Prevotella in Pigs: The Positive and Negative Associations with Production and Health
Abstract
:1. Introduction
2. Microbial Ecology along the Pig Gastrointestinal Tract
2.1. Microbial Composition and Diversity
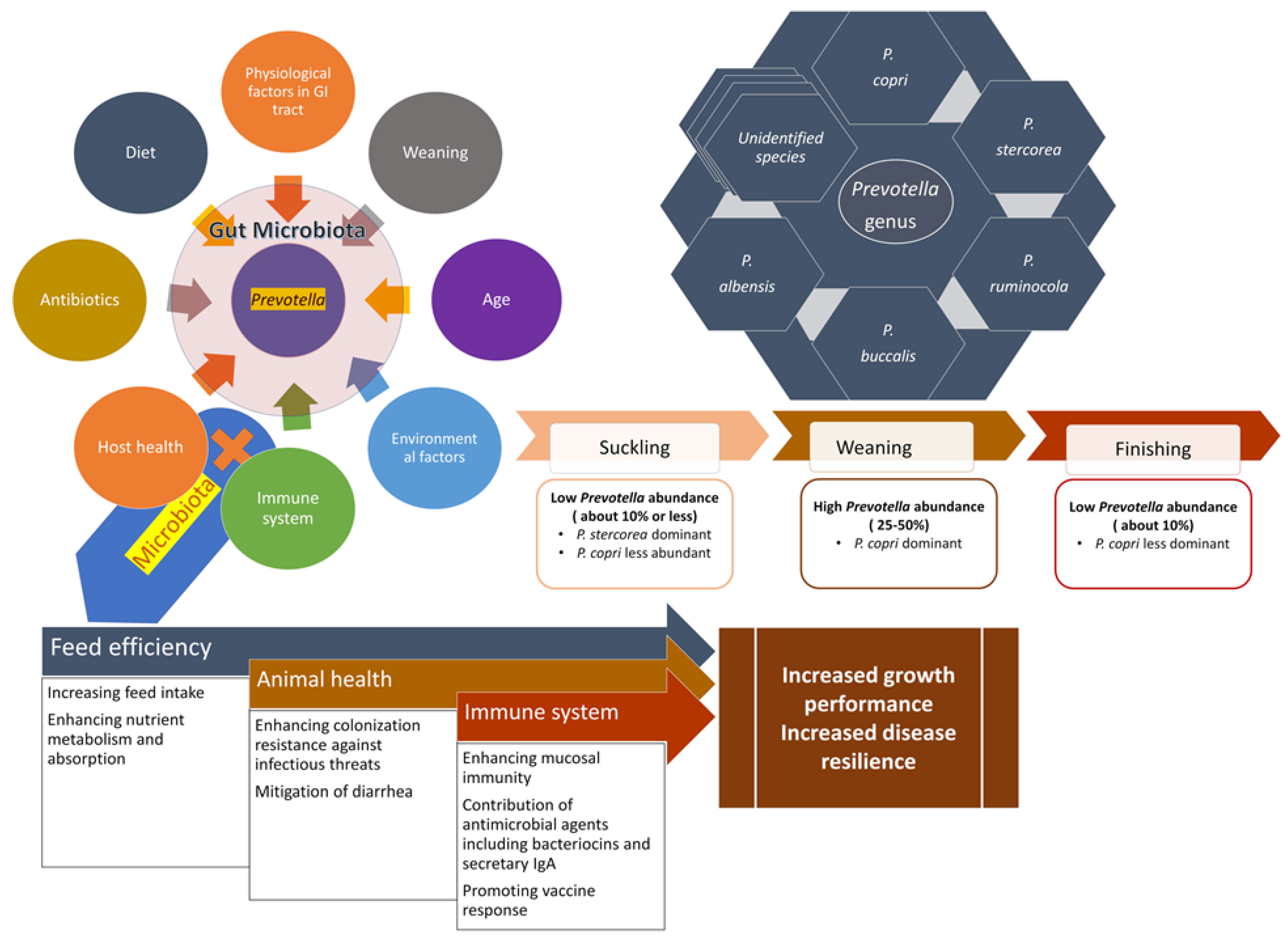
2.2. Factors Shaping the Bacterial Microbiota in the Pig Gastrointestinal Tract
3. Prevotella within the Gut Microbial Community of Pigs
3.1. Taxonomy and a Brief History of the Genus Prevotella
3.2. Presence and Abundance of Prevotella along the Pig Gastrointestinal Tract
3.3. Different Prevotella Species Present in the Pig Gastrointestinal Tract
4. Prevotella in Feed Efficiency and Growth Performance
5. Prevotella and Diarrhea in Pigs
6. Prevotella and the Intestinal Immune System
7. Prevotella and Vaccine Response
8. Prevotella and Bacterial Interactions
9. Challenges and Future Opportunities Associated with Harnessing Prevotella to Improve Pig Health
9.1. Challenges
9.2. Future Opportunities
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Patil, Y.; Gooneratne, R.; Ju, X.H. Interactions between host and gut microbiota in domestic pigs: A review. Gut Microbes 2020, 11, 310–334. [Google Scholar] [CrossRef] [PubMed]
- McCormack, U.M.; Curião, T.; Buzoianu, S.G.; Prieto, M.L.; Ryan, T.; Varley, P.; Crispie, F.; Magowan, E.; Metzler-Zebeli, B.U.; Berry, D.; et al. Exploring a Possible Link between the Intestinal Microbiota and Feed Efficiency in Pigs. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouhse, J.; Zijlstra, R.; Willing, P. The role of gut microbiota in the health and disease of pigs. Anim. Front. 2016, 6, 30–36. [Google Scholar] [CrossRef] [Green Version]
- Willing, B.P.; Russell, S.L.; Finlay, B.B. Shifting the balance: Antibiotic effects on host-microbiota mutualism. Nat. Rev. Microbiol. 2011, 9, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, A.; Mazmanian, S.K. Disruption of the gut microbiome as a risk factor for microbial infections. Curr. Opin. Microbiol. 2013, 16, 221–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- Fouhse, J.M.; Yang, K.; More-Bayona, J.; Gao, Y.; Goruk, S.; Plastow, G.; Field, C.J.; Barreda, D.R.; Willing, B.P. Neonatal Exposure to Amoxicillin Alters Long-Term Immune Response Despite Transient Effects on Gut-Microbiota in Piglets. Front. Immunol. 2019, 10, 2059. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Yang, K.; Ju, T.; Ho, T.; McKay, C.A.; Gao, Y.; Forget, S.K.; Gartner, S.R.; Field, C.J.; Chan, C.B.; et al. Early life antibiotic exposure affects pancreatic islet development and metabolic regulation. Sci. Rep. 2017, 7, 41778. [Google Scholar] [CrossRef] [Green Version]
- Schachtschneider, K.M.; Yeoman, C.J.; Isaacson, R.E.; White, B.A.; Schook, L.B.; Pieters, M. Modulation of systemic immune responses through commensal gastrointestinal microbiota. PLoS ONE 2013, 8, e53969. [Google Scholar] [CrossRef]
- Dou, S.; Gadonna-Widehem, P.; Rome, V.; Hamoudi, D.; Rhazi, L.; Lakhal, L.; Larcher, T.; Bahi-Jaber, N.; Pinon-Quintana, A.; Guyonvarch, A.; et al. Characterisation of Early-Life Fecal Microbiota in Susceptible and Healthy Pigs to Post-Weaning Diarrhoea. PLoS ONE 2017, 12, e0169851. [Google Scholar] [CrossRef]
- Canibe, N.; O’Dea, M.; Abraham, S. Potential relevance of pig gut content transplantation for production and research. J. Anim. Sci. Biotechnol. 2019, 10, 55. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tsai, T.; Deng, F.; Wei, X.; Chai, J.; Knapp, J.; Apple, J.; Maxwell, C.V.; Lee, J.A.; Li, Y.; et al. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome 2019, 7, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bin, P.; Tang, Z.; Liu, S.; Chen, S.; Xia, Y.; Liu, J.; Wu, H.; Zhu, G. Intestinal microbiota mediates Enterotoxigenic Escherichia coli-induced diarrhea in piglets. BMC Vet. Res. 2018, 14, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holman, D.B.; Brunelle, B.W.; Trachsel, J.; Allen, H.K. Meta-analysis To Define a Core Microbiota in the Swine Gut. mSystems 2017, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Looft, T.; Allen, H.K.; Cantarel, B.L.; Levine, U.Y.; Bayles, D.O.; Alt, D.P.; Henrissat, B.; Stanton, T.B. Bacteria, phages and pigs: The effects of in-feed antibiotics on the microbiome at different gut locations. ISME J. 2014, 8, 1566–1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, S.; Fang, S.; He, M.; Huang, X.; Yang, H.; Yang, B.; Chen, C.; Huang, L. Age-based dynamic changes of phylogenetic composition and interaction networks of health pig gut microbiome feeding in a uniformed condition. BMC Vet. Res. 2019, 15, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramayo-Caldas, Y.; Mach, N.; Lepage, P.; Levenez, F.; Denis, C.; Lemonnier, G.; Leplat, J.J.; Billon, Y.; Berri, M.; Doré, J.; et al. Phylogenetic network analysis applied to pig gut microbiota identifies an ecosystem structure linked with growth traits. ISME J. 2016, 10, 2973–2977. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Yang, M.; Fang, S.; Huang, X.; He, M.; Ke, S.; Gao, J.; Wu, J.; Zhou, Y.; Fu, H.; et al. Evaluating the profound effect of gut microbiome on host appetite in pigs. BMC Microbiol. 2018, 18, 215. [Google Scholar] [CrossRef] [Green Version]
