Development of Monoclonal Antibodies and Antigen-Capture ELISA for Human Parechovirus Type 3
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of Expression Vector
2.2. Cell-Free Protein Synthesis
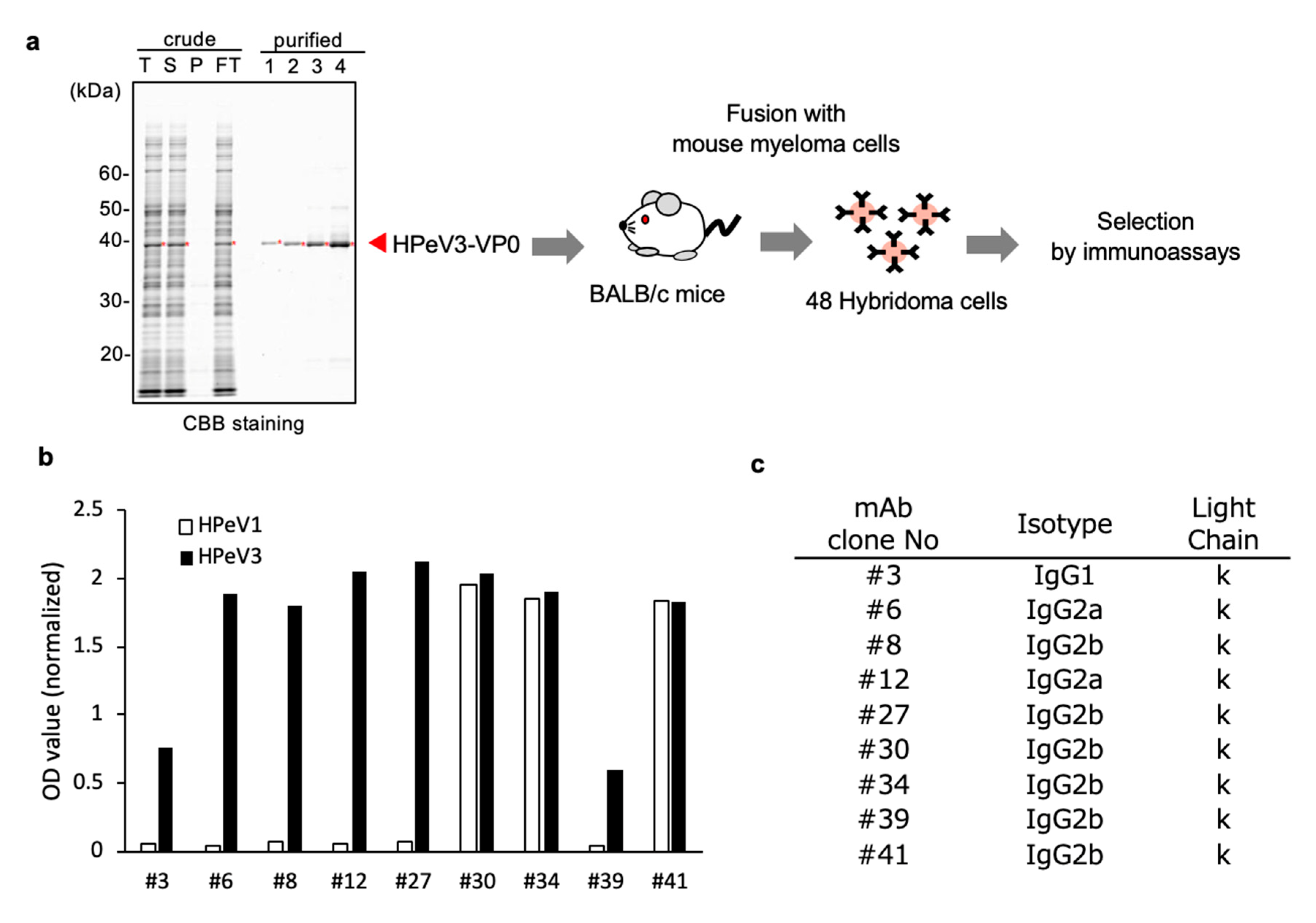
2.3. Monoclonal Antibody Production
2.4. Cell and Virus Culture
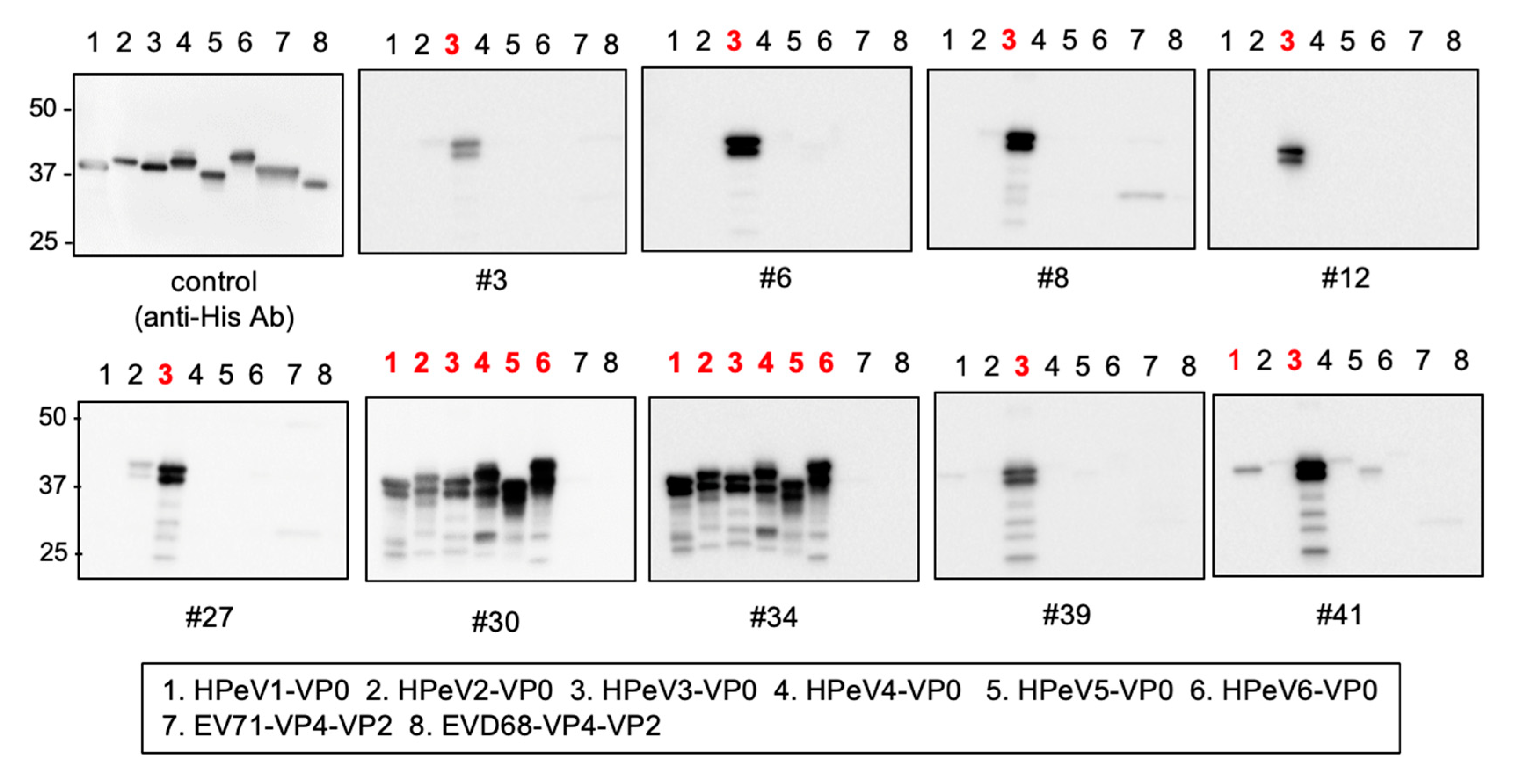
2.5. Immunoblotting
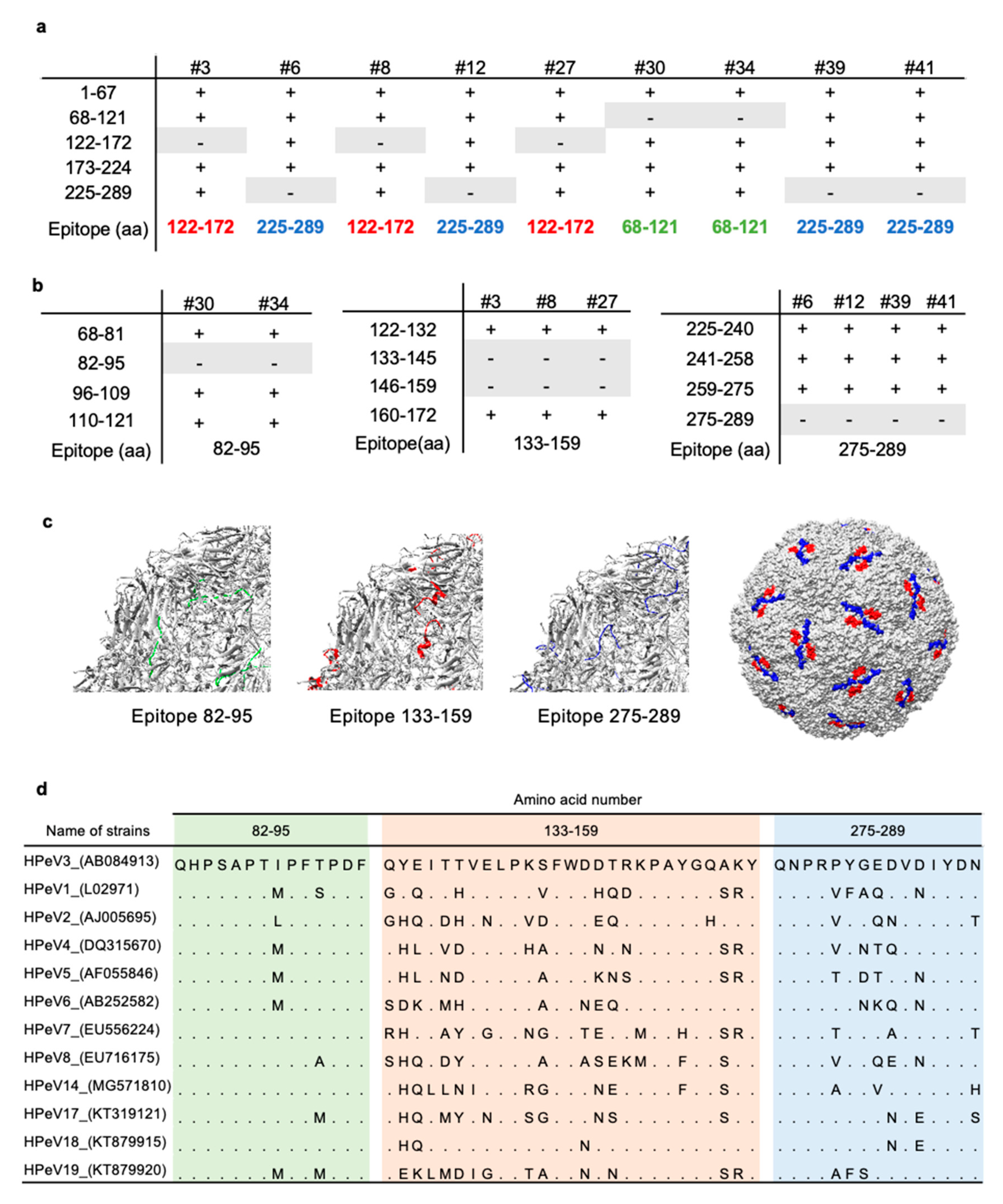
2.6. Epitope Mapping
2.7. Multiple Sequence Alignment
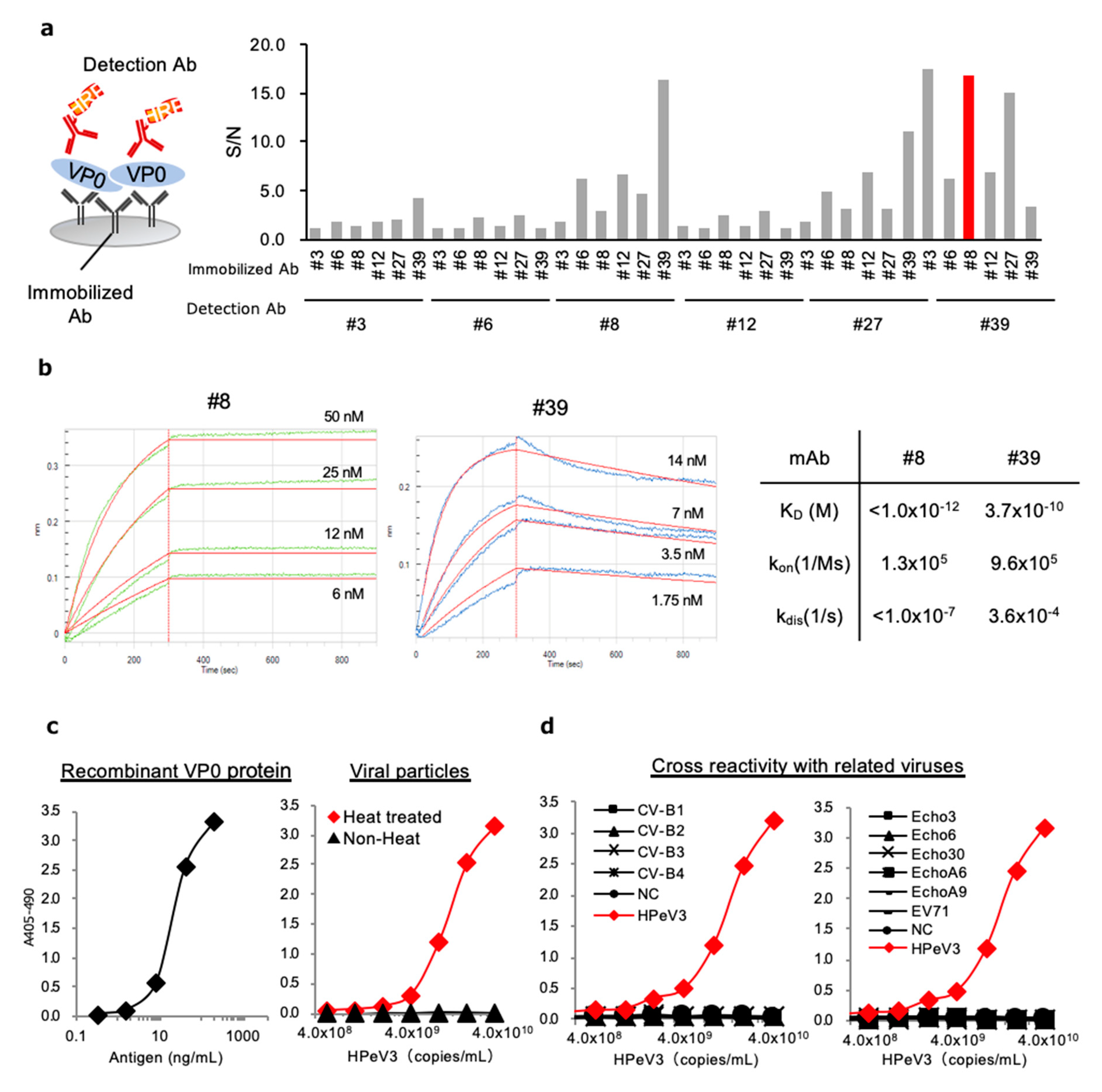
2.8. Affinity Measurement of mAbs
2.9. Selection of the Optimal Pair of mAbs for Sandwich ELISA
3. Results
3.1. Generation of mAbs to Target HPeV3-VP0 Protein
3.2. Epitope Mapping of Anti-HPeV3-VP0 mAb
3.3. Cross-Reactivity of Anti-HPeV3-VP0 mAbs
3.4. Development of Antigen-Capture ELISA for HPeV3 VP0
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Olijve, L.; Jennings, L.; Walls, T. Human Parechovirus: An Increasingly Recognized Cause of Sepsis-Like Illness in Young Infants. Clin. Microbiol. Rev. 2018, 31. [Google Scholar] [CrossRef] [PubMed]
- Chuchaona, W.; Khamrin, P.; Yodmeeklin, A.; Saikruang, W.; Kongsricharoern, T.; Ukarapol, N.; Okitsu, S.; Hayakawa, S.; Ushijima, H.; Maneekarn, N. Detection and characterization of a novel human parechovirus genotype in Thailand. Infect. Genet. Evol. 2015, 31, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Yamashita, T.; Tsuzuki, H.; Takeda, N.; Sakae, K. Isolation and identification of a novel human parechovirus. J. Gen. Virol. 2004, 85. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Garcia, M.D.; Simon-Loriere, E.; Kebe, O.; Sakuntabhai, A.; Ndiaye, K. Identification and molecular characterization of the first complete genome sequence of Human Parechovirus type 15. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- de Crom, S.C.; Rossen, J.W.; van Furth, A.M.; Obihara, C.C. Enterovirus and parechovirus infection in children: A brief overview. Eur. J. Pediatr. 2016, 175, 1023–1029. [Google Scholar] [CrossRef]
- Khatami, A.; McMullan, B.J.; Webber, M.; Stewart, P.; Francis, S.; Timmers, K.J.; Rodas, E.; Druce, J.; Mehta, B.; Sloggett, N.A.; et al. Sepsis-like disease in infants due to human parechovirus type 3 during an outbreak in Australia. Clin. Infect. Dis. 2015, 60, 228–236. [Google Scholar] [CrossRef]
- Karelehto, E.; van der Sanden, S.; Geraets, J.A.; Domanska, A.; van der Linden, L.; Hoogendoorn, D.; Koen, G.; van Eijk, H.; Shakeel, S.; Beaumont, T.; et al. Strain-dependent neutralization reveals antigenic variation of human parechovirus 3. Sci. Rep. 2017, 7, 12075. [Google Scholar] [CrossRef]
- Selvarangan, R.; Nzabi, M.; Selvaraju, S.B.; Ketter, P.; Carpenter, C.; Harrison, C.J. Human parechovirus 3 causing sepsis-like illness in children from midwestern United States. Pediatr. Infect. Dis. J. 2011, 30, 238–242. [Google Scholar] [CrossRef]
- Boivin, G.; Abed, Y.; Boucher, F.D. Human parechovirus 3 and neonatal infections. Emerg. Infect. Dis. 2005, 11, 103–105. [Google Scholar] [CrossRef]
- Levorson, R.E.; Jantausch, B.A.; Wiedermann, B.L.; Spiegel, H.M.; Campos, J.M. Human parechovirus-3 infection: Emerging pathogen in neonatal sepsis. Pediatr. Infect. Dis. J. 2009, 28, 545–547. [Google Scholar] [CrossRef]
