Discovery of Predictors of Mycoplasma hyopneumoniae Vaccine Response Efficiency in Pigs: 16S rRNA Gene Fecal Microbiota Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Considerations
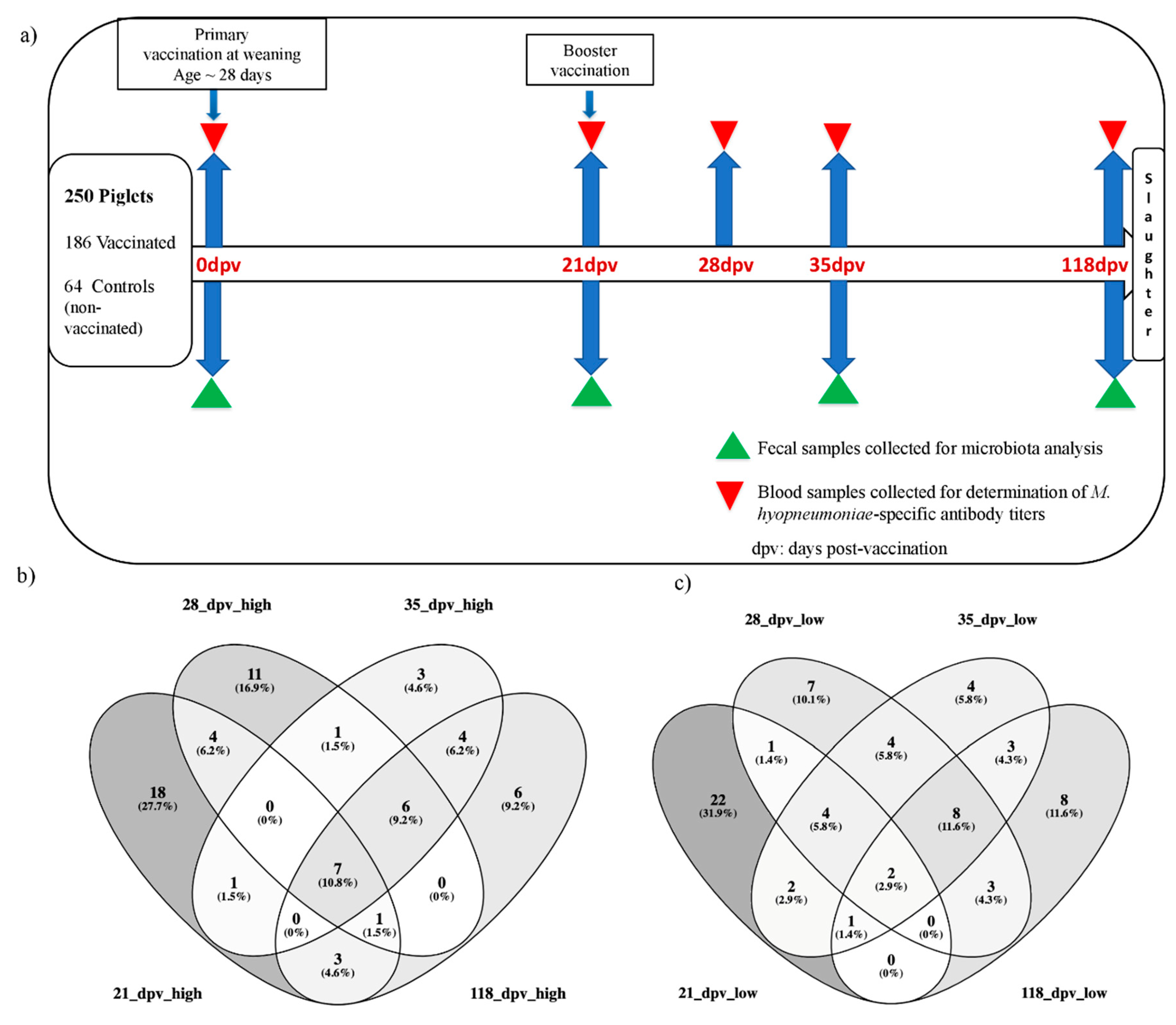
2.2. Animal Design, Vaccination and Sampling Protocol
2.3. Determination of Antibody Response against M. hyo and Selection of High and Low Responder Pigs
2.4. Fecal DNA Preparation
2.5. 16S rRNA Gene Sequencing and Bioinformatics Analyses
2.6. Biostatistical Analyses
2.7. Data Availability
3. Results
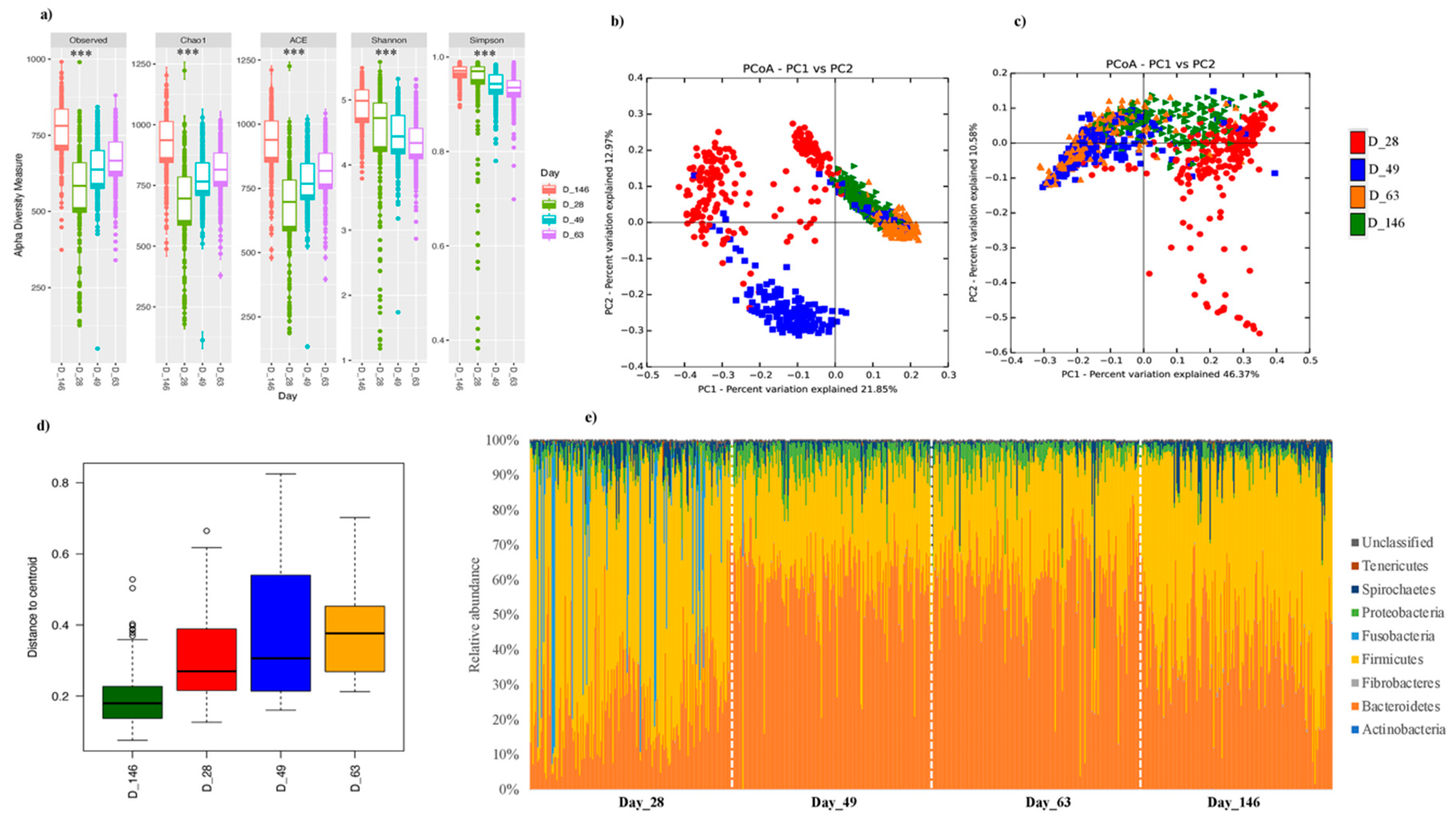
3.1. Longitudinal Investigation of Swine Fecal Microbiota Confirms Compositional Differences at Different Ages
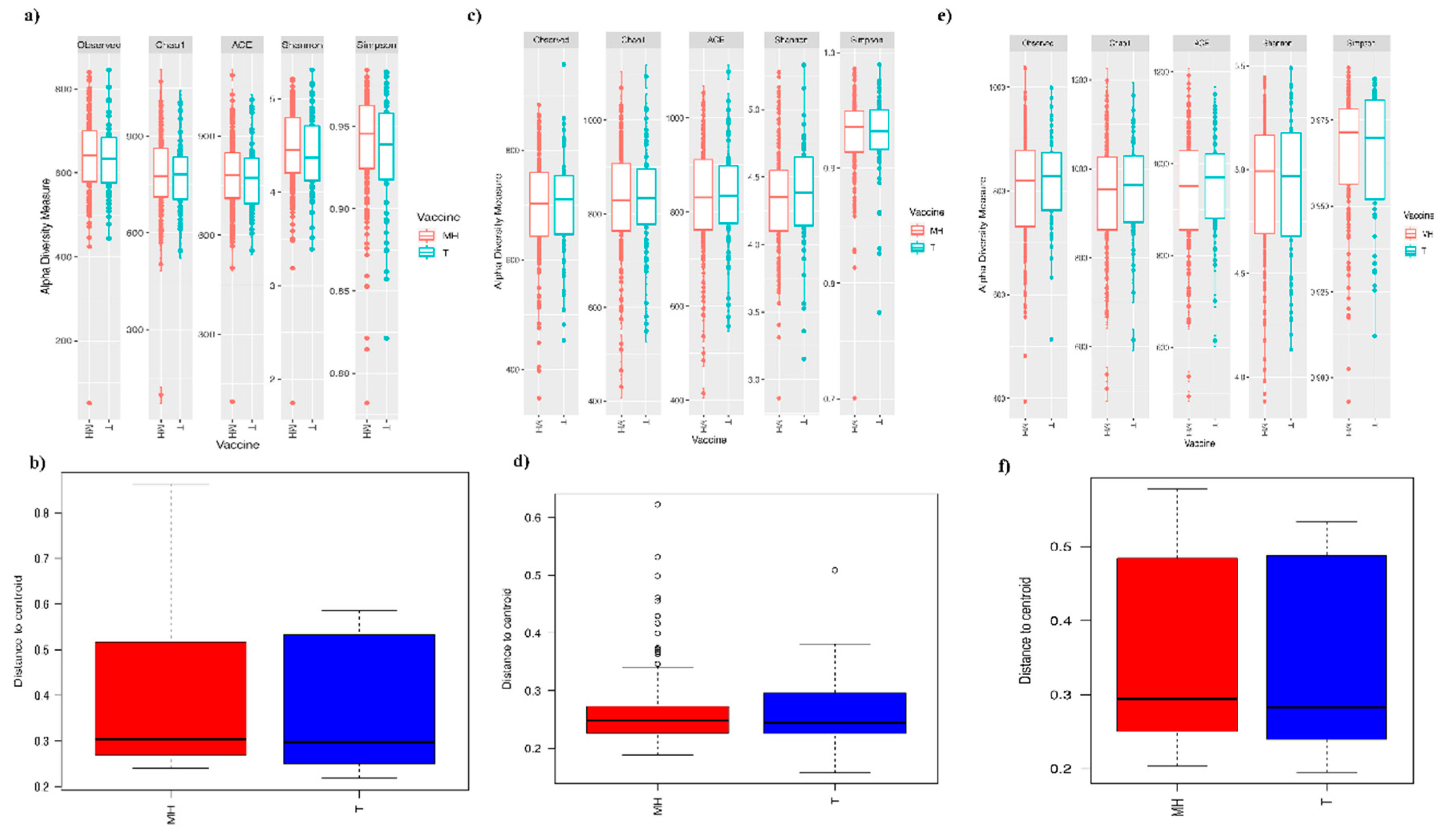
3.2. Fecal Microbiota Composition of the Vaccinated and Control Pigs Did Not Significantly Differ Post-Vaccination
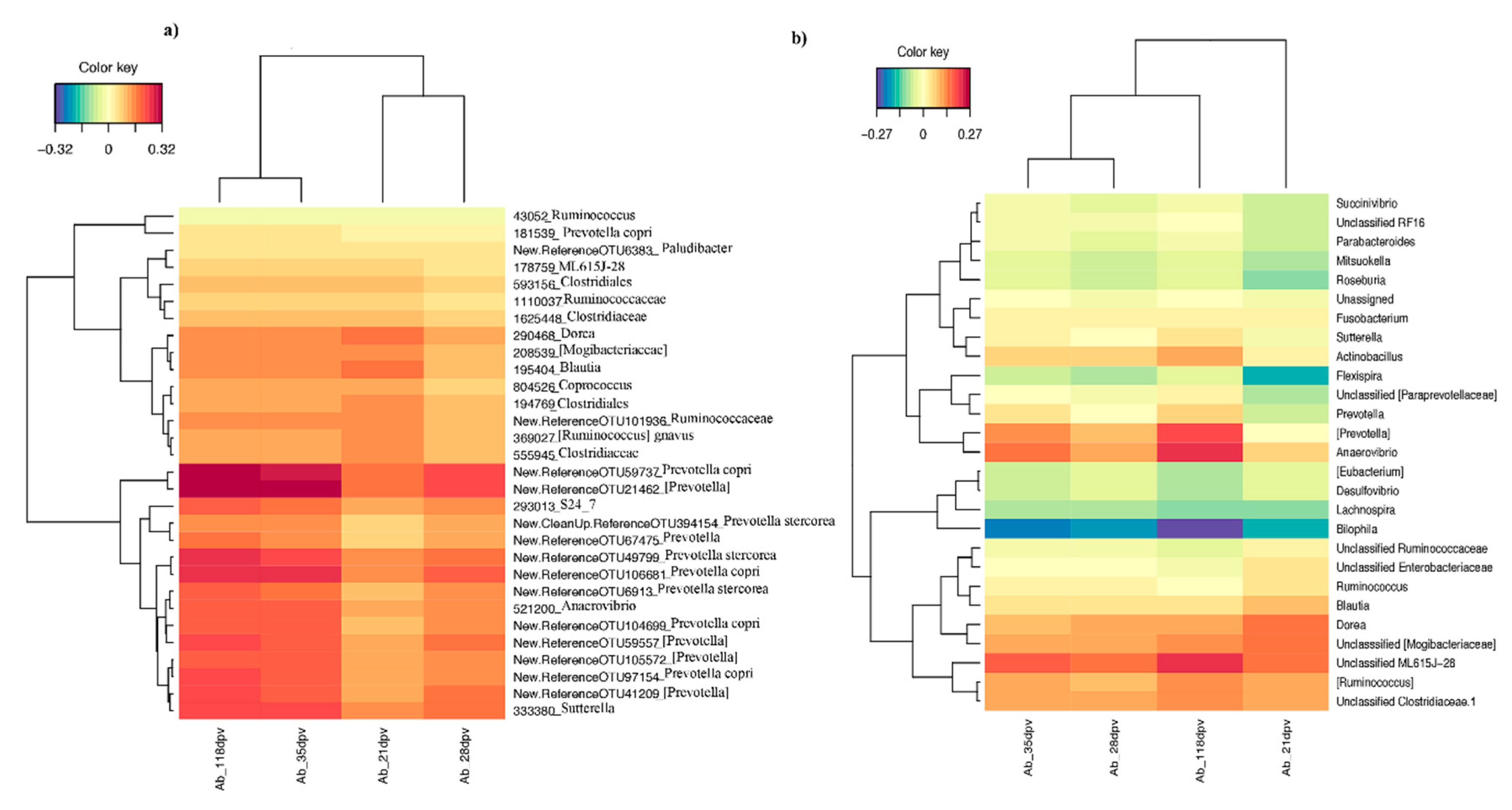
3.3. Early-Life Fecal Microbiota Composition before Vaccination as a Predictor of M. hyo Vaccine Response
3.3.1. sPLS Prediction Analysis
3.3.2. Differential Abundance Analysis at Different Taxonomic Levels
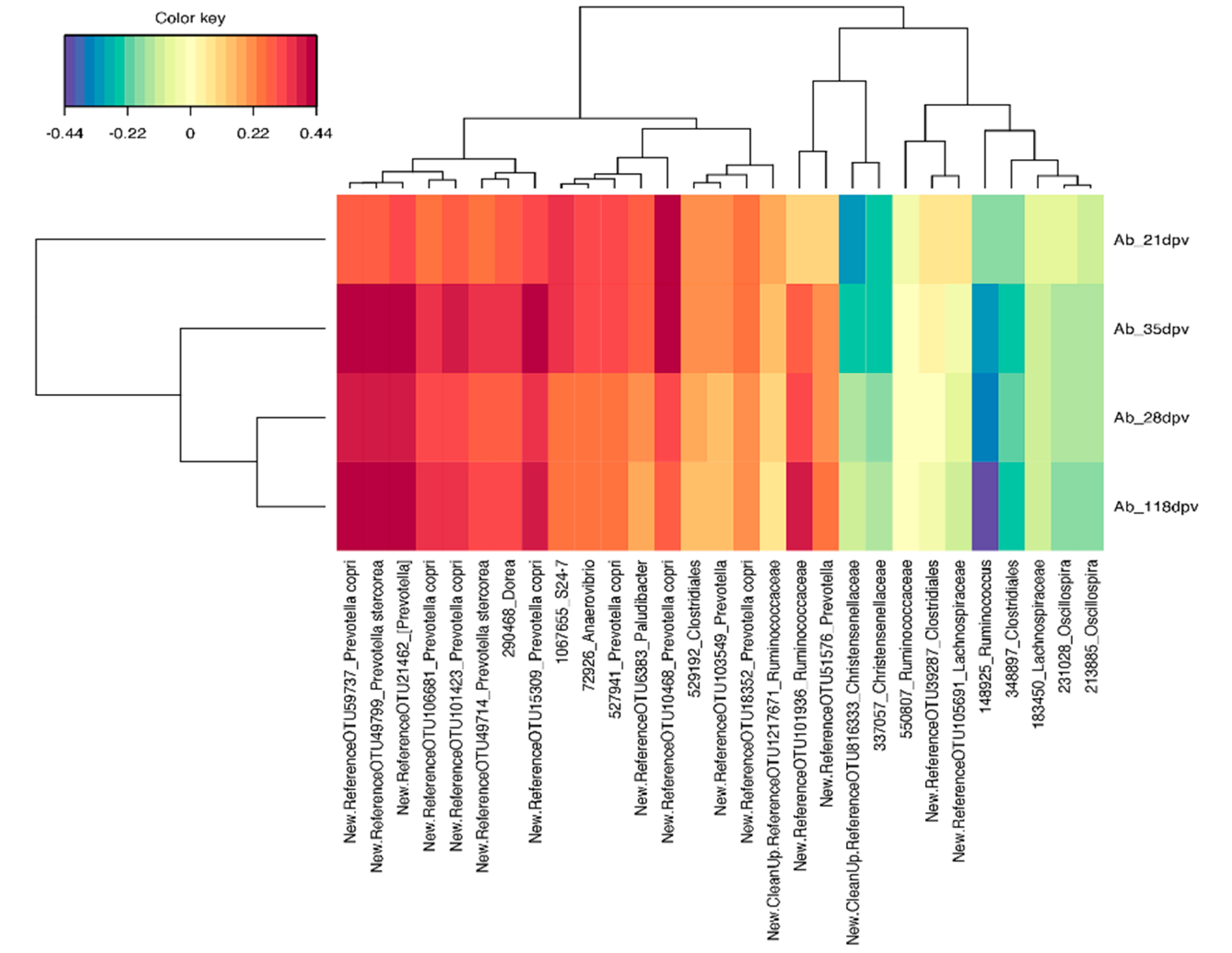
3.4. Contrasted Antibody Response Was Not Accompanied by Significant Changes in Microbiota Composition at 21, 35, and 118 dpv
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hoen, A.G.; Li, J.; Moulton, L.A.; O’Toole, G.A.; Housman, M.L.; Koestler, D.C.; Guill, M.F.; Moore, J.H.; Hibberd, P.L.; Morrison, H.G.; et al. Associations between gut microbial colonization in early life and respiratory outcomes in cystic fibrosis. Proc. J. Pediatr. 2015, 167, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Dou, S.; Gadonna-Widehem, P.; Rome, V.; Hamoudi, D.; Rhazi, L.; Lakhal, L.; Larcher, T.; Bahi-Jaber, N.; Pinon-Quintana, A.; Guyonvarch, A.; et al. Characterisation of Early-Life Fecal Microbiota in Susceptible and Healthy Pigs to Post-Weaning Diarrhoea. PLoS ONE 2017, 12, e0169851. [Google Scholar] [CrossRef] [PubMed]
- Ranucci, G.; Buccigrossi, V.; De Freitas, M.B.; Guarino, A.; Giannattasio, A. Early-Life Intestine Microbiota and Lung Health in Children. J. Immunol. Res. 2017, 2017, 8450496. [Google Scholar] [CrossRef] [PubMed]
- Fouhse, J.M.; Yang, K.; More-Bayona, J.; Gao, Y.; Goruk, S.; Plastow, G.; Field, C.J.; Barreda, D.R.; Willing, B.P. Neonatal Exposure to Amoxicillin Alters Long-Term Immune Response Despite Transient Effects on Gut-Microbiota in Piglets. Front. Immunol. 2019, 10, 2059. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Núñez, G. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology 2014, 146, 1477–1488. [Google Scholar] [CrossRef]
- Buffie, C.G.; Pamer, E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013, 11, 780–801. [Google Scholar] [CrossRef]
- Bauer, H.; Horowitz, R.E.; Levenson, S.M.; Popper, H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am. J. Pathol. 1963, 42, 471–483. [Google Scholar]