- Mach, N.; Berri, M.; Estelle, J.; Levenez, F.; Lemonnier, G.; Denis, C.; Leplat, J.J.; Chevaleyre, C.; Billon, Y.; Dore, J.; et al. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 2015, 7, 554–569. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Precup, G.; Vodnar, D.C. Gut. Br. J. Nutr. 2019, 122, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Amaral, W.Z.; Lubach, G.R.; Proctor, A.; Lyte, M.; Phillips, G.J.; Coe, C.L. Social Influences on Prevotella and the Gut Microbiome of Young Monkeys. Psychosom. Med. 2017, 79, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lang, T.; Shen, J.; Dai, J.; Tian, L.; Wang, X. Core Gut Bacteria Analysis of Healthy Mice. Front. Microbiol. 2019, 10, 887. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Chai, J.; Diao, Q.; Huang, W.; Zhuang, Y.; Zhang, N. The Signature Microbiota Drive Rumen Function Shifts in Goat Kids Introduced to Solid Diet Regimes. Microorganisms 2019, 7, 516. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Ni, J.; Li, H. Effect of green tea and mulberry leaf powders on the gut microbiota of chicken. BMC Vet. Res. 2019, 15, 77. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, D.M.; Weimer, P.J. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 2007, 75, 165–174. [Google Scholar] [CrossRef]
- Kim, J.N.; Méndez-García, C.; Geier, R.R.; Iakiviak, M.; Chang, J.; Cann, I.; Mackie, R.I. Metabolic networks for nitrogen utilization in Prevotella ruminicola 23. Sci. Rep. 2017, 7, 7851. [Google Scholar] [CrossRef]
- Wirth, R.; Kádár, G.; Kakuk, B.; Maróti, G.; Bagi, Z.; Szilágyi, Á.; Rákhely, G.; Horváth, J.; Kovács, K.L. The Planktonic Core Microbiome and Core Functions in the Cattle Rumen by Next Generation Sequencing. Front. Microbiol. 2018, 9, 2285. [Google Scholar] [CrossRef] [Green Version]
- Gorvitovskaia, A.; Holmes, S.P.; Huse, S.M. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome 2016, 4, 15. [Google Scholar] [CrossRef] [Green Version]
- Larsen, J.M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef] [Green Version]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019, 4, 623–632. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Pasolli, E.; Tett, A.; Tarallo, S.; Naccarati, A.; De Angelis, M.; Neviani, E.; Cocolin, L.; Gobbetti, M.; Segata, N.; et al. Distinct Genetic and Functional Traits of Human Intestinal Prevotella copri Strains Are Associated with Different Habitual Diets. Cell Host Microbe 2019, 25, 444–453. [Google Scholar] [CrossRef] [Green Version]
- de Aquino, S.G.; Abdollahi-Roodsaz, S.; Koenders, M.I.; van de Loo, F.A.; Pruijn, G.J.; Marijnissen, R.J.; Walgreen, B.; Helsen, M.M.; van den Bersselaar, L.A.; de Molon, R.S.; et al. Periodontal pathogens directly promote autoimmune experimental arthritis by inducing a TLR2- and IL-1-driven Th17 response. J. Immunol. 2014, 192, 4103–4111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagaoka, K.; Yanagihara, K.; Morinaga, Y.; Nakamura, S.; Harada, T.; Hasegawa, H.; Izumikawa, K.; Ishimatsu, Y.; Kakeya, H.; Nishimura, M.; et al. Prevotella intermedia induces severe bacteremic pneumococcal pneumonia in mice with upregulated platelet-activating factor receptor expression. Infect. Immun. 2014, 82, 587–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.; Zhou, B.; Xia, X.; Chen, S.; Deng, Y.; Wang, Y.; Wu, L.; Tian, Y.; Zhao, B.; Xu, H.; et al. Prevotella copri is associated with carboplatin-induced gut toxicity. Cell Death Dis. 2019, 10, 714. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Donovan, S.M. Human microbiota-associated swine: Current progress and future opportunities. ILAR J. 2015, 56, 63–73. [Google Scholar] [CrossRef]
- Walters, E.M.; Wells, K.D.; Bryda, E.C.; Schommer, S.; Prather, R.S. Swine models, genomic tools and services to enhance our understanding of human health and diseases. Lab Anim. 2017, 46, 167–172. [Google Scholar] [CrossRef]
- Kinder, H.A.; Baker, E.W.; West, F.D. The pig as a preclinical traumatic brain injury model: Current models, functional outcome measures, and translational detection strategies. Neural Regen. Res. 2019, 14, 413–424. [Google Scholar] [CrossRef]
- Hillman, E.T.; Lu, H.; Yao, T.; Nakatsu, C.H. Microbial Ecology along the Gastrointestinal Tract. Microbes Environ. 2017, 32, 300–313. [Google Scholar] [CrossRef] [Green Version]
- Richards, J.; Gong, J.; de Lange, C. The gastrointestinal microbiota and its role in monogastric nutrition and health with an emphasis on pigs: Current understanding, possible modulations, and new technologies for ecological studies. Can. J. Anim. Sci. 2005, 85, 421–435. [Google Scholar] [CrossRef]
- Isaacson, R.; Kim, H.B. The intestinal microbiome of the pig. Anim. Health Res. Rev. 2012, 13, 100–109. [Google Scholar] [CrossRef]
- Leser, T.D.; Amenuvor, J.Z.; Jensen, T.K.; Lindecrona, R.H.; Boye, M.; Møller, K. Culture-independent analysis of gut bacteria: The pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 2002, 68, 673–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, D.; Tiezzi, F.; Schillebeeckx, C.; McNulty, N.P.; Schwab, C.; Shull, C.; Maltecca, C. Host contributes to longitudinal diversity of fecal microbiota in swine selected for lean growth. Microbiome 2018, 6, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crespo-Piazuelo, D.; Estellé, J.; Revilla, M.; Criado-Mesas, L.; Ramayo-Caldas, Y.; Óvilo, C.; Fernández, A.I.; Ballester, M.; Folch, J.M. Characterization of bacterial microbiota compositions along the intestinal tract in pigs and their interactions and functions. Sci. Rep. 2018, 8, 12727. [Google Scholar] [CrossRef]