- Sano, K.; Hamada, H.; Hirose, S.; Sugiura, K.; Harada, S.; Koizumi, M.; Hara, M.; Nishijima, H.; Taira, M.; Ogura, A.; et al. Prevalence and characteristics of human parechovirus and enterovirus infection in febrile infants. Pediatr. Int. 2018, 60, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Harvala, H.; Calvert, J.; Van Nguyen, D.; Clasper, L.; Gadsby, N.; Molyneaux, P.; Templeton, K.; McWilliams Leitch, C.; Simmonds, P. Comparison of diagnostic clinical samples and environmental sampling for enterovirus and parechovirus surveillance in Scotland, 2010 to 2012. Euro. Surveill. 2014, 19. [Google Scholar] [CrossRef] [PubMed]
- Britton, P.N.; Dale, R.C.; Nissen, M.D.; Crawford, N.; Elliott, E.; Macartney, K.; Khandaker, G.; Booy, R.; Jones, C.A.; Investigators, P.-A. Parechovirus Encephalitis and Neurodevelopmental Outcomes. Pediatrics 2016, 137, e20152848. [Google Scholar] [CrossRef] [PubMed]
- Sedmak, G.; Nix, W.A.; Jentzen, J.; Haupt, T.E.; Davis, J.P.; Bhattacharyya, S.; Pallansch, M.A.; Oberste, M.S. Infant deaths associated with human parechovirus infection in Wisconsin. Clin. Infect. Dis. 2010, 50, 357–361. [Google Scholar] [CrossRef]
- Verboon-Maciolek, M.A.; Groenendaal, F.; Hahn, C.D.; Hellmann, J.; van Loon, A.M.; Boivin, G.; de Vries, L.S. Human parechovirus causes encephalitis with white matter injury in neonates. Ann. Neurol. 2008, 64, 266–273. [Google Scholar] [CrossRef]
