Detailed Molecular Interactions of Favipiravir with SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza Virus Polymerases In Silico
Abstract
1. Introduction
2. Materials and Methods
2.1. Structural Modeling
2.2. Docking Simulation
3. Results
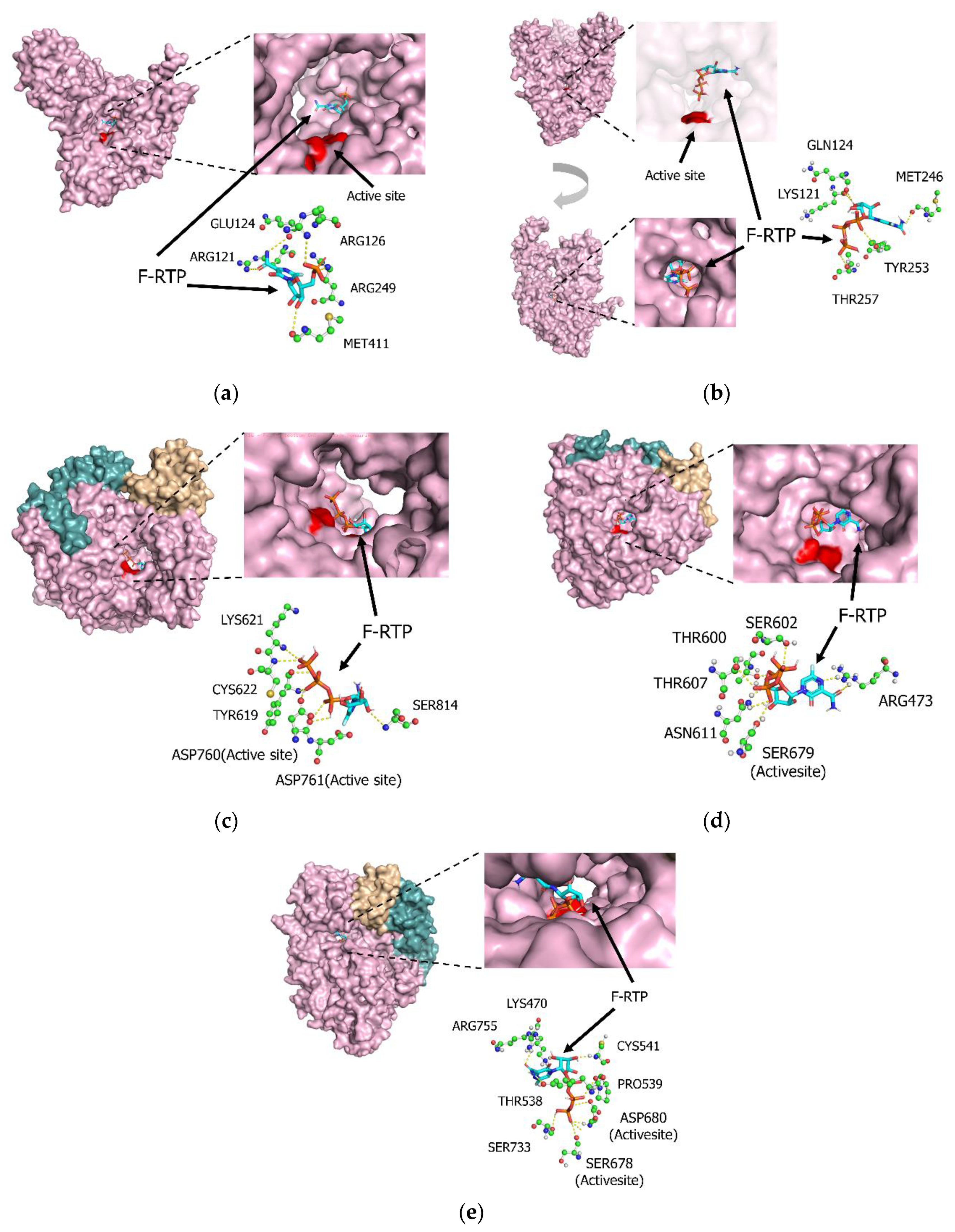
3.1. Molecular Interactions between Favipiravir and F-RTP with Influenza PB1 Proteins
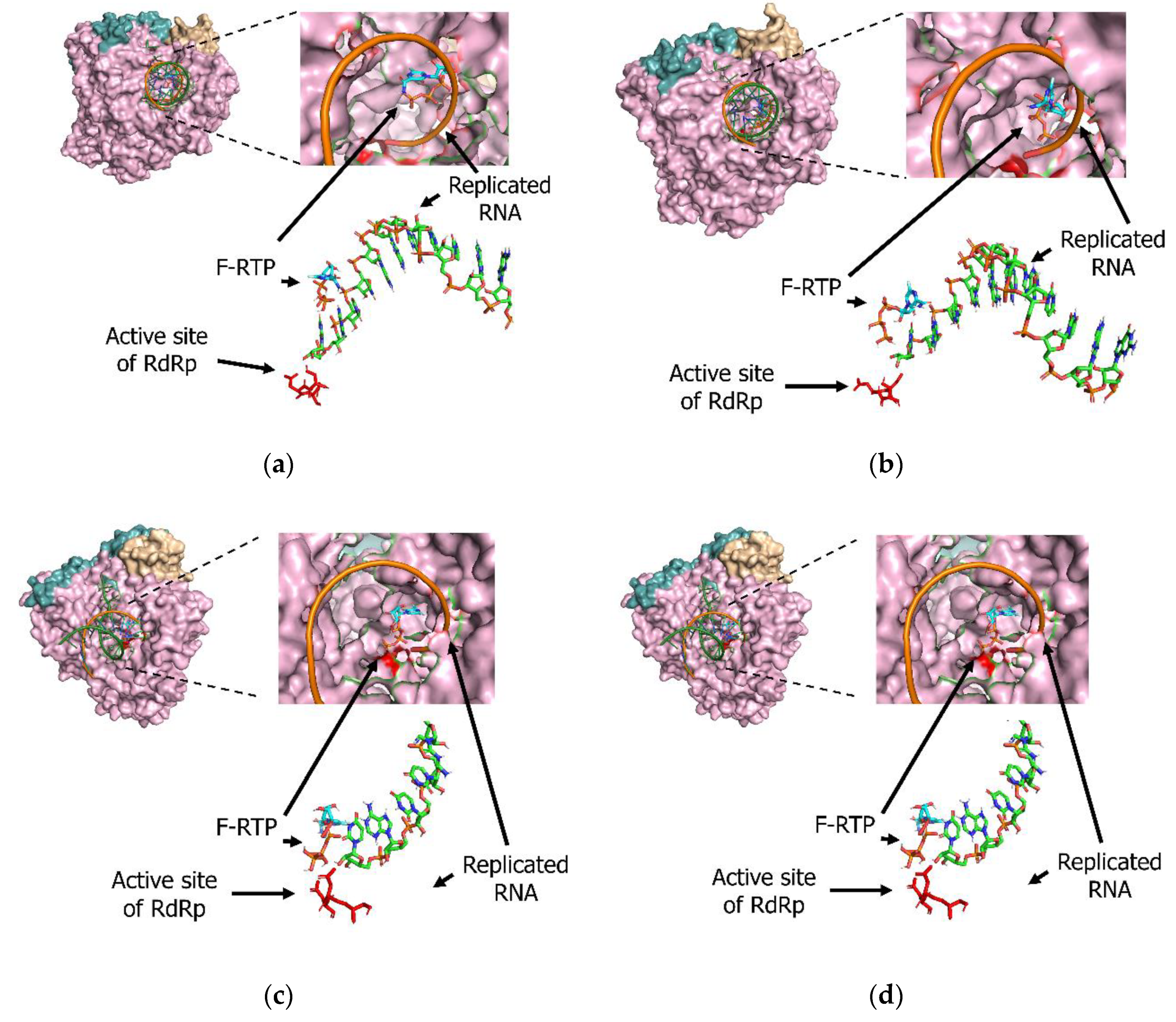
3.2. Molecular Interactions between Favipiravir and F-RTP with SARS-CoV-2/SARS-CoV/MERS-CoV Replication-Associated Proteins RdRp and NSP-15 Endonuclease Alone
3.3. Molecular Interactions among Various Polymerases, Nucleotide Triphosphate (NTP), Magnesium Ion, and Favipiravir/F-RTP
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Zhu, N.A.; Zhang, D.; Wang, W.; Li, X.; Yang, B.O.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Coronavirus Disease (COVID-19). Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 30 September 2020).