- van Winsen, R.L.; Urlings, B.A.; Lipman, L.J.; Snijders, J.M.; Keuzenkamp, D.; Verheijden, J.H.; van Knapen, F. Effect of fermented feed on the microbial population of the gastrointestinal tracts of pigs. Appl. Environ. Microbiol. 2001, 67, 3071–3076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez, E.; Sánchez, B.; Farina, A.; Margolles, A.; Rodríguez, J.M. Characterization of the bile and gall bladder microbiota of healthy pigs. Microbiologyopen 2014, 3, 937–949. [Google Scholar] [CrossRef]
- Begley, M.; Gahan, C.G.; Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, A.F.; Eckmann, L. How bile acids confer gut mucosal protection against bacteria. Proc. Natl. Acad. Sci. USA 2006, 103, 4333–4334. [Google Scholar] [CrossRef] [Green Version]
- Corfield, A.P. The Interaction of the Gut Microbiota with the Mucus Barrier in Health and Disease in Human. Microorganisms 2018, 6, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroeder, B.O. Fight them or feed them: How the intestinal mucus layer manages the gut microbiota. Gastroenterol. Rep. 2019, 7, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Wright, D.P.; Rosendale, D.I.; Robertson, A.M. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol. Lett. 2000, 190, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Mathias, A.; Pais, B.; Favre, L.; Benyacoub, J.; Corthésy, B. Role of secretory IgA in the mucosal sensing of commensal bacteria. Gut Microbes 2014, 5, 688–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catanzaro, J.R.; Strauss, J.D.; Bielecka, A.; Porto, A.F.; Lobo, F.M.; Urban, A.; Schofield, W.B.; Palm, N.W. IgA-deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci. Rep. 2019, 9, 13574. [Google Scholar] [CrossRef] [PubMed]
- Le Sciellour, M.; Renaudeau, D.; Zemb, O. Longitudinal Analysis of the Microbiota Composition and Enterotypes of Pigs from Post-Weaning to Finishing. Microorganisms 2019, 7, 622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zheng, Z.; Yu, L.; Wu, S.; Sun, L.; Xu, Q.; Cai, S.; Qin, N.; Bao, W. Examination of the temporal and spatial dynamics of the gut microbiome in newborn piglets reveals distinct microbial communities in six intestinal segments. Sci. Rep. 2019, 9, 3453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeineldin, M.; Aldridge, B.; Lowe, J. Antimicrobial Effects on Swine Gastrointestinal Microbiota and Their Accompanying Antibiotic Resistome. Front. Microbiol. 2019, 10, 1035. [Google Scholar] [CrossRef] [PubMed]
- Lekagul, A.; Tangcharoensathien, V.; Yeung, S. Patterns of antibiotic use in global pig production: A systematic review. Vet. Anim. Sci. 2019, 7, 100058. [Google Scholar] [CrossRef]
- Megahed, A.; Zeineldin, M.; Evans, K.; Maradiaga, N.; Blair, B.; Aldridge, B.; Lowe, J. Impacts of environmental complexity on respiratory and gut microbiome community structure and diversity in growing pigs. Sci. Rep. 2019, 9, 13773. [Google Scholar] [CrossRef] [Green Version]
- McCormack, U.M.; Curião, T.; Wilkinson, T.; Metzler-Zebeli, B.U.; Reyer, H.; Ryan, T.; Calderon-Diaz, J.A.; Crispie, F.; Cotter, P.D.; Creevey, C.J.; et al. Fecal Microbiota Transplantation in Gestating Sows and Neonatal Offspring Alters Lifetime Intestinal Microbiota and Growth in Offspring. mSystems 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Shah, H.N.; Collins, D.M. Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int. J. Syst. Bacteriol. 1990, 40, 205–208. [Google Scholar] [CrossRef] [Green Version]
- Wybo, I.; Soetens, O.; De Bel, A.; Echahidi, F.; Vancutsem, E.; Vandoorslaer, K.; Piérard, D. Species identification of clinical Prevotella isolates by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2012, 50, 1415–1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alauzet, C.; Marchandin, H.; Lozniewski, A. New insights into Prevotella diversity and medical microbiology. Future Microbiol. 2010, 5, 1695–1718. [Google Scholar] [CrossRef] [PubMed]
- Nograšek, B.; Accetto, T.; Fanedl, L.; Avguštin, G. Description of a novel pectin-degrading bacterial species Prevotella pectinovora sp. nov., based on its phenotypic and genomic traits. J. Microbiol. 2015, 53, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Iljazovic, A.; Roy, U.; Gálvez, E.J.C.; Lesker, T.R.; Zhao, B.; Gronow, A.; Amend, L.; Will, S.E.; Hofmann, J.D.; Pils, M.C.; et al. Perturbation of the gut microbiome by Prevotella spp. enhances host susceptibility to mucosal inflammation. Mucosal Immunol. 2020. [Google Scholar] [CrossRef]