- Vergnano, S.; Kadambari, S.; Whalley, K.; Menson, E.N.; Martinez-Alier, N.; Cooper, M.; Sanchez, E.; Heath, P.T.; Lyall, H. Characteristics and outcomes of human parechovirus infection in infants (2008–2012). Eur. J. Pediatr. 2015, 174, 919–924. [Google Scholar] [CrossRef]
- Aizawa, Y.; Izumita, R.; Saitoh, A. Human parechovirus type 3 infection: An emerging infection in neonates and young infants. J. Infect. Chemother. 2017, 23, 419–426. [Google Scholar] [CrossRef]
- Mizuta, K.; Yamakawa, T.; Kurokawa, K.; Chikaoka, S.; Shimizu, Y.; Itagaki, T.; Katsushima, F.; Katsushima, Y.; Ito, S.; Aoki, Y.; et al. Epidemic myalgia and myositis associated with human parechovirus type 3 infections occur not only in adults but also in children: Findings in Yamagata, Japan, 2014. Epidemiol. Infect. 2016, 144. [Google Scholar] [CrossRef]
- Miyazaki, M.; Hara, K.; Takayoshi, T.; Kawase, T.; Nakagawa, Y.; Arai, T.; Sugimoto, T.; Nishiyama, K.; Gonzalez, G.; Hanaoka, N.; et al. Epidemic Myalgia Associated with Human Parechovirus Type 3 Infection. Intern. Med. 2020, 59. [Google Scholar] [CrossRef]
- Romero, J.R.; Selvarangan, R. The human Parechoviruses: An overview. Adv. Pediatr. 2011, 58, 65–85. [Google Scholar] [CrossRef]
- Mizuta, K.; Yamakawa, T.; Nagasawa, H.; Itagaki, T.; Katsushima, F.; Katsushima, Y.; Shimizu, Y.; Ito, S.; Aoki, Y.; Ikeda, T.; et al. Epidemic myalgia associated with human parechovirus type 3 infection among adults occurs during an outbreak among children: Findings from Yamagata, Japan, in 2011. J. Clin. Virol. 