- Lamousé-Smith, E.S.; Tzeng, A.; Starnbach, M.N. The intestinal flora is required to support antibody responses to systemic immunization in infant and germ free mice. PLoS ONE 2011, 6, e27662. [Google Scholar] [CrossRef]
- Zimmermann, P.; Curtis, N. The influence of the intestinal microbiome on vaccine responses. Vaccine 2018, 36, 4433–4439. [Google Scholar] [CrossRef]
- Lynn, D.J.; Pulendran, B. The potential of the microbiota to influence vaccine responses. J. Leukoc. Biol. 2017, 103, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Velasquez, D.E.; Parashar, U.; Jiang, B. Decreased performance of live attenuated, oral rotavirus vaccines in low-income settings: Causes and contributing factors. Expert Rev. Vaccines 2018, 17, 145–161. [Google Scholar] [CrossRef] [PubMed]
- Valdez, Y.; Brown, E.M.; Finlay, B.B. Influence of the microbiota on vaccine effectiveness. Trends Immunol. 2014, 35, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.B.R.; Antunes, L.C.M.; Brett Finlay, B. Should the human microbiome be considered when developing vaccines? PLoS Pathog. 2010, 6, e1001190. [Google Scholar] [CrossRef]
- Collins, N.; Belkaid, Y. Do the microbiota influence vaccines and protective immunity to pathogens?: Engaging our endogenous adjuvants. Cold Spring Harb. Perspect. Biol. 2018, 10, a028860. [Google Scholar] [CrossRef]
- Littman, D.R. Do the microbiota influence vaccines and protective immunity to pathogens?: If so, is there potential for efficacious microbiota-based vaccines? Cold Spring Harb. Perspect. Biol. 2018, 10, a029355. [Google Scholar] [CrossRef]
- Macpherson, A.J. Do the Microbiota Influence Vaccines and Protective Immunity to Pathogens? Cold Spring Harb. Perspect. Biol. 2018, 10, a029363. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Z.; Ravindran, R.; Chassaing, B.; Carvalho, F.A.; Maddur, M.S.; Bower, M.; Hakimpour, P.; Gill, K.P.; Nakaya, H.I.; Yarovinsky, F.; et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 2014, 41, 478–492. [Google Scholar] [CrossRef]
- Huda, M.N.; Lewis, Z.; Kalanetra, K.M.; Rashid, M.; Ahmad, S.M.; Raqib, R.; Qadri, F.; Underwood, M.A.; Mills, D.A.; Stephensen, C.B. Stool microbiota and vaccine responses of infants. Pediatrics 2014, 134, e362–e372. [Google Scholar] [CrossRef]
- Harris, V.C.; Armah, G.; Fuentes, S.; Korpela, K.E.; Parashar, U.; Victor, J.C.; Tate, J.; de Weerth, C.; Giaquinto, C.; Wiersinga, W.J.; et al. Significant Correlation between the Infant Gut Microbiome and Rotavirus Vaccine Response in Rural Ghana. J. Infect. Dis. 2017, 215, 34–41. [Google Scholar] [CrossRef]
- Lynn, M.A.; Tumes, D.J.; Choo, J.M.; Sribnaia, A.; Blake, S.J.; Leong, L.E.X.; Young, G.P.; Marshall, H.S.; Wesselingh, S.L.; Rogers, G.B.; et al. Early-Life Antibiotic-Driven Dysbiosis Leads to Dysregulated Vaccine Immune Responses in Mice. Cell Host Microbe 2018, 23, 653–660. [Google Scholar] [CrossRef]