- Avgustin, G.; Wallace, R.J.; Flint, H.J. Phenotypic diversity among ruminal isolates of Prevotella ruminicola: Proposal of Prevotella brevis sp. nov., Prevotella bryantii sp. nov., and Prevotella albensis sp. nov. and redefinition of Prevotella ruminicola. Int. J. Syst. Bacteriol. 1997, 47, 284–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulger Toprak, N.; Alida, C.M.V.; Urban, E.; Wybo, I.; Justesen, U.S.; Jean-Pierre, H.; Morris, T.; Akgul, O.; Kulekci, G.; Soyletir, G.; et al. Performance of mass spectrometric identification of clinical Prevotella species using the VITEK MS system: A prospective multi-center study. Anaerobe 2018, 54, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Kumada, H.; Hamada, N.; Takahashi, Y.; Okamoto, M.; Bakir, M.A.; Benno, Y. Prevotella falsenii sp. nov., a Prevotella intermedia-like organism isolated from monkey dental plaque. Int J. Syst. Evol. Microbiol. 2009, 59, 319–322. [Google Scholar] [CrossRef] [Green Version]
- Downes, J.; Wade, W.G. Prevotella fusca sp. nov. and Prevotella scopos sp. nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 2011, 61, 854–858. [Google Scholar] [CrossRef]
- Berger, P.; Adékambi, T.; Mallet, M.N.; Drancourt, M. Prevotella massiliensis sp. nov. isolated from human blood. Res. Microbiol. 2005, 156, 967–973. [Google Scholar] [CrossRef]
- Downes, J.; Tanner, A.C.R.; Dewhirst, F.E.; Wade, W.G. Prevotella saccharolytica sp. nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 2010, 60, 2458–2461. [Google Scholar] [CrossRef] [Green Version]
- Jousimies-Somer, H.; Summanen, P.R. Recent taxonomic changes and terminology update of clinically significant anaerobic gram-negative bacteria (excluding spirochetes). Clin. Infect. Dis. 2002, 35, S17–S21. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Kurakawa, T.; Umemoto, E.; Motooka, D.; Ito, Y.; Gotoh, K.; Hirota, K.; Matsushita, M.; Furuta, Y.; Narazaki, M.; et al. Dysbiosis Contributes to Arthritis Development via Activation of Autoreactive T Cells in the Intestine. Arthritis Rheumatol. 2016, 68, 2646–2661. [Google Scholar] [CrossRef]
- Mann, E.; Schmitz-Esser, S.; Zebeli, Q.; Wagner, M.; Ritzmann, M.; Metzler-Zebeli, B.U. Mucosa-associated bacterial microbiome of the gastrointestinal tract of weaned pigs and dynamics linked to dietary calcium-phosphorus. PLoS ONE 2014, 9, e86950. [Google Scholar] [CrossRef] [PubMed]
- Guevarra, R.B.; Hong, S.H.; Cho, J.H.; Kim, B.R.; Shin, J.; Lee, J.H.; Kang, B.N.; Kim, Y.H.; Wattanaphansak, S.; Isaacson, R.E.; et al. The dynamics of the piglet gut microbiome during the weaning transition in association with health and nutrition. J. Anim. Sci. Biotechnol. 2018, 9, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, G.G.; Lee, J.Y.; Jin, G.D.; Park, J.; Choi, Y.H.; Chae, B.J.; Kim, E.B.; Choi, Y.J. Evaluating the association between body weight and the intestinal microbiota of weaned piglets via 16S rRNA sequencing. Appl. Microbiol. Biotechnol. 2017, 101, 5903–5911. [Google Scholar] [CrossRef] [PubMed]
- Munyaka, P.M.; Blanc, F.; Estellé, J.; Lemonnier, G.; Leplat, J.J.; Rossignol, M.N.; Jardet, D.; Plastow, G.; Billon, Y.; Willing, B.P.; et al. Discovery of Predictors of Mycoplasma hyopneumoniae Vaccine Response Efficiency in Pigs: 16S rRNA Gene Fecal Microbiota Analysis. Microorganisms 2020, 8, 1151. [Google Scholar] [CrossRef] [PubMed]
- Frese, S.A.; Parker, K.; Calvert, C.C.; Mills, D.A. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome 2015, 3, 28. [Google Scholar] [CrossRef] [Green Version]
- Lamendella, R.; Domingo, J.W.; Ghosh, S.; Martinson, J.; Oerther, D.B. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 2011, 11, 103. [Google Scholar] [CrossRef] [Green Version]
- Tan, Z.; Yang, T.; Wang, Y.; Xing, K.; Zhang, F.; Zhao, X.; Ao, H.; Chen, S.; Liu, J.; Wang, C. Metagenomic Analysis of Cecal Microbiome Identified Microbiota and Functional Capacities Associated with Feed Efficiency in Landrace Finishing Pigs. Front. Microbiol. 2017, 8, 1546. [Google Scholar] [CrossRef] [Green Version]
- Flint, H.J.; Bayer, E.A. Plant cell wall breakdown by anaerobic microorganisms from the Mammalian digestive tract. Ann. N. Y. Acad. Sci. 2008, 1125, 280–288. [Google Scholar] [CrossRef]
- Pollock, J.; Gally, D.L.; Glendinning, L.; Tiwari, R.; Hutchings, M.R.; Houdijk, J.G.M. Analysis of temporal fecal microbiota dynamics in weaner pigs with and without exposure to enterotoxigenic Escherichia coli1,2. J. Anim. Sci. 2018, 96, 3777–3790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Huang, X.; Zhao, S.; Sun, W.; Yan, Z.; Wang, P.; Li, S.; Huang, W.; Zhang, S.; Liu, L.; et al. Structure and Function of the Fecal Microbiota in Diarrheic Neonatal Piglets. Front. Microbiol. 2017, 8, 502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiros, T.G.; Luise, D.; Derakhshani, H.; Petri, R.; Trevisi, P.; D’Inca, R.; Auclair, E.; van Kessel, A.G. Effect of live yeast Saccharomyces cerevisiae supplementation on the performance and cecum microbial profile of suckling piglets. PLoS ONE 2019, 14, e0219557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quan, J.; Cai, G.; Ye, J.; Yang, M.; Ding, R.; Wang, X.; Zheng, E.; Fu, D.; Li, S.; Zhou, S.; et al. A global comparison of the microbiome compositions of three gut locations in commercial pigs with extreme feed conversion ratios. Sci. Rep. 2018, 8, 4536. [Google Scholar] [CrossRef] [Green Version]