2013, 58, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Saito, K.; Hara, Y.; Aoyagi, T.; Kitakawa, K.; Abe, Y.; Takemura, H.; Ikeda, F.; Kaku, M.; Kanemitsu, K. Severe epidemic myalgia with an elevated level of serum interleukin-6 caused by human parechovirus type 3: A case report and brief review of the literature. BMC Infect. Dis. 2018, 18, 381. [Google Scholar] [CrossRef]
- van der Sanden, S.M.; Koopmans, M.P.; van der Avoort, H.G. Detection of human enteroviruses and parechoviruses as part of the national enterovirus surveillance in the Netherlands, 1996-2011. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1525–1531. [Google Scholar] [CrossRef]
- Domanska, A.; Flatt, J.W.; Jukonen, J.J.J.; Geraets, J.A.; Butcher, S.J. A 2.8-Angstrom-Resolution Cryo-Electron Microscopy Structure of Human Parechovirus 3 in Complex with Fab from a Neutralizing Antibody. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, S.; Westerhuis, B.M.; Domanska, A.; Koning, R.I.; Matadeen, R.; Koster, A.J.; Bakker, A.Q.; Beaumont, T.; Wolthers, K.C.; Butcher, S.J. Multiple capsid-stabilizing interactions revealed in a high-resolution structure of an emerging picornavirus causing neonatal sepsis. Nat. Commun. 2016, 7, 11387. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, S.; Dykeman, E.C.; White, S.J.; Ora, A.; Cockburn, J.J.B.; Butcher, S.J.; Stockley, P.G.; Twarock, R. Genomic RNA folding mediates assembly of human parechovirus. Nat. Commun. 2017, 8, 5. [Google Scholar] [CrossRef]
- Chen, B.C.; Chang, J.T.; Huang, T.S.; Chen, J.J.; Chen, Y.S.; Jan, M.W.; Chang, T.H. Parechovirus A Detection by a Comprehensive Approach in a Clinical Laboratory. Viruses 2018, 10, 711. [Google Scholar] [CrossRef]
