Simultaneous Detection of Bluetongue Virus Serotypes Using xMAP Technology
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA Extraction
2.2. BTV Serotyping Primers for RT-PCR Stage of BTV xMAP Assay
2.3. RT-PCR
2.4. Coupling of Microsphere to Capture Probes
2.5. Detection of BTV Serotypes Using xMAP Assay
2.6. BTV xMAP Data Collection and Analysis
2.7. BTV Serotype-Specific RT-qPCR
2.8. BTV Group-Specific RT-qPCR
2.9. Assay Optimization
2.10. Panel of Samples for Evaluation of BTV xMAP
2.11. Limit of Detection
2.12. Repeatability of BTV xMAP
2.13. Price Comparison of BTV xMAP with Serotype-Specific RT-PCR Assays
3. Results
3.1. Assay Optimization
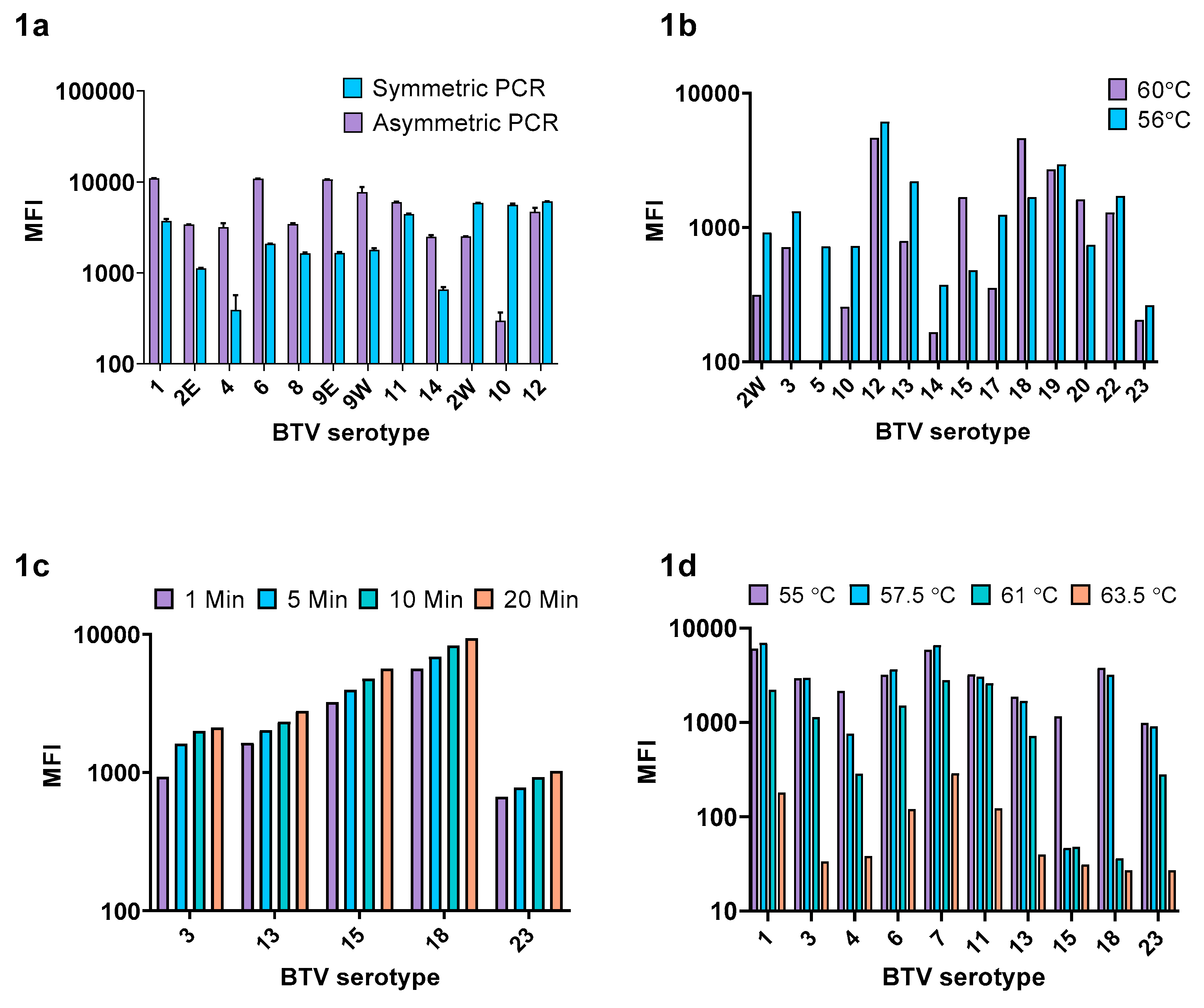
3.1.1. Ratio of Biotinylated:Non Biotinylated Primer
3.1.2. xMAP PCR Annealing Temperature
3.1.3. Biotin/Microsphere Complex Incubation Temperature and SAPE Incubation Duration
3.1.4. Design and Optimization of the BTV xMAP Serotyping Panels
3.2. Detection of BTV Reference Strain Serotypes 1–24
3.3. Performance of the BTV xMAP Assay on Diagnostic Samples
3.4. Limit of Detection
3.5. Repeatability
3.6. Price Comparison with RT-qPCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Batten, C.A.; Henstock, M.R.; Bin-Tarif, A.; Steedman, H.M.; Waddington, S.; Edwards, L.; Oura, C.A.L. Bluetongue virus serotype 26: Infection kinetics and pathogenesis in Dorset Poll sheep. Vet. Microbiol. 2012, 157, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Darpel, K.E.; Monaghan, P.; Anthony, S.J.; Takamatsu, H.-H.; Mertens, P.P.C. Bluetongue virus in the mammalian host and the induced immune response. In Bluetongue, 1st ed.; Mellor, P.S., Baylis, M., Mertens, P.P.C., Eds.; Academic Press: London, UK, 2009; pp. 265–284. [Google Scholar]
- Carpenter, S.; Groschup, M.H.; Garros, C.; Felippe-Bauer, M.L.; Purse, B.V. Culicoides biting midges, arboviruses and public health in Europe. Antivir. Res. 2013, 100, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Ratinier, M.; Shaw, A.E.; Barry, G.; Gu, Q.; Di Gialleonardo, L.; Janowicz, A.; Varela, M.; Randall, R.E.; Caporale, M.; Palmarini, M. Bluetongue virus NS4 Protein Is an Interferon Antagonist and a Determinant of Virus Virulence. J. Virol. 2016, 90, 5427–5439. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.; Hardy, A.; Barry, G.; Pinto, R.M.; Caporale, M.; Melzi, E.; Hughes, J.; Taggart, A.; Janowicz, A.; Varela, M.; et al. Characterization of a second open reading frame in genome segment 10 of Bluetongue virus. J. Gen. Virol. 2015, 96, 3280–3293. [Google Scholar] [CrossRef] [PubMed]