- Unno, T.; Choi, J.H.; Hur, H.G.; Sadowsky, M.J.; Ahn, Y.T.; Huh, C.S.; Kim, G.B.; Cha, C.J. Changes in human gut microbiota influenced by probiotic fermented milk ingestion. J. Dairy Sci. 2015, 98, 3568–3576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Du, L.; Li, X.; Zhong, H.; Ding, Y.; Liu, Z.; Ge, L. Identification of the core bacteria in rectums of diarrheic and non-diarrheic piglets. Sci. Rep. 2019, 9, 18675. [Google Scholar] [CrossRef] [PubMed]
- Koh, H.W.; Kim, M.S.; Lee, J.S.; Kim, H.; Park, S.J. Changes in the Swine Gut Microbiota in Response to Porcine Epidemic Diarrhea Infection. Microbes Environ. 2015, 30, 284–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Huang, X.; Wang, P.; Yan, Z.; Sun, W.; Zhao, S.; Gun, S. Longitudinal development of the gut microbiota in healthy and diarrheic piglets induced by age-related dietary changes. Microbiologyopen 2019, 8, e923. [Google Scholar] [CrossRef] [Green Version]
- Shirkey, T.W.; Siggers, R.H.; Goldade, B.G.; Marshall, J.K.; Drew, M.D.; Laarveld, B.; Van Kessel, A.G. Effects of commensal bacteria on intestinal morphology and expression of proinflammatory cytokines in the gnotobiotic pig. Exp. Biol. Med. 2006, 231, 1333–1345. [Google Scholar] [CrossRef]
- Stokes, C.R. The development and role of microbial-host interactions in gut mucosal immune development. J. Anim. Sci. Biotechnol. 2017, 8, 12. [Google Scholar] [CrossRef] [Green Version]
- Duncan, S.H.; Louis, P.; Flint, H.J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 2004, 70, 5810–5817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratajczak, W.; Rył, A.; Mizerski, A.; Walczakiewicz, K.; Sipak, O.; Laszczyńska, M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim. Pol. 2019, 66, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franke, T.; Deppenmeier, U. Physiology and central carbon metabolism of the gut bacterium Prevotella copri. Mol. Microbiol. 2018, 109, 528–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cushing, K.; Alvarado, D.M.; Ciorba, M.A. Butyrate and Mucosal Inflammation: New Scientific Evidence Supports Clinical Observation. Clin. Transl. Gastroenterol. 2015, 6, e108. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Kelly, C.J.; Battista, K.D.; Schaefer, R.; Lanis, J.M.; Alexeev, E.E.; Wang, R.X.; Onyiah, J.C.; Kominsky, D.J.; Colgan, S.P. Microbial-Derived Butyrate Promotes Epithelial Barrier Function through IL-10 Receptor-Dependent Repression of Claudin-2. J. Immunol. 2017, 199, 2976–2984. [Google Scholar] [CrossRef] [Green Version]
- Pabst, O.; Slack, E. IgA and the intestinal microbiota: The importance of being specific. Mucosal Immunol. 2020, 13, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Donaldson, G.P.; Ladinsky, M.S.; Yu, K.B.; Sanders, J.G.; Yoo, B.B.; Chou, W.C.; Conner, M.E.; Earl, A.M.; Knight, R.; Bjorkman, P.J.; et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 2018, 360, 795–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.; Yan, H.; Diao, H.; Yu, B.; He, J.; Yu, J.; Zheng, P.; Mao, X.; Luo, Y.; Chen, D. Early Gut Microbiota Intervention Suppresses DSS-Induced Inflammatory Responses by Deactivating TLR/NLR Signalling in Pigs. Sci. Rep. 2017, 7, 3224. [Google Scholar] [CrossRef] [Green Version]
- Lucke, K.; Miehlke, S.; Jacobs, E.; Schuppler, M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J. Med. Microbiol. 2006, 55, 617–624. [Google Scholar] [CrossRef]
- Maeda, Y.; Takeda, K. Host-microbiota interactions in rheumatoid arthritis. Exp. Mol. Med. 2019, 51, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Piñeiro, A.M.; Johansson, M.E. The colonic mucus protection depends on the microbiota. Gut Microbes 2015, 6, 326–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rho, J.H.; Wright, D.P.; Christie, D.L.; Clinch, K.; Furneaux, R.H.; Roberton, A.M. A novel mechanism for desulfation of mucin: Identification and cloning of a mucin-desulfating glycosidase (sulfoglycosidase) from Prevotella strain RS2. J. Bacteriol. 2005, 187, 1543–1551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolhion, N.; Chassaing, B.; Nahori, M.A.; de Bodt, J.; Moura, A.; Lecuit, M.; Dussurget, O.; Bérard, M.; Marzorati, M.; Fehlner-Peach, H.; et al. A Listeria monocytogenes Bacteriocin Can Target the Commensal Prevotella copri and Modulate Intestinal Infection. Cell Host Microbe 2019, 26, 691–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciabattini, A.; Olivieri, R.; Lazzeri, E.; Medaglini, D. Role of the Microbiota in the Modulation of Vaccine Immune Responses. Front. Microbiol. 2019, 10, 1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlasova, A.N.; Takanashi, S.; Miyazaki, A.; Rajashekara, G.; Saif, L.J. How the gut microbiome regulates host immune responses to viral vaccines. Curr. Opin. Virol. 2019, 37, 16–25. [Google Scholar] [CrossRef]
- Hagan, T.; Cortese, M.; Rouphael, N.; Boudreau, C.; Linde, C.; Maddur, M.S.; Das, J.; Wang, H.; Guthmiller, J.; Zheng, N.Y.; et al. Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell 2019, 178, 1313–1328. [Google Scholar] [CrossRef]
- Lamousé-Smith, E.S.; Tzeng, A.; Starnbach, M.N. The intestinal flora is required to support antibody responses to systemic immunization in infant and germ free mice. PLoS ONE 2011, 6, e27662. [Google Scholar] [CrossRef] [Green Version]