- Yitbarek, A.; Astill, J.; Hodgins, D.C.; Parkinson, J.; Nagy, É.; Sharif, S. Commensal gut microbiota can modulate adaptive immune responses in chickens vaccinated with whole inactivated avian influenza virus subtype H9N2. Vaccine 2019, 37, 6640–6647. [Google Scholar] [CrossRef] [PubMed]
- Munyaka, P.M.; Kommadath, A.; Fouhse, J.; Wilkinson, J.; Diether, N.; Stothard, P.; Estellé, J.; Rogel-Gaillard, C.; Plastow, G.; Willing, B.P. Characterization of whole blood transcriptome and early-life fecal microbiota in high and low responder pigs before, and after vaccination for Mycoplasma hyopneumoniae. Vaccine 2019, 37, 1743–1755. [Google Scholar] [CrossRef] [PubMed]
- Blanc, F.; Maroilley, T.; Revilla, M.; Lemonnier, G.; Leplat, J.; Billon, Y.; Bouchez, O.; Bidanel, J.; Bed’Hom, B.; Pinard-van der Laan, M.; et al. Influence of genetics and pre-vaccination blood transcriptome on the variability of antibody levels after vaccination against Mycoplasma hyopneumoniae in pigs. Unpublished work. 2020. [Google Scholar]
- GenESI, INRAE, Pig Phenotyping and Innovative Breeding Facility. Available online: https://doi.org/10.15454/1.5572415481185847E12 (accessed on 28 July 2020).
- Godon, J.J.; Zumstein, E.; Dabert, P.; Habouzit, F.; Moletta, R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 1997, 63, 2802–2813. [Google Scholar] [CrossRef] [PubMed]
- Massacci, F.R.; Berri, M.; Lemonnier, G.; Guettier, E.; Blanc, F.; Jardet, D.; Rossignol, M.N.; Mercat, M.-J.; Doré, J.; Lepage, P.; et al. Late weaning is associated with increased microbial diversity and Faecalibacterium prausnitzii abundance in the fecal microbiota of piglets. Anim. Microbiome 2020, 2, 2. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pẽa, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Crespo-Piazuelo, D.; Migura-Garcia, L.; Estellé, J.; Criado-Mesas, L.; Revilla, M.; Castelló, A.; Muñoz, M.; García-Casco, J.M.; Fernández, A.I.; Ballester, M.; et al. Association between the pig genome and its gut microbiota composition. Sci. Rep. 2019, 9, 8791. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2013, 2, 16. [Google Scholar] [CrossRef]
- Cao, K.A.L.; Costello, M.E.; Lakis, V.A.; Bartolo, F.; Chua, X.Y.; Brazeilles, R.; Rondeau, P. MixMC: A multivariate statistical framework to gain insight into microbial communities. PLoS ONE 2016, 11, e0160169. [Google Scholar]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Lê Cao, K.A.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics 2011, 12, 253. [Google Scholar] [CrossRef] [PubMed]
- Paulson, J.N.; Colin Stine, O.; Bravo, H.C.; Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 2013, 10, 1200–1202. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, r60. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, r79. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.A.; Bouladoux, N.; Sun, C.M.; Wohlfert, E.A.; Blank, R.B.; Zhu, Q.; Grigg, M.E.; Berzofsky, J.A.; Belkaid, Y. Commensal DNA Limits Regulatory T Cell Conversion and Is a Natural Adjuvant of Intestinal Immune Responses. Immunity 2008, 29, 637–649. [Google Scholar] [CrossRef]