- White, J.R.; Eaton, B.T. Conformation of the VP2 protein of Bluetongue virus (BTV) determines the involvement in virus neutralization of highly conserved epitopes within the BTV serogroup. J. Gen. Virol. 1990, 71, 1325–1332. [Google Scholar] [CrossRef]
- Hofmann, M.A.; Renzullo, S.; Mader, M.; Chaignat, V.; Worwa, G.; Thuer, B. Genetic characterization of toggenburg Orbivirus, a new Bluetongue virus, from goats, Switzerland. Emerg. Infect. Dis. 2008, 14, 1855–1861. [Google Scholar] [CrossRef] [PubMed]
- Zientara, S.; Sailleau, C.; Viarouge, C.; Hoeper, D.; Beer, M.; Jenckel, M.; Hoffmann, B.; Romey, A.; Bakkali-Kassimi, L.; Fablet, A.; et al. Novel Bluetongue virus in Goats, Corsica, France, 2014. Emerg. Infect. Dis. 2014, 20, 2123–2125. [Google Scholar] [CrossRef]
- Bumbarov, V.; Golender, N.; Jenckel, M.; Wernike, K.; Beer, M.; Khinich, E.; Zalesky, O.; Erster, O. Characterization of Bluetongue virus serotype 28. Transbound. Emerg. Dis. 2020, 67, 171–182. [Google Scholar] [CrossRef]
- Wright, I.M. Serological and genetic characterisation of putative new serotypes of Bluetongue virus and Epizootic haemorrhagic disease virus isolated from an Alpaca. Magister Scientiae in. Biochemistry Dissertation, North-West University, Potchefstroom, South Africa, 2014. [Google Scholar]
- Lorusso, A.; Sghaier, S.; Di Domenico, M.; Barbria, M.E.; Zaccaria, G.; Megdich, A.; Portanti, O.; Ben Seliman, I.; Spedicato, M.; Pizzurro, F.; et al. Analysis of bluetongue serotype 3 spread in Tunisia and discovery of a novel strain related to the Bluetongue virus isolated from a commercial sheep pox vaccine. Infect. Genet. Evol. 2018, 59, 63–71. [Google Scholar] [CrossRef]
- Rajko-Nenow, P.; Golender, N.; Bumbarov, V.; Brown, H.; Frost, L.; Darpel, K.; Tennakoon, C.; Flannery, J.; Batten, C. Complete genome sequence of a novel Bluetongue virus isolated from a commercial sheeppox vaccine. Microbiol. Resour. Announc. 2020, 9, e01539-19. [Google Scholar] [CrossRef]
- Sun, E.; Huang, L.; Xu, Q.; Wang, H.; Xue, X.; Lu, P.; Li, W.; Liu, W.; Bu, Z.; Wu, D. Emergence of a novel Bluetongue virus serotype, China 2014. Transbound. Emerg. Dis. 2016, 63, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Savini, G.; Puggioni, G.; Meloni, G.; Marcacci, M.; Di Domenico, M.; Rocchigiani, A.M.; Spedicato, M.; Oggiano, A.; Manunta, D.; Teodori, L.; et al. Novel putative Bluetongue virus in healthy goats from Sardinia, Italy. Infect. Genet. Evol. 2017, 51, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Belbis, G.; Zientara, S.; Breard, E.; Sailleau, C.; Caignard, G.; Vitour, D.; Attoui, H. Bluetongue virus: From BTV-1 to BTV-27. Adv. Virus Res. 2017, 99, 161–197. [Google Scholar] [CrossRef]