- Twitchell, E.L.; Tin, C.; Wen, K.; Zhang, H.; Becker-Dreps, S.; Azcarate-Peril, M.A.; Vilchez, S.; Li, G.; Ramesh, A.; Weiss, M.; et al. Modeling human enteric dysbiosis and rotavirus immunity in gnotobiotic pigs. Gut Pathog. 2016, 8, 51. [Google Scholar] [CrossRef] [Green Version]
- Desselberger, U. The Mammalian Intestinal Microbiome: Composition, Interaction with the Immune System, Significance for Vaccine Efficacy, and Potential for Disease Therapy. Pathogens 2018, 7, 57. [Google Scholar] [CrossRef] [Green Version]
- Munyaka, P.M.; Kommadath, A.; Fouhse, J.; Wilkinson, J.; Diether, N.; Stothard, P.; Estelle, J.; Rogel-Gaillard, C.; Plastow, G.; Willing, B.P. Characterization of whole blood transcriptome and early-life fecal microbiota in high and low responder pigs before, and after vaccination for Mycoplasma hyopneumoniae. Vaccine 2019, 37, 1743–1755. [Google Scholar] [CrossRef]
- Chilton, P.M.; Hadel, D.M.; To, T.T.; Mitchell, T.C.; Darveau, R.P. Adjuvant activity of naturally occurring monophosphoryl lipopolysaccharide preparations from mucosa-associated bacteria. Infect. Immun. 2013, 81, 3317–3325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takada, K.; Hirasawa, M.; Ikeda, T. Isolation and purification of bacteriocin from Prevotella intermedia (Bacteroides intermedius). J. Periodontol. 1991, 62, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Kaewsrichan, J.; Douglas, C.W.; Teanpaisan, R. Characterization of minimal bacteriocin operon from Prevotella nigrescens ATCC 25261. Lett. Appl. Microbiol. 2005, 40, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Dicks, L.M.T.; Dreyer, L.; Smith, C.; van Staden, A.D. Corrigendum: A Review: The Fate of Bacteriocins in the Human Gastro-Intestinal Tract: Do They Cross the Gut-Blood Barrier? Front. Microbiol. 2018, 9, 2938. [Google Scholar] [CrossRef] [PubMed]
- Gould, A.L.; Zhang, V.; Lamberti, L.; Jones, E.W.; Obadia, B.; Korasidis, N.; Gavryushkin, A.; Carlson, J.M.; Beerenwinkel, N.; Ludington, W.B. Microbiome interactions shape host fitness. Proc. Natl. Acad. Sci. USA 2018, 115, E11951–E11960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tett, A.; Huang, K.D.; Asnicar, F.; Fehlner-Peach, H.; Pasolli, E.; Karcher, N.; Armanini, F.; Manghi, P.; Bonham, K.; Zolfo, M.; et al. The Prevotella copri Complex Comprises Four Distinct Clades Underrepresented in Westernized Populations. Cell Host Microbe 2019, 26, 666–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clavel, T.; Gomes-Neto, J.C.; Lagkouvardos, I.; Ramer-Tait, A.E. Deciphering interactions between the gut microbiota and the immune system via microbial cultivation and minimal microbiomes. Immunol. Rev. 2017, 279, 8–22. [Google Scholar] [CrossRef]
- Salipante, S.J.; SenGupta, D.J.; Cummings, L.A.; Land, T.A.; Hoogestraat, D.R.; Cookson, B.T. Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J. Clin. Microbiol. 2015, 53, 1072–1079. [Google Scholar] [CrossRef] [Green Version]
- Balloux, F.; Brønstad Brynildsrud, O.; van Dorp, L.; Shaw, L.P.; Chen, H.; Harris, K.A.; Wang, H.; Eldholm, V. From Theory to Practice: Translating Whole-Genome Sequencing (WGS) into the Clinic. Trends Microbiol. 2018, 26, 1035–1048. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef] [Green Version]
- Ghimire, S.; Roy, C.; Wongkuna, S.; Antony, L.; Maji, A.; Keena, M.C.; Foley, A.; Scaria, J. Identification of Clostridioides difficile-Inhibiting Gut Commensals Using Culturomics, Phenotyping, and Combinatorial Community Assembly. mSystems 2020, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenske, G.J.; Ghimire, S.; Antony, L.; Christopher-Hennings, J.; Scaria, J. Integration of culture-dependent and independent methods provides a more coherent picture of the pig gut microbiome. FEMS Microbiol. Ecol. 2020, 96. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Shibata, K.; Sakamoto, M.; Tomita, S.; Benno, Y. Prevotella copri sp. nov. and Prevotella stercorea sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2007, 57, 941–946. [Google Scholar] [CrossRef] [Green Version]
- Varel, V.H.; Bryant, M.P. Nutritional features of Bacteroides fragilis subsp. fragilis. Appl. Microbiol. 1974, 28, 251–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Li, L.; Butcher, J.; Stintzi, A.; Figeys, D. Advancing functional and translational microbiome research using meta-omics approaches. Microbiome 2019, 7, 154. [Google Scholar] [CrossRef] [PubMed]
- Chiquette, J.; Allison, M.J.; Rasmussen, M.A. Prevotella bryantii 25A used as a probiotic in early-lactation dairy cows: Effect on ruminal fermentation characteristics, milk production, and milk composition. J. Dairy Sci. 2008, 91, 3536–3543. [Google Scholar] [CrossRef] [PubMed]
- Fraga, M.; Fernández, S.; Perelmuter, K.; Pomiés, N.; Cajarville, C.; Zunino, P. The use of Prevotella bryantii 3C5 for modulation of the ruminal environment in an ovine model. Braz J. Microbiol. 2018, 49 (Suppl. 1), 101–106. [Google Scholar] [CrossRef]
| Prevotella Species | Associated with Human Infections 2 | Presence in Pig GIT 3 | Comment | References |
|---|---|---|---|---|
| P. albensis | - | Yes | Initially isolated from ruminants | [65] |
| P. amnii | - | - | - | [61] |
| P. baronia | Yes | Yes | - | [60] |
| P. bergensis | Yes | Yes | - | [61] |