- Salk, H.M.; Simon, W.L.; Lambert, N.D.; Kennedy, R.B.; Grill, D.E.; Kabat, B.F.; Poland, G.A. Taxa of the Nasal Microbiome Are Associated with Influenza-Specific IgA Response to Live Attenuated Influenza Vaccine. PLoS ONE 2016, 11, e0162803. [Google Scholar] [CrossRef]
- Huda, M.; Ahmad, S.; Kalanetra, K.; Taft, D.; Lewis, Z.; Raqib, R.; Mills, D.; Stephensen, C. Infant stool microbiota at the time of vaccination at 6 w of age is associated with vaccine responses measured at 2 y of age. Immunology 2017, 198, 139. [Google Scholar]
- Fix, J.; Chandrashekhar, K.; Perez, J.; Bucardo, F.; Hudgens, M.G.; Yuan, L.; Twitchell, E.; Azcarate-Peril, M.A.; Vilchez, S.; Becker-Dreps, S. Association between Gut Microbiome Composition and Rotavirus Vaccine Response among Nicaraguan Infants. Am. J. Trop. Med. Hyg. 2019, 102, 213–219. [Google Scholar] [CrossRef]
- Parker, E.P.K.; Praharaj, I.; Zekavati, A.; Lazarus, R.P.; Giri, S.; Operario, D.J.; Liu, J.; Houpt, E.; Iturriza-Gómara, M.; Kampmann, B.; et al. Influence of the intestinal microbiota on the immunogenicity of oral rotavirus vaccine given to infants in south India. Vaccine 2018, 36, 264–272. [Google Scholar] [CrossRef]
- Argüello, H.; Estellé, J.; Leonard, F.C.; Crispie, F.; Cotter, P.D.; O’Sullivan, O.; Lynch, H.; Walia, K.; Duffy, G.; Lawlor, P.G.; et al. Influence of the Intestinal Microbiota on Colonization Resistance to Salmonella and the Shedding Pattern of Naturally Exposed Pigs. mSystems 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Bearson, S.M.D.; Allen, H.K.; Bearson, B.L.; Looft, T.; Brunelle, B.W.; Kich, J.D.; Tuggle, C.K.; Bayles, D.O.; Alt, D.; Levine, U.Y.; et al. Profiling the gastrointestinal microbiota in response to Salmonella: Low versus high Salmonella shedding in the natural porcine host. Infect. Genet. Evol. 2013, 16, 330–340. [Google Scholar] [CrossRef]
- Surendran Nair, M.; Eucker, T.; Martinson, B.; Neubauer, A.; Victoria, J.; Nicholson, B.; Pieters, M. Influence of pig gut microbiota on Mycoplasma hyopneumoniae susceptibility. Vet. Res. 2019, 50, 86. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D. Human gut microbiome: Hopes, threats and promises. Gut 2018, 67, 1716–1725. [Google Scholar] [CrossRef] [PubMed]
- Ley, R. Gut microbiota in 2015: Prevotella in the gut: Choose carefully. Nat Rev Gastroenterol Hepatol. 2016, 13, 69–70. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Pellegrini, N.; Laghi, L.; Gobbetti, M.; Ercolini, D. Unusual sub-genus associations of faecal Prevotella and Bacteroides with specific dietary patterns. Microbiome 2016, 4, 57. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Chilton, P.M.; Hadelq, D.M.; To, T.T.; Mitchell, T.C.; Darveau, R.P. Adjuvant activity of naturally occurring monophosphoryl lipopolysaccharide preparations from mucosa-associated bacteria. Infect. Immun. 2013, 81, 3317–3325. [Google Scholar] [CrossRef]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Björck, I.; Bäckhed, F. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef]