- The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [CrossRef]
- Mahumud, R.A.; Kamara, J.K.; Renzaho, A.M.N. The epidemiological burden and overall distribution of chronic comorbidities in coronavirus disease-2019 among 202,005 infected patients: Evidence from a systematic review and meta-analysis. Infection 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, B.O.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Noviello, S.; Pagliano, P. Update on treatment of COVID-19: Ongoing studies between promising and disappointing results. Le Infez. Med. 2020, 28, 198–211. [Google Scholar]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef]
- Sangawa, H.; Komeno, T.; Nishikawa, H.; Yoshida, A.; Takahashi, K.; Nomura, N.; Furuta, Y. Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob. Agents Chemother. 2013, 57, 5202–5208. [Google Scholar] [CrossRef]
- Takashita, E. Influenza Polymerase Inhibitors: Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2020, a038687. [Google Scholar] [CrossRef]
- Goldhill, D.H.; Te Velthuis, A.J.W.; Fletcher, R.A.; Langat, P.; Zambon, M.; Lackenby, A.; Barclay, W.S. The mechanism of resistance to favipiravir in influenza. Proc. Natl. Acad. Sci. USA 2018, 115, 11613–11618. [Google Scholar] [CrossRef]
- Te Velthuis, A.J.W.; Fodor, E. Influenza virus RNA polymerase: Insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016, 14, 479–493. [Google Scholar] [CrossRef]
- Bai, C.-Q.; Mu, J.-S.; Kargbo, D.; Song, Y.-B.; Niu, W.-K.; Nie, W.-M.; Kanu, A.; Liu, W.-W.; Wang, Y.-P.; Dafae, F.; et al. Clinical and Virological Characteristics of Ebola Virus Disease Patients Treated with Favipiravir (T-705)-Sierra Leone, 2014. Clin. Infect. Dis. 2016, 63, 1288–1294. [Google Scholar] [CrossRef] [PubMed]
- Raabe, V.N.; Kann, G.; Ribner, B.S.; Morales, A.; Varkey, J.B.; Mehta, A.K.; Lyon, G.M.; Vanairsdale, S.; Faber, K.; Becker, S.; et al. Favipiravir and Ribavirin Treatment of Epidemiologically Linked Cases of Lassa Fever. Clin. Infect. Dis. 2017, 65, 855–859. [Google Scholar] [CrossRef] [PubMed]
- Arias, A.; Thorne, L.; Goodfellow, I. Favipiravir elicits antiviral mutagenesis during virus replication in vivo. eLife 2014, 3, e03679. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2014, 47, 5.6.1–5.6.32. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Raborn, R.T.; Brendel, V.P. Using RAMPAGE to Identify and Annotate Promoters in Insect Genomes. Methods Mol. Biol. 2019, 1858, 99–116. [Google Scholar]
- Kimura, H.; Kurusu, H.; Sada, M.; Kurai, D.; Murakami, K.; Kamitani, W.; Tomita, H.; Katayama, K.; Ryo, A. Molecular pharmacology of ciclesonide against SARS-CoV-2. J. Allergy Clin. Immunol. 2020, 146, 330–331. [Google Scholar] [CrossRef]
- Smith, T.J. MolView: A program for analyzing and displaying atomic structures on the Macintosh personal computer. J. Mol. Graph. 1995, 13, 122–125. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wu, A.; Peng, Y.; Wang, J.; Guo, Y.; Chen, Z.; Zhang, H.; Wang, Y.; Dong, J.; Wang, L.; et al. Integrating computational modeling and functional assays to decipher the structure-function relationship of influenza virus PB1 protein. Sci. Rep. 2014, 4, 7192. [Google Scholar] [CrossRef] [PubMed]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Furuta, Y.; Takahashi, K.; Fukuda, Y.; Kuno, M.; Kamiyama, T.; Kozaki, K.; Nomura, N.; Egawa, H.; Minami, S.; Watanabe, Y.; et al. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob. Agents Chemother. 2002, 46, 977–981. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Y.; Huang, J.; Yin, P.; Cheng, Z.; Wu, J.; Chen, S.; Zhang, Y.; Chen, B.; Lu, M.; et al. Favipiravir versus Arbidol for COVID-19: A Randomized Clinical Trial. medRxiv 2020. [Google Scholar] [CrossRef]