- Flannery, J.; Frost, L.; Fay, P.; Hicks, H.; Henstock, M.; Smreczak, M.; Orlowska, A.; Rajko-Nenow, P.; Darpel, K.; Batten, C. BTV-14 infection in sheep elicits viraemia with mild clinical symptoms. Microorganisms 2020, 8, 892. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Restirction Zones Established by the Member States. Available online: https://ec.europa.eu/food/animals/animal-diseases/control-measures/bluetongue_en (accessed on 1 September 2020).
- Kundlacz, C.; Caignard, G.; Sailleau, C.; Viarouge, C.; Postic, L.; Vitour, D.; Zientara, S.; Breard, E. Bluetongue virus in France: An Illustration of the European and Mediterranean Context since the 2000s. Viruses 2019, 11, 672. [Google Scholar] [CrossRef]
- Lorusso, A.; Sghaier, S.; Ancora, M.; Marcacci, M.; Di Gennaro, A.; Portanti, O.; Mangone, I.; Teodori, L.; Leone, A.; Camma, C.; et al. Molecular epidemiology of Bluetongue virus serotype 1 circulating in Italy and its connection with northern Africa. Infect. Genet. Evol. 2014, 28, 144–149. [Google Scholar] [CrossRef]
- Lorusso, A.; Guercio, A.; Purpari, G.; Camma, C.; Calistri, P.; D’Alterio, N.; Hammami, S.; Sghaier, S.; Savini, G. Bluetongue virus serotype 3 in Western Sicily, November 2017. Vete. Ital. 2017, 53, 273–275. [Google Scholar] [CrossRef]
- De Clercq, K.; Mertens, P.; De Leeuw, I.; Oura, C.; Houdart, P.; Potgieter, A.C.; Maan, S.; Hooyberghs, J.; Batten, C.; Vandemeulebroucke, E.; et al. Emergence of Bluetongue Serotypes in Europe, Part 2: The Occurrence of a BTV-11 Strain in Belgium. Transbound. Emerg. Dis. 2009, 56, 355–361. [Google Scholar] [CrossRef]
- Orlowska, A.; Trebas, P.; Smreczak, M.; Marzec, A.; Zmudzinski, J.F. First detection of Bluetongue virus serotype 14 in Poland. Arch. Virol. 2016, 161, 1969–1972. [Google Scholar] [CrossRef]
- Eschbaumer, M.; Hoffmann, B.; Moss, A.; Savini, G.; Leone, A.; König, P.; Zemke, J.; Conraths, F.; Beer, M. Emergence of Bluetongue virus serotype 6 in Europe-German field data and experimental infection of cattle. Vet. Microbiol. 2010, 143, 189–195. [Google Scholar] [CrossRef]
- Angeloni, S.; Cordes, R.; Dunbar, S.; Garcia, C.; Gibson, G.; Martin, C.; Stone, V. xMAP Cookbook: A Collection of Methods and Protocols for Developing Multiplex Assays with xMAP Technology; Luminex: Austin, TX, USA, 2016. [Google Scholar]
