,175 | AP018045/6 | [35] |
| Characteristic | Vibrio anguillarum J360 |
|---|---|
| Growth at: | |
| 4 °C | + |
| 15 °C | + |
| 28 °C | + |
| 37 °C | − |
| LB NaCl 0% | − |
| LB NaCl 0.5% | + |
| LB NaCl 2% | + |
| Plate Count Agar 50% seawater | + |
| TCBS | − |
| Motility | + |
| Fimbria Type I | + |
| Siderophores synthesis | + |
| Catalase | + |
| Oxidase | + |
| Antibiogram: | Halo diameter (mm) |
| Tetracycline (10 mg/mL) | 31 (Susceptible) |
| Oxytetracycline (30 mg/mL) | 34 (Susceptible) |
| Ampicillin (10 mg/mL) | 0 (Resistant) |
| Sulfamethoxazole (25 mg/mL) | 33 (Susceptible) |
| Chloramphenicol (30 mg/mL) | 35 (Susceptible) |
| Colistin sulphate (10 mg/mL) | 15 (Susceptible) |
| Oxalinic acid (2 mg/mL) | 39 (Susceptible) |
| O-129 | 39 (susceptible) |
| Labels | Size (Mb) | Topology | RefSeq ID | INSDC Identifier |
|---|---|---|---|---|
| Chromosome-I | 3.32 | Circular | NZ_CP034672 | CP034672 |
| Chromosome-II | 1.17 | Circular | NZ_CP034673 | CP034673 |
| Plasmid pVaJ360-I | 0.06 | Circular | NZ_CP034674 | CP034674 |
| Plasmid pVaJ360-II | 0.012 | Circular | NZ_MT050454 | MT050454 |
| Chromosome-I | Chromosome-II | Plasmid pVaJ360-I | Plasmid pVasJ360-II | |
|---|---|---|---|---|
| Genome size (bp) | 3,320,860 | 1,172,081 | 56,630 | 11,995 |
| G+C content (%) | 44.6 | 44.1 | 43.7 | 47 |
| Number of subsystems | 441 | 88 | 2 | 1 |
| Number of coding sequences | 3149 | 1143 | 96 | 24 |
| Number of RNAs | 129 | 5 | - | 1 |
| Attribute | Data Provided |
|---|---|
| Annotation pipeline | NCBI |
| Annotation method | Best placed reference protein set; GeneMarks v4.6 |
| Genes (total) | 4371 |
| CDSs (total) | 4234 |
| Genes (coding) | 3966 |
| Genes (RNA) | 137 |
| rRNAs | 10, 9, 9 (5S, 16S, 23S) |
| Complete rRNAs | 10, 9, 9 (5S, 16S, 23S) |
| tRNAs | 105 |
| ncRNAs | 4 |
| Pseudogenes (total) | 268 |
| Pseudogenes (ambiguous residues) | 0 of 268 |
| Pseudogenes (frameshifted) | 105 of 268 |
| Pseudogenes (incomplete) | 163 of 268 |
| Pseudogenes (internal stop) | 45 of 268 |
| Pseudogenes (multiple problems) | 41 of 268 |
| Gene Subsystem Category and Name | Presence/Absence of Gene in V. anguillarum J360 | GenBank Accession N° | |
|---|---|---|---|
| Chromosome-I | Chromosome-II | ||
| Iron transport and regulation | |||
| iron-regulated protein A | X | DYL72_00705 | |
| tonB2, exbD2 | X | DYL72_00755, DYL72_00745 | |
| tonB1, exbB, exbD1 | X | DYL72_00260, DYL72_00265, DYL72_00270 | |
| fur | X | DYL72_03070 | |
| iron ABC transport permease | X | DYL72_00175, DYL72_00765 | |
| Heme transport | |||
| Ton-B-dependent hemoglobin receptor | X | X | DYL72_17445, DYL72_00730, DYL72_20920 |
| hutXZ | X | DYL72_00735, DYL72_00740 | |
| heme ABC transporter protein | X | DYL72_00770, DYL72_02835 | |
| heme exporter protein ccmBD | X | DYL72_02830, DYL72_02840 | |
| Ferrous and ferric transport | |||