- Abed, Y.; Wolf, D.; Dagan, R.; Boivin, G. Development of a serological assay based on a synthetic peptide selected from the VP0 capsid protein for detection of human parechoviruses. J. Clin. Microbiol. 2007, 45. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Matsuyama, S.; Fukushi, S.; Matsunaga, S.; Matsushima, Y.; Kuroyama, H.; Kimura, H.; Takeda, M.; Chimuro, T.; Ryo, A. Development of Monoclonal Antibody and Diagnostic Test for Middle East Respiratory Syndrome Coronavirus Using Cell-Free Synthesized Nucleocapsid Antigen. Front. Microbiol. 2016, 7, 509. [Google Scholar] [CrossRef]
- Khatun, H.; Yamaoka, Y.; Matsushima, Y.; Matsunaga, S.; Kimura, H.; Ho, J.; Shuda, M.; Ryo, A. Production and characterization of monoclonal antibodies specific for major capsid VP1 protein of trichodysplasia spinulosa-associated polyomavirus. Microbiol. Immunol. 2018, 62, 763–773. [Google Scholar] [CrossRef]
- Wildenbeest, J.G.; Benschop, K.S.; Minnaar, R.P.; Bouma-de Jongh, S.; Wolthers, K.C.; Pajkrt, D. Clinical relevance of positive human parechovirus type 1 and 3 PCR in stool samples. Clin. Microbiol. Infect. 2014, 20, O640–O647. [Google Scholar] [CrossRef] [PubMed]
- Kadambari, S.; Harvala, H.; Simmonds, P.; Pollard, A.J.; Sadarangani, M. Strategies to improve detection and management of human parechovirus infection in young infants. Lancet Infect. Dis. 2019, 19, e51–e58. [Google Scholar] [CrossRef]