- Reslova, N.; Michna, V.; Kasny, M.; Mikel, P.; Kralik, P. xMAP technology: Applications in detection of pathogens. Front. Microbiol. 2017, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Ayouba, A.; Touré, A.; Butel, C.; Binetruy, F.; Sow, M.S.; Foulongne, V.; Delaporte, E.; Peeters, M. Development of a sensitive and specific serological assay based on Luminex technology for detection of antibodies to Zaire Ebola virus. J. Clin. Microbiol. 2017, 55, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Pavliakova, D.; Giardina, P.C.; Moghazeh, S.; Sebastian, S.; Koster, M.; Pavliak, V.; McKeen, A.; French, R.; Jansen, K.U.; Pride, M. Development and validation of 13-plex Luminex-based assay for measuring human serum antibodies to Streptococcus pneumoniae capsular polysaccharides. mSphere 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Wu, S.Q.; Jiang, L.; Xiao, R.H.; Li, T.; Mei, L.; Lv, J.Z.; Liu, J.J.; Lin, X.M.; Han, X.Q. Establishment and optimization of a liquid bead array for the simultaneous detection of ten insect-borne pathogens. Parasit. Vectors 2018, 11, 442. [Google Scholar] [CrossRef] [PubMed]
- Munro, S.B.; Kuypers, J.; Jerome, K.R. Comparison of a multiplex real-time PCR assay with a multiplex Luminex assay for influenza virus detection. J. Clin. Microbiol. 2013, 51, 1124–1129. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, Y.; Xu, Z.-q.; Zhang, Q.; Jin, M.; Yu, J.-m.; Li, J.-s.; Liu, N.; Cui, S.-x.; Kong, X.-y.; Wang, H. Simultaneous detection of seven enteric viruses associated with acute gastroenteritis by a multiplexed Luminex-based assay. J. Clin. Microbiol. 2012, 50, 2384–2389. [Google Scholar] [CrossRef] [PubMed]
- Hamza, I.A.; Jurzik, L.; Wilhelm, M. Development of a Luminex assay for the simultaneous detection of human enteric viruses in sewage and river water. J. Virol. Methods 2014, 204, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Yu, X.-L.; Gao, X.-B.; Xue, C.-Y.; Song, C.-X.; Li, Y.; Cao, Y.-C. Bead-based suspension array for simultaneous differential detection of five major swine viruses. Appl. Microbiol. Biotechnol. 2015, 99, 919–928. [Google Scholar] [CrossRef]
- Perry, M.D.; Corden, S.A.; Howe, R.A. Evaluation of the Luminex xTAG Gastrointestinal Pathogen Panel and the Savyon Diagnostics Gastrointestinal Infection Panel for the detection of enteric pathogens in clinical samples. J. Med. Microbiol. 2014, 63, 1419–1426. [Google Scholar] [CrossRef]
- Maan, S.; Maan, N.S.; Belaganahalli, M.N.; Potgieter, A.C.; Kumar, V.; Batra, K.; Wright, I.M.; Kirkland, P.D.; Mertens, P.P.C. Development and Evaluation of Real Time RT-PCR Assays for Detection and Typing of Bluetongue virus. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Hofmann, M.; Griot, C.; Chaignat, V.; Perler, L.; Thur, B. Blauzungenkrankheit erreicht die Schweiz. Schweiz. Arch. Tierheilkd. 