- De Vadder, F.; Kovatcheva-Datchary, P.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-Produced Succinate Improves Glucose Homeostasis via Intestinal Gluconeogenesis. Cell Metab. 2016, 24, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Vitaglione, P.; Mennella, I.; Ferracane, R.; Rivellese, A.A.; Giacco, R.; Ercolini, D.; Gibbons, S.M.; La Storia, A.; Gilbert, J.A.; Jonnalagadda, S.; et al. Whole-grain wheat consumption reduces inflammation in a randomized controlled trial on overweight and obese subjects with unhealthy dietary and lifestyle behaviors: Role of polyphenols bound to cereal dietary fiber. Am. J. Clin. Nutr. 2015, 101, 251–261. [Google Scholar] [CrossRef]
- Angelis, M.D.; Montemurno, E.; Vannini, L.; Cosola, C.; Cavallo, N.; Gozzi, G.; Maranzano, V.; Di Cagno, R.; Gobbetti, M.; Gesualdo, L. Effect of whole-grain barley on the human fecal microbiota and metabolome. Appl. Environ. Microbiol. 2015, 81, 7945–7956. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Rhodes, M.E.; Neff, C.P.; Fontenot, A.P.; Campbell, T.B.; Palmer, B.E. HIV-induced alteration in gut Microbiota: Driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes 2014, 5, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Elinav, E.; Strowig, T.; Kau, A.L.; Henao-Mejia, J.; Thaiss, C.A.; Booth, C.J.; Peaper, D.R.; Bertin, J.; Eisenbarth, S.C.; Gordon, J.I.; et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011, 145, 745–757. [Google Scholar] [CrossRef]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013, 2, e01202. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Kurakawa, T.; Umemoto, E.; Motooka, D.; Ito, Y.; Gotoh, K.; Hirota, K.; Matsushita, M.; Furuta, Y.; Narazaki, M.; et al. Dysbiosis Contributes to Arthritis Development via Activation of Autoreactive T Cells in the Intestine. Arthritis Rheumatol. 2016, 68, 2646–2661. [Google Scholar] [CrossRef]
- Rolhion, N.; Chassaing, B.; Nahori, M.A.; de Bodt, J.; Moura, A.; Lecuit, M.; Dussurget, O.; Bérard, M.; Marzorati, M.; Fehlner-Peach, H.; et al. A Listeria monocytogenes Bacteriocin Can Target the Commensal Prevotella copri and Modulate Intestinal Infection. Cell Host Microbe 2019, 26, 691–701. [Google Scholar] [CrossRef]
- Alpizar-Rodriguez, D.; Lesker, T.R.; Gronow, A.; Gilbert, B.; Raemy, E.; Lamacchia, C.; Gabay, C.; Finckh, A.; Strowig, T. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann. Rheum. Dis. 2019, 78, 590–593. [Google Scholar] [CrossRef]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.H.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef]
- Gupta, V.K.; Chaudhari, N.M.; Iskepalli, S.; Dutta, C. Divergences in gene repertoire among the reference Prevotella genomes derived from distinct body sites of human. BMC Genomics 2015, 16, 153. [Google Scholar] [CrossRef][Green Version]