| ferric ABC transporter | X | DYL72_00180 | |
| feoABC | X | DYL72_02945, DYL72_02940, DYL72_02935 | |
| Ferrichrome | |||
| fhuACBD | X | DYL72_06770, DYL72_10590 | |
| Hemolysins | |||
| hemolysin genes | X | X | DYL72_01800, DYL72_07805, DYL72_12295, DYL72_17765 |
| thermolabile hemolysin | X | DYL72_17760 | |
| Toxins-associated genes | |||
| toxins and pseudogenes | X | DYL72_00035, DYL72_00045, DYL72_14975, DYL72_14985 | |
| rtxA, hipA | X | DYL72_01180, DYL72_03440 | |
| ampC | X | DYL72_17850 | |
| type II toxin–antitoxin system RelBE/ParDE/DinJ family | X | DYL72_18560, DYL72_18565, DYL72_18715, DYL72_18750, DYL72_19140, DYL72_19250, DYL72_19960 | |
| type II toxin–antitoxin system prevent-host-death family antitoxin | X | DYL72_18805 | |
| Txe/YoeB family addiction module | X | DYL72_18810 | |
| type II toxin–antitoxin system Phd/YefM family antitoxin | X | DYL72_19255 | |
| type II toxin–antitoxin system YafQ family toxin | X | DYL72_19965 | |
| toxin–antitoxin system subunit antitoxin | X | DYL72_20370 | |
| Metalloproteases | |||
| CPBP family intramembrane metalloprotease | X | DYL72_00295 | |
| pmbA, tldD, ftsH | X | DYL72_05300, DYL72_09550, DYL72_10735 | |
| SprT family zinc-dependent metalloprotease | X | DYL72_09830 | |
| M6 family metalloprotease domain-containing protein | X | DYL72_17780 | |
| Secreted enzymes | |||
| phospholipase gene | X | DYL72_16230 | |
| lipase | X | DYL72_17465, DYL72_18050, DYL72_20375 | |
| Motility and Chemotaxis | |||
| fliRQPONMLKJIHGFE, fliSTD, fliL | X | DYL72_03140-DYL72_03205, DYL72_03225-DYL72_03235, DYL72_08330 | |
| motYA, motB | X | DYL72_12660, DYL72_00275, DYL72_12090 | |
| flhFAB | X | DYL72_02900, DYL72_02905, DYL72_03135, | |
| flag, flaCA | X | DYL72_03240, DYL72_03670, DYL72_03675 | |
| flagellin | X | DYL72_03245, DYL72_03250, DYL72_03255 | |
| flgLKJIHGFEDCB, flgAMNP | X | DYL72_03685-DYL72_03735, DYL72_03750-DYL72_03765 | |
| flagellar basal-body protein | X | DYL72_03775 | |
| pomA | X | DYL72_12085 | |
| flagellar brake protein | X | DYL72_17355, DYL72_20195 | |
| methyl-accepting chemotaxis protein | X (17) | X (16) | |
| chemotaxis response regulator protein-glutamate methylesterase | X | X | DYL72_00595, DYL72_02870, DYL72_16590, |
| cheD, cheW, cheA, cheV, cheY, cheX, cheC | X | X | DYL72_00600, DYL72_00610, DYL72_00620, DYL72_01045, DYL72_02885, DYL72_09235, DYL72_20035 |
| cheV2, cheW2, cheW3, cheA2, cheV3, cheD2, cheW4, cheW5, cheA3, cheV4 | X | X | DYL72_02435, DYL72_02855, DYL72_02860, DYL72_02875, DYL72_03745, DYL72_16595, DYL72_16610, DYL72_16615, DYL72_16620, DYL72_21345 |
| chemotaxis protein | X | X | DYL72_00635, DYL72_16275, DYL72_16460 |
| chemotaxis protein methyltransferase | X | DYL72_03740 | |
| Type IV pilus | |||
| pilQ, pilW, pilM, pilT, pilB | X | DYL72_05800, DYL72_01100, DYL72_05820, DYL72_09795, DYL72_10350 | |
| Type VI secretion system (T6SS) | |||