2008, 150, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Deregt, D.; Gilbert, S.A.; Dudas, S.; Pasick, J.; Baxi, S.; Burton, K.M.; Baxi, M.K. A multiplex DNA suspension microarray for simultaneous detection and differentiation of Classical swine fever virus and other pestiviruses. J. Virol. Methods 2006, 136, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Reed, G.F.; Lynn, F.; Meade, B.D. Use of coefficient of variation in assessing variability of quantitative assays. Clin. Diagn. Lab. Immunol. 2002, 9, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Flannery, J.; Rajko-Nenow, P.; Hicks, H.; Hill, H.; Gubbins, S.; Batten, C. Evaluating the most appropriate pooling ratio for EDTA blood samples to detect Bluetongue virus using real-time RT-PCR. Vet. Microbiol. 2018, 217, 58–63. [Google Scholar] [CrossRef]
- Hemadri, D.; Maan, S.; Chanda, M.M.; Rao, P.P.; Putty, K.; Krishnajyothi, Y.; Reddy, G.H.; Kumar, V.; Batra, K.; Reddy, Y.V.; et al. Dual Infection with Bluetongue virus Serotypes and First-Time Isolation of Serotype 5 in India. Transbound. Emerg. Dis. 2017. [Google Scholar] [CrossRef]
- Brenner, J.; Batten, C.; Yadin, H.; Bumbarov, V.; Friedgut, O.; Rotenberg, D.; Golender, N.; Oura, C.A.L. Clinical syndromes associated with the circulation of multiple serotypes of Bluetongue virus in dairy cattle in Israel. Vet. Rec. 2011, 169, 389. [Google Scholar] [CrossRef]
- Brown-Joseph, T.; Batten, C.; Harrup, L.E.; Frost, L.; Flannery, J.; Hicks, H.; Ramkissoon, V.; Ramdeen, R.; Carrington, C.V.; Oura, C.A.L. Bluetongue virus infection in naive cattle: Identification of circulating serotypes and associated Culicoides biting midge species in Trinidad. Vet. Microbiol. 2017, 211, 1–5. [Google Scholar] [CrossRef]
- Golender, N.; Eldar, A.; Ehrlich, M.; Khinich, Y.; Kenigswald, G.; Varsano, J.S.; Ertracht, S.; Abramovitz, I.; Assis, I.; Shlamovitz, I.; et al. Emergence of a Novel Reassortant Strain of Bluetongue Serotype 6 in Israel, 2017: Clinical Manifestations of the Disease and Molecular Characterization. Viruses 2019, 11, 633. [Google Scholar] [CrossRef]
- Ries, C.; Beer, M.; Hoffmann, B. BlueTYPE—A low density TaqMan-RT-qPCR array for the identification of all 24 classical Bluetongue virus serotypes. J. Virol. Methods 2020, 113881. [Google Scholar] [CrossRef]

| Serotyping Panel | ORC ID | Serotype | RT-qPCR CT Value | xMAP MFI |
|---|---|---|---|---|
| RSArrrr/02 | 2W | 28.6 | 1391.3 | |
| RSArrrr/05 | 5 | 20.8 | 574.5 | |
| A | RSArrrr/19 | 19 | 17.2 | 1628.8 |
| RSArrrr/22 | 22 | 15.5 | 4963.0 | |
| RSArrrr/24 | 24 | 19.8 | 2249.4 | |
| RSArrrr/08 | 8 | 20.1 | 4959.0 | |
| RSArrrr/09 | 9 | 13.7 | 6624.0 | |
| B | RSArrrr/10 | 10 | 17.6 | 4904.0 |