- De Filippis, F.; Pasolli, E.; Tett, A.; Tarallo, S.; Naccarati, A.; De Angelis, M.; Neviani, E.; Cocolin, L.; Gobbetti, M.; Segata, N.; et al. Distinct Genetic and Functional Traits of Human Intestinal Prevotella copri Strains Are Associated with Different Habitual Diets. Cell Host Microbe 2019, 25, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Fehlner-Peach, H.; Magnabosco, C.; Raghavan, V.; Scher, J.U.; Tett, A.; Cox, L.M.; Gottsegen, C.; Watters, A.; Wiltshire-Gordon, J.D.; Segata, N.; et al. Distinct Polysaccharide Utilization Profiles of Human Intestinal Prevotella copri Isolates. Cell Host Microbe 2019, 26, 680–690. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tsai, T.; Deng, F.; Wei, X.; Chai, J.; Knapp, J.; Apple, J.; Maxwell, C.V.; Lee, J.A.; Li, Y.; et al. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome 2019, 7, 109. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zheng, Z.; Yu, L.; Wu, S.; Sun, L.; Wu, S.; Xu, Q.; Cai, S.; Qin, N.; Bao, W. Examination of the temporal and spatial dynamics of the gut microbiome in newborn piglets reveals distinct microbial communities in six intestinal segments. Sci. Rep. 2019, 9, 3453. [Google Scholar] [CrossRef] [PubMed]
- De Rodas, B.; Youmans, B.P.; Danzeisen, J.L.; Tran, H.; Johnson, T.J. Microbiome profiling of commercial pigs from farrow to finish. J. Anim. Sci. 2018, 96, 1778–1794. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, Y.; Liu, S.; Huang, J.; Zhai, Z.; He, C.; Ding, J.; Wang, J.; Wang, H.; Fan, W.; et al. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS ONE 2015, 10, e0117441. [Google Scholar] [CrossRef]
- Mach, N.; Berri, M.; Estellé, J.; Levenez, F.; Lemonnier, G.; Denis, C.; Leplat, J.J.; Chevaleyre, C.; Billon, Y.; Doré, J.; et al. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 2015, 7, 554–569. [Google Scholar] [CrossRef]
- Guevarra, R.B.; Lee, J.H.; Lee, S.H.; Seok, M.J.; Kim, D.W.; Kang, B.N.; Johnson, T.J.; Isaacson, R.E.; Kim, H.B. Piglet gut microbial shifts early in life: Causes and effects. J. Anim. Sci. Biotechnol. 2019, 10, 1. [Google Scholar] [CrossRef]


| Differentially Abundant (DA) OTUs | |||||
|---|---|---|---|---|---|
| Ab Response Time Course | dpv | Classification | Number of DA OTUs | Higher Abundance in “High Responders” | Higher Abundance in “Low Responders” |
| Early (before Booster Vaccination) | 21 | high vs. low | 14 | New.ReferenceOTU6913_Prevotella stercorea, 1105328_Coprococcus, New.ReferenceOTU41209_[Prevotella], New.ReferenceOTU10520_[Prevotella], 308786_Ruminococcus, 157772_Oscillospira | New.ReferenceOTU25778_S24−7, New.ReferenceOTU21882_Butyricimonas, New.ReferenceOTU17960_Bacteroidales, New.ReferenceOTU10995_Bacteroidales, New.ReferenceOTU101339_Bacteroidales, New.CleanUp.ReferenceOTU1337388_Bacteroidales, 592649_Clostridiales, 335670_Ruminococcaceae |