- Matsunaga, S.; Kawakami, S.; Matsuo, I.; Okayama, A.; Tsukagoshi, H.; Kudoh, A.; Matsushima, Y.; Shimizu, H.; Okabe, N.; Hirano, H.; et al. Wheat germ cell-free system-based production of hemagglutinin-neuraminidase glycoprotein of human parainfluenza virus type 3 for generation and characterization of monoclonal antibody. Front. Microbiol. 2014, 5, 208. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.N.; Reynolds, G.M.; Waldron, E.E.; Ward, E.; Giannopoulos, K.; Murray, P.G. Monoclonal antibodies. Mol. Pathol. 2000, 53, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Sirivichayakul, C.; Limkittikul, K.; Chanthavanich, P.; Yoksan, S.; Ratchatatat, A.; Lim, J.K.; Arunsodsai, W.; Sabchareon, A. Monoclonal antibody-based capture ELISA in the diagnosis of previous dengue infection. Virol. J. 2019, 16, 125. [Google Scholar] [CrossRef] [PubMed]
- Pikora, C.; Wittish, C.; Desrosiers, R.C. Identification of two N-linked glycosylation sites within the core of the simian immunodeficiency virus glycoprotein whose removal enhances sensitivity to soluble CD4. J. Virol. 2005, 79, 12575–12583. [Google Scholar] [CrossRef][Green Version]
- Stove, V.; Ramos, P.A.; Wallemacq, P.; Vogeser, M.; Schuetzenmeister, A.; Schmiedel, C.; Shipkova, M. Measurement of sirolimus concentrations in human blood using an automated electrochemiluminescence immunoassay (ECLIA): A multicenter evaluation. Clin. Chem. Lab. Med. 2018, 56, 764–775. [Google Scholar] [CrossRef]
- Wada, A.; Sakoda, Y.; Oyamada, T.; Kida, H. Development of a highly sensitive immunochromatographic detection kit for H5 influenza virus hemagglutinin using silver amplification. J. Virol. Methods 2011, 178, 82–86. [Google Scholar] [CrossRef]