| RSArrrr/16 | 16 | 20.7 | 2644.0 | |
| RSArrrr/21 | 21 | 14.5 | 1812.3 | |
| Ind1994/01 | 2E | 14.7 | 4904.5 | |
| RSArrrr/12 | 12 | 14.6 | 4604.8 | |
| C | RSArrrr/14 | 14 | 20.5 | 438.5 |
| RSArrrr/17 | 17 | 15.2 | 5061.5 | |
| RSArrrr/20 | 20 | 16.2 | 6513.5 | |
| RSArrrr/01 | 1 | 15.2 | 9667.3 | |
| RSArrrr/04 | 4 | 15.7 | 4385.3 | |
| D | RSArrrr/06 | 6 | 18.9 | 8483.5 |
| RSArrrr/07 | 7 | 16.4 | 8038.5 | |
| RSArrrr/11 | 11 | 15.4 | 3313.8 | |
| RSArrrr/03 | 3 | 15.5 | 6701.8 | |
| RSArrrr/13 | 13 | 13.6 | 6326.0 | |
| E | RSArrrr/15 | 15 | 13.9 | 7983.8 |
| RSArrrr/18 | 18 | 16.0 | 10662.3 | |
| RSArrrr/23 | 23 | 21.3 | 1027.0 |
| RT-qPCR | xMAP | RT-qPCR | xMAP | RT-qPCR | xMAP | RT-qPCR | xMAP | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample (Serotype) | CT Value | MFI | Sample (Serotype) | CT Value | MFI | Sample (Serotype) | CT Value | MFI | Sample (Serotype) | CT Value | MFI |
| OS 1 (4) | 29.6 | 1038.5 | OS 27 (4) | 28.6 | 1120.5 | PTS 13 (1) | 28.2 | 4749.0 | PTS 32 (N) | N.D. | N.D. |
| OS 3 (4) | 26.5 | 3321.0 | OS 28 (4) | 25.6 | 1621.5 | PTS 14 (1) | N.D. | 4127.5 | PTS 33 (?) | N.D. | N.D. |
| OS 4 (4) | 25.1 | 3477.0 | UKS 1 (N) | N.D. | N.D. | PTS 15 (1) | N.D. | 742.0 | PTS 34 (?) | N.D. | N.D. |
| OS 5 (11) | 35.9 | 2934.0 | UKS 2 (N) | N.D. | N.D. | PTS 16 (1) | 27.9 | 5258.0 | PTS 35 (3) | 29.9 | 1175.8 |
| OS 8 (12) | 25.2 | 260.5 | UKS 3 (N) | N.D. | N.D. | PTS 17 (1) | 24.1 | 5787.5 | PTS 36 (3) | 26.2 | 1803.3 |
| OS 9 (4) | 26.9 | 2144.0 | UKS 4 (N) | N.D. | N.D. | PTS 18 (8) | 36.4 | 183.0 | PTS 37 (3) | 22.6 | 2394.3 |
| OS 10 (4) | 28.4 | 2340.0 | PTS 1 (1) | 27.3 | 4886.0 | PTS 19 (8) | 28.2 | 1318.0 | PTS 38 (?) | N.D. | N.D. |
| OS 11 (4) | 23.0 | 4055.5 | PTS 2 (1) | 27.5 | 5074.0 | PTS 20 (N) | N.D. | N.D. | PTS 39 (4) | 28.8 | 630.0 |
| OS 12 (4) | 27.6 | 3111.0 | PTS 3 (4) | 26.3 | 3878.5 | PTS 21 (1) | 27.0 | 4392.0 | PTS 40 (?) | N.D. | N.D. |
| OS 13 (4) | 24.0 | 3942.0 | PTS 4 (4) | 27.7 | 3556.0 | PTS 22 (1) | 34.0 | 1107.5 | PTS 41 (1) | 26.2 | 3142.8 |
| OS 14 (8) | 34.4 | 396.0 | PTS 5 (8) | 30.9 | 1935.8 | PTS 23 (N) | N.D. | N.D. | PTS 42 (4) | N.D. | 1257.5 |
| OS 17 (11) | 31.0 | 2973.5 | PTS 6 (8) | 33.5 | 1201.0 | PTS 24 (4) | 29.5 | 2415.5 | PTS 43 (?) | N.D. | N.D. |
| OS 18 (4) | 30.4 | 2323.0 | PTS 7 (4) | 34.9 | 502.0 | PTS 25 (1) | 34.9 | 214.0 | PTS 44 (2) | 33.6 | 250.0 |
| OS 19 (?) | N.D. | N.D. | PTS 8 (4) | 34.6 | 262.0 | PTS 26 (1) | 30.1 | 3776.5 | PTS 45 (1) | 34.0 | 1003.8 |
| OS 21 (?) | N.D. | N.D. | PTS 9 (3) | 30.8 | 1541.3 | PTS 28 (N) | N.D. | N.D. | PTS 46 (1) | 30.1 | 2537.8 |