| Maximum Intensity (Post Booster Vaccination) | 28 | high vs. low | 44 | New.ReferenceOTU89450_Anaerovibrio, New.ReferenceOTU69704_Prevotella copri, New.ReferenceOTU6913_Prevotella stercorea, New.ReferenceOTU67475_Prevotella, New.ReferenceOTU6383_Paludibacter, New.ReferenceOTU59737_Prevotella copri, New.ReferenceOTU57668_unclassified, New.ReferenceOTU41209_[Prevotella], New.ReferenceOTU21462_[Prevotella], New.ReferenceOTU15309_Prevotella copri, New.ReferenceOTU11599_Prevotella copri, New.ReferenceOTU110597_Prevotella stercorea, New.ReferenceOTU108770_[Prevotella], New.ReferenceOTU106681_Prevotella copri, New.ReferenceOTU101936_Ruminococcaceae, New.ReferenceOTU101423_Prevotella copri, New.ReferenceOTU10106_Prevotella copri, New.ReferenceOTU10063_Anaerovibrio, New.CleanUp.ReferenceOTU891171_Prevotella copri, New.CleanUp.ReferenceOTU816333_Christensenellaceae, New.CleanUp.ReferenceOTU729842_Prevotella copri, 1067655_S24−7, 589329_Prevotella copri, 579750_Lachnospiraceae, 333195_Ruminococcaceae, 301678_Ruminococcaceae, 301012_Clostridiales, 300859_Prevotella, 208949_Bacteroides, 172163_Clostridiales, 72926_Anaerovibrio, 22466_Prevotella | New.ReferenceOTU95104_Lachnospiraceae, New.ReferenceOTU25356_Clostridiales, 827702_Clostridiales, 772972_Clostridiales, 443620_Oscillospira, 369827_Ruminococcaceae, 346686_Oscillospira, 314639_Clostridiales, 263546_Oscillospira, 231028_Oscillospira, 213885_Oscillospira, 152612_Clostridiales |
| 35 | high vs. low | 29 | 72926_Anaerovibrio, 550807_Ruminococcaceae, New.ReferenceOTU49714_Prevotella stercorea, New.ReferenceOTU49799_Prevotella stercorea, New.ReferenceOTU15309_Prevotella copri, New.ReferenceOTU59737_Prevotella copri, New.ReferenceOTU106681_Prevotella copri, New.ReferenceOTU51576_Prevotella, New.ReferenceOTU103549_Prevotella, New.ReferenceOTU18352_Prevotella copri, New.ReferenceOTU101423_Prevotella copri, New.ReferenceOTU10468_Prevotella copri, 527941_Prevotella copri, 290468_Dorea, New.ReferenceOTU21462_[Prevotella], New.ReferenceOTU6383_Paludibacter, 1067655_S24−7, New.cleanUp.ReferenceOTU1217671_Ruminococcaceae, New.ReferenceOTU101936_Ruminococcaceae, 529192_Clostridiales | New.ReferenceOTU39287_Clostridiales, 231028_Oscillospira, 213885_Oscillospira, 148925_Ruminococcus, 348897_Clostridiales, New.cleanUp.ReferenceOTU816333_Christensenellaceae, 337057_Christensenellaceae, New.ReferenceOTU105691_Lachnospiraceae, 183450_Lachnospiraceae | |
| Persistence (Before Slaughtering) | 118 | high vs. low | 69 | New.ReferenceOTU9154_S24−7, New.ReferenceOTU75092_Treponema, New.ReferenceOTU6913_Prevotella stercorea, New.ReferenceOTU67475_Prevotella, New.ReferenceOTU59737_Prevotella copri, New.ReferenceTU63368_Treponema, New.ReferenceOTU57668_Unassigned, New.ReferenceOTU49714_Prevotella stercorea, New.ReferenceOTU39287_Clostridiales, New.ReferenceOTU46053_Christensenellaceae, New.ReferenceOTU21462_[Prevotella], New.ReferenceOTU24010_Prevotella copri, New.ReferenceOTU15309_Prevotella copri, New.ReferenceOTU1235_Bacteroidales, New.ReferenceOTU110599_Prevotella copri, New.ReferenceOTU107301_Prevotella, New.ReferenceOTU106681_Prevotella copri, New.ReferenceOTU106222_Parabacteroides, New.ReferenceOTU10468_Prevotella