| P. bivia | Yes | Yes | - | [61] |
| P. brevis | - | - | Initially isolated from ruminants | [65] |
| P. bryantii | - | Yes | Initially isolated from ruminants | [65] |
| P. buccae | Yes | Yes | - | [61] |
| P. buccalis | Yes | Yes | - | [61] |
| P. conceptionensis | Yes | - | - | [66] |
| P. coporis | Yes | - | - | [61] |
| P. copri | Yes | Yes | Highly abundant pig gut after weaning | [61] |
| P. dentalis | Yes | Yes | - | [61] |
| P. denticola | Yes | Yes | - | [61] |
| P. disiens | Yes | Yes | - | [61] |
| P. enoeca | Yes | Yes | - | [60] |
| P. heparinolytica | Yes | - | - | [61] |
| P. histicola | Yes | - | - | [61] |
| P. falsenii | - | - | Initially isolated from monkey | [67] |
| P. fusca | - | Yes | Human oral cavity | [68] |
| P. intermedia | Yes | - | - | [61] |
| P. intestinalis | - | - | Mouse colonic content | [64] |
| P. loescheii | Yes | - | - | [61] |
| P. massilliensis | - | - | - | [60] |
| P. maculosa | - | Yes | - | [61] |
| P. marshii | - | - | - | [61] |
| P. melaninogenica | Yes | Yes | - | [60] |
| P. micans | - | Yes | - | [61] |
| P. multiformis | - | - | - | [60] |
| P. multisaccharivorax | - | - | - | [61] |
| P. muris | - | - | Mouse colonic content | [64] |
| P. nanceiensis | Yes | - | - | [66] |
| P. nigreceiensis | - | - | - | [61] |
| P. nigrescens | Yes | Yes | - | [60] |
| P. oralis | Yes | Yes | - | [60] |
| P. oris | Yes | Yes | - | [60] |
| P. oulora | - | - | - | [69] |
| P. oulorum | Yes | Yes | - | [69] |
| P. pallens | Yes | - | - | [61] |
| P. pectinovora | - | - | Initially isolated from pig feces | [63] |
| P. pleuritidis | - | - | - | [61] |
| P. rodentium | - | - | Mouse colonic content | [64] |
| P. ruminicola | Yes | - | Initially isolated from ruminants | [65] |
| P. saccharolytica | Yes | Human oral cavity | [70] | |
| P. salivae | Yes | Yes | - | [61] |
| P. scopos | - | - | Human oral cavity | [68] |
| P. shahii | - | Yes | - | [61] |
| P. stercorea | - | Yes | More abundant in suckling piglet gut | [61] |
| P. tannerae | - | - | - | [61] |
| P. timonensis | Yes | - | - | [61] |
| P. veroralis | Yes | - | - | [61] |
| P. zoogleoformans | Yes | - | - | [60] |
| Study Categories | Animals | Country of Origin | Collected Samples | Point of Sample Collection | Samples Processed for | Prevotella Abundance | Main Findings | References |
|---|---|---|---|---|---|---|---|---|
| Prevotella and enterotypes of pig gut microbiota and host performance | A cohort of 953 pigs from a F6 population of heterogeneous pig cross | China | Fecal | At the ages of 25, 120 and 240 days, which represented the time of preweaning, mid-stage of fattening and slaughtering (weaned at d 28) | 16S rRNA gene sequencing (V3-V4); Illumina MiSeq | Day 25 (preweaning): Fusobacterium vs. Prevotella dominant enterotypes Days 80, 120 and 240: Treponema vs. Prevotella-dominant enterotypes | Besides the piglets, even some adult pigs switched putative enterotypes between ages | [16] |
| At all sampling time points, Prevotella was most abundant and served as one of the two main network hubs | The topological features of phylogenetic cooccurrence networks, including scale, stability and complexity were increased along with the age | |||||||
| A total of 575 Large White pigs | France | Fecal | At the age of 60 days (weaned at d 28) | 16S rRNA gene sequencing (V3-V4); Roche 454 GS FLX Titanium | Ruminococcus and Treponema vs. Prevotella and Mitsuokella-driven enterotype (PEA vs. PEB) | Diversity analysis revealed a significantly higher level of alpha-diversity and richness for PEA than for PEB | [17] | |
| Animals that clustered with the PEB were on average 850 g heavier and had an extra average daily gain (ADG) of 17.9 g per day than those that clustered with the PEA | ||||||||
| Showed the link between microbial ecosystems and pig growth traits | ||||||||
| 280 commercial Duroc pigs | China | Fecal | At the age of 140 days (weaned at d 28) | 16S rRNA gene sequencing (V4); Illumina MiSeq | Prevotella vs. Treponema- predominant enterotypes | 12 out of the 18 OTUs positively associated with the average daily feed intake (ADFI) were annotated to Prevotella, and Prevotella was the hub bacteria in the co-abundance network. These results suggest that Prevotella might be a keystone bacterial taxon for increasing host feed intake. | [18] | |
| 1039 pigs | USA | Rectal swabs | At weaning (18.6 ± 1.09 days), week 15 (118.2 ± 1.18) days, and off-test (196.4 ± 7.86 days) | 16S rRNA gene sequencing (V4); Illumina MiSeq | At weaning: Prevotella (6.78%) and the 7th predominant genus; week 15: Prevotella (13.1%), and the 1st predominant genus; off-test (at slaughtering): Prevotella (6.74%), and the 2nd predominant genus | Prevotella dominant enterotype was observed at weaning stage. However, no significant correlations between any enterotypes at weaning and average daily gain were detected | [43] | |