| OS 24 (?) | N.D. | N.D. | PTS 10 (3) | 31.1 | 1714.3 | PTS 29 (N) | N.D. | N.D. | PTS 47 (?) | N.D. | N.D. |
| OS 25 (4) | 28.1 | 977.0 | PTS 11 (1) | 21.6 | 5406.0 | PTS 30 (N) | N.D. | N.D. | - | - | |
| OS 26 (4) | 20.6 | 2296.5 | PTS 12 (1) | 25.1 | 5595.0 | PTS 31 (N) | N.D. | N.D. | - | - |
| Sample | Serotypes Detected | RT-qPCR CT Value | xMAP MFI |
|---|---|---|---|
| OS 2 | 4 | 28.1 | 3204.5 |
| 9 | N.D. | 370.0 | |
| OS 6 | 4 | 22.9 | 3864.0 |
| 9 | N.D. | 200.5 | |
| OS 7 | 9 | 24.3 | 3437.5 |
| 11 | 27.8 | 3363.0 | |
| 4 | 33.6 | N.D. | |
| OS 15 | 9 | 29.5 | 1790.0 |
| 11 | 31.9 | 1693.0 | |
| OS 16 | 9 | 26.5 | 2989.5 |
| 11 | 35.3 | 312.0 | |
| OS 20 | 9 | 31.6 | 1681.5 |
| 11 | N.D. | 248.5 | |
| OS 22 | 9 | 23.2 | 3976.5 |
| 11 | 33.3 | 1430.5 | |
| OS 23 | 4 | 21.5 | 3717.0 |
| 9 | N.D. | 216.5 | |
| PTS 27 | 15 | 34.5 | 556.0 |
| 4 | N.D. | 164.0 |
| BTV-3 (ZIM2002/03) | BTV-15 (ISR2006/11) | BTV-22 (MAR2005/05) | ||||
|---|---|---|---|---|---|---|
| Dilution Factor | xMA PMFI (%CV) | RT-qPCR CT (%CV) | xMAP MFI (%CV) | RT-qPCR CT (%CV) | xMAP MFI (%CV) | RT-qPCR CT (%CV) |
| 10−1 | 5902.3 (3.33) | 20.1 (0.95) | 7289.0 (4.05) | 21.1 (0.19) | 2613.3 (1.81) | 15.5 (0.54) |
| 10−2 | 7050.5 (3.25) | 24.1 (2.01) | 6496.5 (3.24) | 23.7 (0.77) | 2767.0 (7.48) | 18.5 (0.76) |
| 10−3 | 6912.5 (0.77) | 26.9 (0.49) | 4629.5 (4.87) | 27.4 (0.26) | 3051.2 (7.82) | 22.1 (1.35) |
| 10−4 | 2919.3 (3.37) | 30.2 (0.31) | 2721.2 (8.74) | 30.8 (0.47) | 2797.6 (0.88) | 26.0 (0.58) |
| 10−5 | 1301.0 (4.54) | 33.7 (0.57) | 1554.2 (6.02) | 34.5 (1.72) | 1801.5 (13.58) | 29.6 (0.11) |
| 10−6 | N.D. | N.D. | 593.0 (35.36) | N.D. | 811.1 (17.35) | 34.3 (1.61) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashby, M.; Rajko-Nenow, P.; Batten, C.; Flannery, J. Simultaneous Detection of Bluetongue Virus Serotypes Using xMAP Technology. Microorganisms 2020, 8, 1564. https://doi.org/10.3390/microorganisms8101564
Ashby M, Rajko-Nenow P, Batten C, Flannery J. Simultaneous Detection of Bluetongue Virus Serotypes Using xMAP Technology. Microorganisms. 2020; 8(10):1564. https://doi.org/10.3390/microorganisms8101564
Chicago/Turabian StyleAshby, Martin, Paulina Rajko-Nenow, Carrie Batten, and John Flannery. 2020. "Simultaneous Detection of Bluetongue Virus Serotypes Using xMAP Technology" Microorganisms 8, no. 10: 1564. https://doi.org/10.3390/microorganisms8101564
APA StyleAshby, M., Rajko-Nenow, P., Batten, C., & Flannery, J. (2020). Simultaneous Detection of Bluetongue Virus Serotypes Using xMAP Technology. Microorganisms, 8(10), 1564. https://doi.org/10.3390/microorganisms8101564