- Cai, Q.; Yang, M.; Liu, D.; Chen, J.; Shu, D.; Xia, J.; Liao, X.; Gu, Y.; Yang, Y.; Shen, C.; et al. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering 2020. [Google Scholar] [CrossRef] [PubMed]
- Shannon, A.; Selisko, B.; Le, N.; Huchting, J.; Touret, F.; Piorkowski, G.; Fattorini, V.; Ferron, F.; Decroly, E.; Meier, C.; et al. Favipiravir strikes the SARS-CoV-2 at its Achilles heel, the RNA polymerase. BioRxiv 2020. [Google Scholar] [CrossRef]
- Shiraki, K.; Daikoku, T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol. Ther. 2020, 209, 107512. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.U.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Smither, S.J.; Eastaugh, L.S.; Steward, J.A.; Nelson, M.; Lenk, R.P.; Lever, M.S. Post-exposure efficacy of oral T-705 (Favipiravir) against inhalational Ebola virus infection in a mouse model. Antivir. Res. 2014, 104, 153–155. [Google Scholar] [CrossRef] [PubMed]
- Oestereich, L.; Lüdtke, A.; Wurr, S.; Rieger, T.; Muñoz Fontela, C.; Günther, S. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antivir. Res. 2014, 105, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Eloy, P.; Solas, C.; Touret, F.; Mentré, F.; Malvy, D.; De Lamballerie, X.; Guedj, J. Dose Rationale for Favipiravir Use in Patients Infected with SARS-CoV-2. Clin. Pharmacol. Ther. 2020, 108, 188. [Google Scholar] [CrossRef]
- Hagar, M.; Ahmed, H.A.; Aljohani, G.; Alhaddad, O.A. Investigation of Some Antiviral N-Heterocycles as COVID 19 Drug: Molecular Docking and DFT Calculations. Int. J. Mol. Sci. 2020, 21, 3922. [Google Scholar] [CrossRef]
- Harismah, K.; Mirzaei, M. Favipiravir: Structural Analysis and Activity against COVID-19. Adv. J. Chem. Sect. B 2020, 2, 55–60. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sada, M.; Saraya, T.; Ishii, H.; Okayama, K.; Hayashi, Y.; Tsugawa, T.; Nishina, A.; Murakami, K.; Kuroda, M.; Ryo, A.; et al. Detailed Molecular Interactions of Favipiravir with SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza Virus Polymerases In Silico. Microorganisms 2020, 8, 1610. https://doi.org/10.3390/microorganisms8101610
Sada M, Saraya T, Ishii H, Okayama K, Hayashi Y, Tsugawa T, Nishina A, Murakami K, Kuroda M, Ryo A, et al. Detailed Molecular Interactions of Favipiravir with SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza Virus Polymerases In Silico. Microorganisms. 2020; 8(10):1610. https://doi.org/10.3390/microorganisms8101610
Chicago/Turabian StyleSada, Mitsuru, Takeshi Saraya, Haruyuki Ishii, Kaori Okayama, Yuriko Hayashi, Takeshi Tsugawa, Atsuyoshi Nishina, Koichi Murakami, Makoto Kuroda, Akihide Ryo, and et al. 2020. "Detailed Molecular Interactions of Favipiravir with SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza Virus Polymerases In Silico" Microorganisms 8, no. 10: 1610. https://doi.org/10.3390/microorganisms8101610
APA StyleSada, M., Saraya, T., Ishii, H., Okayama, K., Hayashi, Y., Tsugawa, T., Nishina, A., Murakami, K., Kuroda, M., Ryo, A., & Kimura, H. (2020). Detailed Molecular Interactions of Favipiravir with SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza Virus Polymerases In Silico. Microorganisms, 8(10), 1610. https://doi.org/10.3390/microorganisms8101610