- Mishra, M.; Tiwari, S.; Gunaseelan, A.; Li, D.; Hammock, B.D.; Gomes, A.V. Improving the sensitivity of traditional Western blotting via Streptavidin containing Poly-horseradish peroxidase (PolyHRP). Electrophoresis 2019, 40, 1731–1739. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goto, K.; Yamaoka, Y.; Khatun, H.; Miyakawa, K.; Nishi, M.; Nagata, N.; Yanaoka, T.; Kimura, H.; Ryo, A. Development of Monoclonal Antibodies and Antigen-Capture ELISA for Human Parechovirus Type 3. Microorganisms 2020, 8, 1437. https://doi.org/10.3390/microorganisms8091437
Goto K, Yamaoka Y, Khatun H, Miyakawa K, Nishi M, Nagata N, Yanaoka T, Kimura H, Ryo A. Development of Monoclonal Antibodies and Antigen-Capture ELISA for Human Parechovirus Type 3. Microorganisms. 2020; 8(9):1437. https://doi.org/10.3390/microorganisms8091437
Chicago/Turabian StyleGoto, Keiko, Yutaro Yamaoka, Hajera Khatun, Kei Miyakawa, Mayuko Nishi, Noriko Nagata, Toshikazu Yanaoka, Hirokazu Kimura, and Akihide Ryo. 2020. "Development of Monoclonal Antibodies and Antigen-Capture ELISA for Human Parechovirus Type 3" Microorganisms 8, no. 9: 1437. https://doi.org/10.3390/microorganisms8091437
APA StyleGoto, K., Yamaoka, Y., Khatun, H., Miyakawa, K., Nishi, M., Nagata, N., Yanaoka, T., Kimura, H., & Ryo, A. (2020). Development of Monoclonal Antibodies and Antigen-Capture ELISA for Human Parechovirus Type 3. Microorganisms, 8(9), 1437. https://doi.org/10.3390/microorganisms8091437