| Prevotella and its positive association with growth performance in pigs | 18 pigs | USA | Rectal swabs | During lactation (days 0, 11, 20), nursery (d 27, 33, 41, 50, 61), growing (d 76, 90, 104, 116), and finishing (d 130, 146, 159, 174) stages | 16S rRNA gene sequencing (V4); Illumina Miseq | Among the top 30 taxa, 11 belong to genus Prevotella, the most diverse and dominant genus throughout most of the stages, especially after the introduction of solid feed | Prevotella spp. (Prevotella copri and several unclassified Prevotella OTUs) were identified as one of the top 50 growth performance-associated taxa at lactation, nursing, growing and finishing stages | [12] |
| 31 healthy piglets | France | Fecal | At the ages of 14, 36, 48, 60 and 70 days (weaned at d 28) | 16S rRNA gene sequencing (V3-V4); Roche 454 GS FLX Titanium | After weaning, the microbiota composition coevolved with their hosts towards two different clusters: unclassified Ruminococcaceae vs. Prevotella | Prevotella cluster was positively correlated with luminal secretory IgA concentrations, and body weight | [19] | |
| A total of 48 piglets (control vs. low vs. high yeast supplemented groups, n = 16) | Canada | Cecum content | At the age of 28 days (at euthanization); body weight measured at 1, 3, 7, 10, 17, 24 and 28 days | 16S rRNA gene sequencing (V1-V3); Roche 454 FLX Titanium | Relative abundance of Prevotella genus in piglets receiving low or high yeast supplementation was 0.46 and 3.07%, respectively | Partial least squares analysis showed that piglet average daily gain (ADG) was positively correlated with genus Prevotella in the high yeast group | [83] | |
| Prevotella and its negative association with diarrhea in pigs | 20 piglets were weaned in poor housing conditions to challenge their susceptibility to post-weaning | France | Fecal | At the age of 7, 14, 21, 30, 38 and 47 days | 16S rRNA gene sequencing (V1-V3); Illumina MiSeq | Prevotellaceae families were increased in healthy pigs compared to diarrheic pigs | The higher abundance of Prevotella may contribute to allowing healthy pigs better adapt to post-weaning dietary conditions, thereby mitigating the risk of developing diarrhea | [10] |
| At the genus level, the higher abundance of Roseburia, Prevotella and genera belonging to Ruminococcaceae was increased in healthy pigs | ||||||||
| 85 commercial piglets | China | Anal swab | During the lactation (0–19 days old), weaning (20–21 days old), and post-weaning periods (22–40 days) | 16S rRNA gene sequencing (V4); Illumina Miseq | Prevotella was the one of the 11 genera whose abundance was significantly higher in non-diarrheic piglets compared to diarrheic piglets | Prevotellacecea UCG-003 was identified as a key node in non-diarrheic piglets upon co-correlation network analysis | [86] | |
| The relative abundances of OTUs belonging to Prevotella2 and Prevotella9 were 0.789% and 0.849% from diarrheic piglets, and 1.787% and 1.692% in the non-diarrheic samples | ||||||||
| 14 piglets from healthy and porcine epidemic diarrhea virus (PEDV) infection-diagnosed group (n = 7) | South Korea | Fecal | Not provided | 16S rRNA gene sequencing (V3); Illumina MiSeq | Relative abundance of most commensal bacteria including Prevotella and Faecalibacterium) in healthy pigs was decreased following dysbiosis induced by PEDV infection | Reduction of these commensal bacteria including Prevotella may have implications in pathogenesis of PVDV-associated diarrhea in pigs | [87] | |
| 51 piglets, and among which 41 piglets were orally challenged with enterotoxigenic Escherichia coli (ETEC) | China | Jejunal and fecal | Fresh feces were collected from day 1 to day 5 (post diarrhea infection (PDI)); while cecum jejunal samples were collected at day 6 PDI | 16S rRNA gene sequencing (not provided); Illumina MiSeq | Healthy piglets had higher abundance of Prevotella in the feces, but lower Lactococcus in the jejunum and lower Escherichia/Shigella in the feces compared to diarrheal piglets | ETEC-induced diarrhea is associated with the alteration of intestinal microbiota, including lower Bacteroidetes: Firmicutes ratio and microbiota diversity in the jejunum and feces, and lower Prevotella in the feces, but higher percentage of Lactococcus in the jejunum and Escherichia/Shigella in the feces | [13] | |
| Prevotella (4.2, 1.7 to 0.2%) decreased as the piglets were transient from pre-diarrheic state to diarrheic state | ||||||||
| Compared to resistant piglets, the diarrheal piglet harbored lower Prevotella |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amat, S.; Lantz, H.; Munyaka, P.M.; Willing, B.P. Prevotella in Pigs: The Positive and Negative Associations with Production and Health. Microorganisms 2020, 8, 1584. https://doi.org/10.3390/microorganisms8101584
Amat S, Lantz H, Munyaka PM, Willing BP. Prevotella in Pigs: The Positive and Negative Associations with Production and Health. Microorganisms. 2020; 8(10):1584. https://doi.org/10.3390/microorganisms8101584
Chicago/Turabian StyleAmat, Samat, Hannah Lantz, Peris M. Munyaka, and Benjamin P. Willing. 2020. "Prevotella in Pigs: The Positive and Negative Associations with Production and Health" Microorganisms 8, no. 10: 1584. https://doi.org/10.3390/microorganisms8101584
APA StyleAmat, S., Lantz, H., Munyaka, P. M., & Willing, B. P. (2020). Prevotella in Pigs: The Positive and Negative Associations with Production and Health. Microorganisms, 8(10), 1584. https://doi.org/10.3390/microorganisms8101584