copri, New.ReferenceOTU101611_Prevotella, New.ReferenceOTU101423_Prevotella copri, New.ReferenceOTU10063_Anaerovibrio, New.CleanUp.ReferenceOTU729842_Prevotella copri, New.CleanUp.ReferenceOTU394154_Prevotella stercorea, 3407052_Bacteroides, 647215_Oscillospira, 584951_Ruminococcaceae, 354599_Treponema, 345114_Fusobacterium, 301678_Ruminococcaceae, 288250_Prevotella, 215963_Clostridium, 172163_Clostridiales | New.ReferenceOTU77656_Bacteroidales, New.ReferenceOTU73263_Bacteroidales, New.ReferenceOTU53970_Christensenellaceae, New.ReferenceOTU51770_Prevotella copri, New.ReferenceOTU46057_Clostridiales, New.ReferenceOTU105562_S24−7, New.ReferenceOTU10341_Bacteroides, New.CleanUp.ReferenceOTU915070_Prevotella copri, New.CleanUp.ReferenceOTU870013_Lachnospiraceae, New.CleanUp.ReferenceOTU133342_Lachnospiracea, 4357657_Clostridiaceae, 1106861_Roseburia, 708680_Roseburia, 621472_Roseburia, 538947_Lachnospiraceae, 531614_Prevotella copri, 354632_Ruminicoccaceae, 519836_Prevotella, 339221_Prevotella copri, 337057_Christensenellaceae, 325842_Ruminococcaceae, 325254_Christensenellaceae, 319659_Clostridiales, 314639_Clostridiales, 312490_Clostridiales, 292242_Ruminococcaceae, 291490_Prevotella, 290253_Oscillospira, 274299_Treponema, 259533_Treponema, 248447_Prevotella, 231028_Oscillospira, 213394_Lachnospiraceae, 211935_Lachnospiraceae, 188079_Ruminococcaceae, 178965_Ruminococcaceae, 148925_Ruminococcus |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munyaka, P.M.; Blanc, F.; Estellé, J.; Lemonnier, G.; Leplat, J.-J.; Rossignol, M.-N.; Jardet, D.; Plastow, G.; Billon, Y.; Willing, B.P.; et al. Discovery of Predictors of Mycoplasma hyopneumoniae Vaccine Response Efficiency in Pigs: 16S rRNA Gene Fecal Microbiota Analysis. Microorganisms 2020, 8, 1151. https://doi.org/10.3390/microorganisms8081151
Munyaka PM, Blanc F, Estellé J, Lemonnier G, Leplat J-J, Rossignol M-N, Jardet D, Plastow G, Billon Y, Willing BP, et al. Discovery of Predictors of Mycoplasma hyopneumoniae Vaccine Response Efficiency in Pigs: 16S rRNA Gene Fecal Microbiota Analysis. Microorganisms. 2020; 8(8):1151. https://doi.org/10.3390/microorganisms8081151
Chicago/Turabian StyleMunyaka, Peris M., Fany Blanc, Jordi Estellé, Gaëtan Lemonnier, Jean-Jacques Leplat, Marie-Noëlle Rossignol, Déborah Jardet, Graham Plastow, Yvon Billon, Benjamin P. Willing, and et al. 2020. "Discovery of Predictors of Mycoplasma hyopneumoniae Vaccine Response Efficiency in Pigs: 16S rRNA Gene Fecal Microbiota Analysis" Microorganisms 8, no. 8: 1151. https://doi.org/10.3390/microorganisms8081151
APA StyleMunyaka, P. M., Blanc, F., Estellé, J., Lemonnier, G., Leplat, J.-J., Rossignol, M.-N., Jardet, D., Plastow, G., Billon, Y., Willing, B. P., & Rogel-Gaillard, C. (2020). Discovery of Predictors of Mycoplasma hyopneumoniae Vaccine Response Efficiency in Pigs: 16S rRNA Gene Fecal Microbiota Analysis. Microorganisms, 8(8), 1151. https://doi.org/10.3390/microorganisms8081151