| tssI, tssBCEFG, tssJ, tssK tssH, tssAM | X | DYL72_16415, DYL72_17000—DYL72_17020, DYL72_17030, DYL72_17045, DYL72_17055, DYL72_17070, DYL72_17075, | |
| tssI2, tssM2, tssKJHFE2, tssC2, tssI3, tssI4, tssI5, tssI6 | X | X | DYL72_17085, DYL72_21115, DYL72_00885, DYL72_00895, DYL72_00900, DYL72_00930, DYL72_00940, DYL72_00945, DYL72_00955, DYL72_00960, DYL72_00975, DYL72_02685, DYL72_10015 |
| tagHO | X | DYL72_17025, DYL72_17065 | |
| Hcp family type VI secretion system effector | X | X | DYL72_16420, DYL72_18695, DYL72_21120, DYL72_02690, DYL72_10010 |
| type VI secretion system tube protein Hcp | X | DYL72_00965 | |
| DotU family type IV/VI secretion system protein | X | X | DYL72_17050, DYL72_00890 |
| type VI secretion protein | X | X | DYL72_17080, DYL72_00950, DYL72_00970, DYL72_00985 |
| type VI secretion system PAAR protein | X | X | DYL72_21095, DYL72_02650, DYL72_02660, DYL72_10045, |
| type VI secretion protein VasB-1 | X | DYL72_00935 | |
| Quorum sensing | |||
| quorum-sensing autoinducer synthase | X | DYL72_17995 | |
| Regulators | |||
| transcriptional regulator LuxR | X | X | DYL72_06050, DYL72_21285 |
| transcriptional regulator LysR | X | DYL72_03060, DYL72_09990 | |
| transcriptional regulator XRE | X | DYL72_10570 | |
| cysB | X | DYL72_13470 | |
| nhaR | X | DYL72_10935 | |
| hfq | X | DYL72_09115 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasquez, I.; Cao, T.; Chakraborty, S.; Gnanagobal, H.; O’Brien, N.; Monk, J.; Boyce, D.; Westcott, J.D.; Santander, J. Comparative Genomics Analysis of Vibrio anguillarum Isolated from Lumpfish (Cyclopterus lumpus) in Newfoundland Reveal Novel Chromosomal Organizations. Microorganisms 2020, 8, 1666. https://doi.org/10.3390/microorganisms8111666
Vasquez I, Cao T, Chakraborty S, Gnanagobal H, O’Brien N, Monk J, Boyce D, Westcott JD, Santander J. Comparative Genomics Analysis of Vibrio anguillarum Isolated from Lumpfish (Cyclopterus lumpus) in Newfoundland Reveal Novel Chromosomal Organizations. Microorganisms. 2020; 8(11):1666. https://doi.org/10.3390/microorganisms8111666
Chicago/Turabian StyleVasquez, Ignacio, Trung Cao, Setu Chakraborty, Hajarooba Gnanagobal, Nicole O’Brien, Jennifer Monk, Danny Boyce, Jillian D. Westcott, and Javier Santander. 2020. "Comparative Genomics Analysis of Vibrio anguillarum Isolated from Lumpfish (Cyclopterus lumpus) in Newfoundland Reveal Novel Chromosomal Organizations" Microorganisms 8, no. 11: 1666. https://doi.org/10.3390/microorganisms8111666
APA StyleVasquez, I., Cao, T., Chakraborty, S., Gnanagobal, H., O’Brien, N., Monk, J., Boyce, D., Westcott, J. D., & Santander, J. (2020). Comparative Genomics Analysis of Vibrio anguillarum Isolated from Lumpfish (Cyclopterus lumpus) in Newfoundland Reveal Novel Chromosomal Organizations. Microorganisms, 8(11), 1666. https://doi.org/10.3390/microorganisms8111666







