Sphingopyxis sp. Strain OPL5, an Isoprene-Degrading Bacterium from the Sphingomonadaceae Family Isolated from Oil Palm Leaves
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, Enrichment Assays, and DNA-SIP Experiments
2.2. Identification of Active Isoprene-Degrading Bacteria from Oil Palm Soils and Leaves
2.3. Enrichment and Isolation of Isoprene-Degrading Bacteria from Oil Palm Samples
2.4. Genome Sequencing and Analysis
2.5. Genome Analysis
2.6. Isoprene Oxidation Assays
3. Results and Discussion
3.1. Testing Methods to Recover Leaf Epiphytes
3.2. Active Isoprene-Degrading Bacteria Associated with an Oil Palm Tree
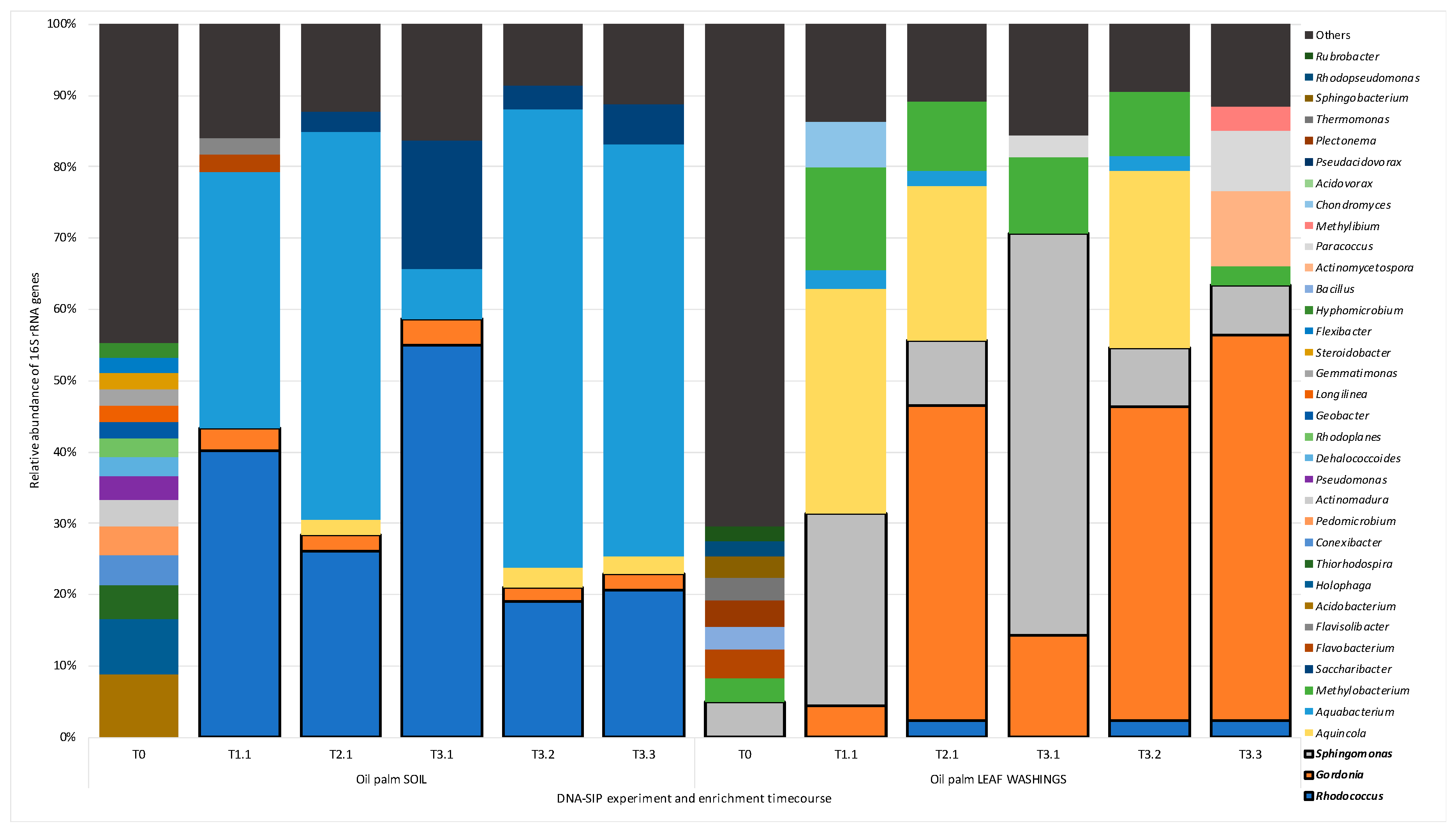
3.3. Bacterial Community Composition in SIP Enrichments
3.4. 13C—Labelling of Putative Isoprene Degraders in Leaf Washings
3.5. 13C—Labelling of Putative Isoprene Degraders in Soil
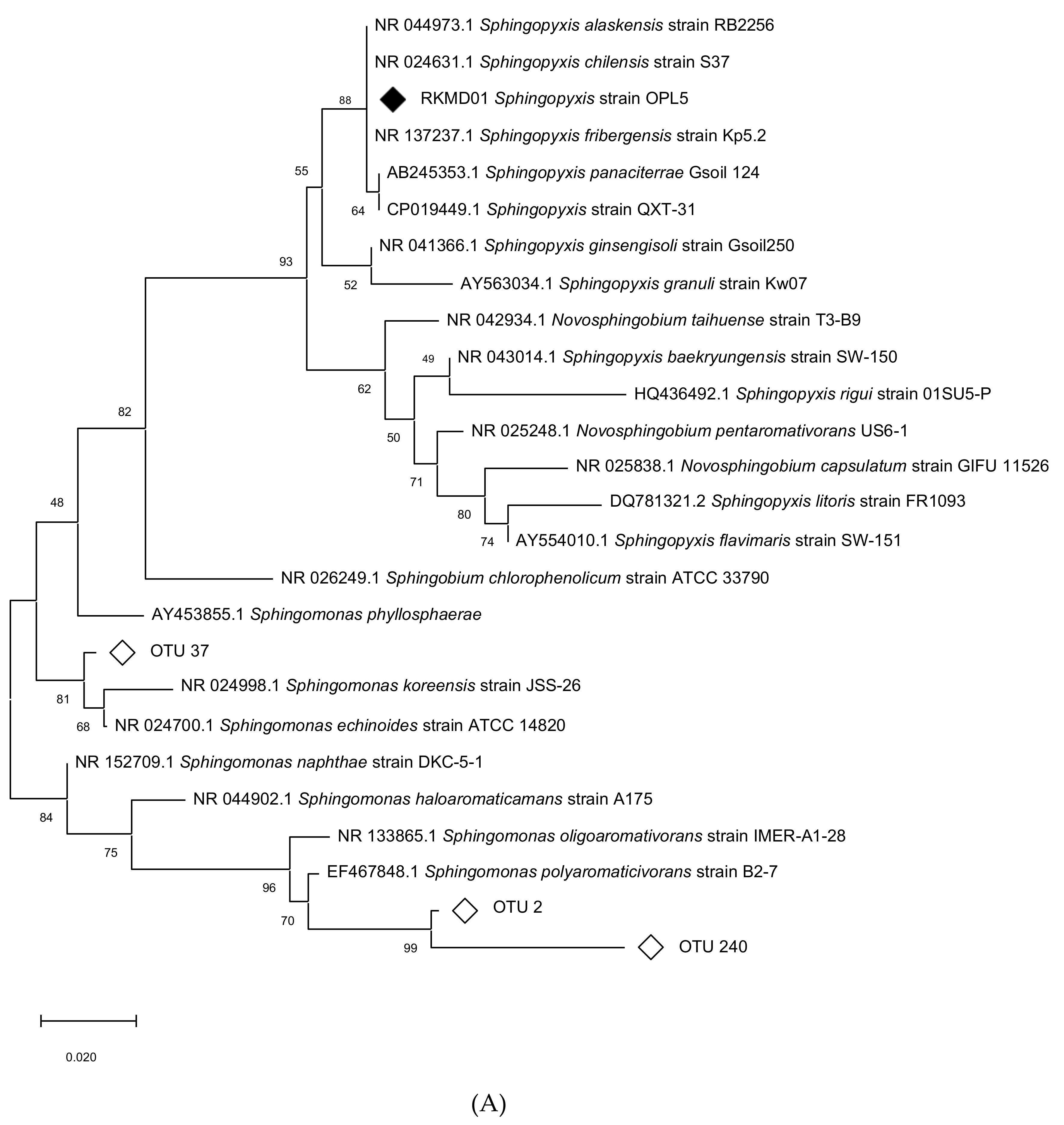
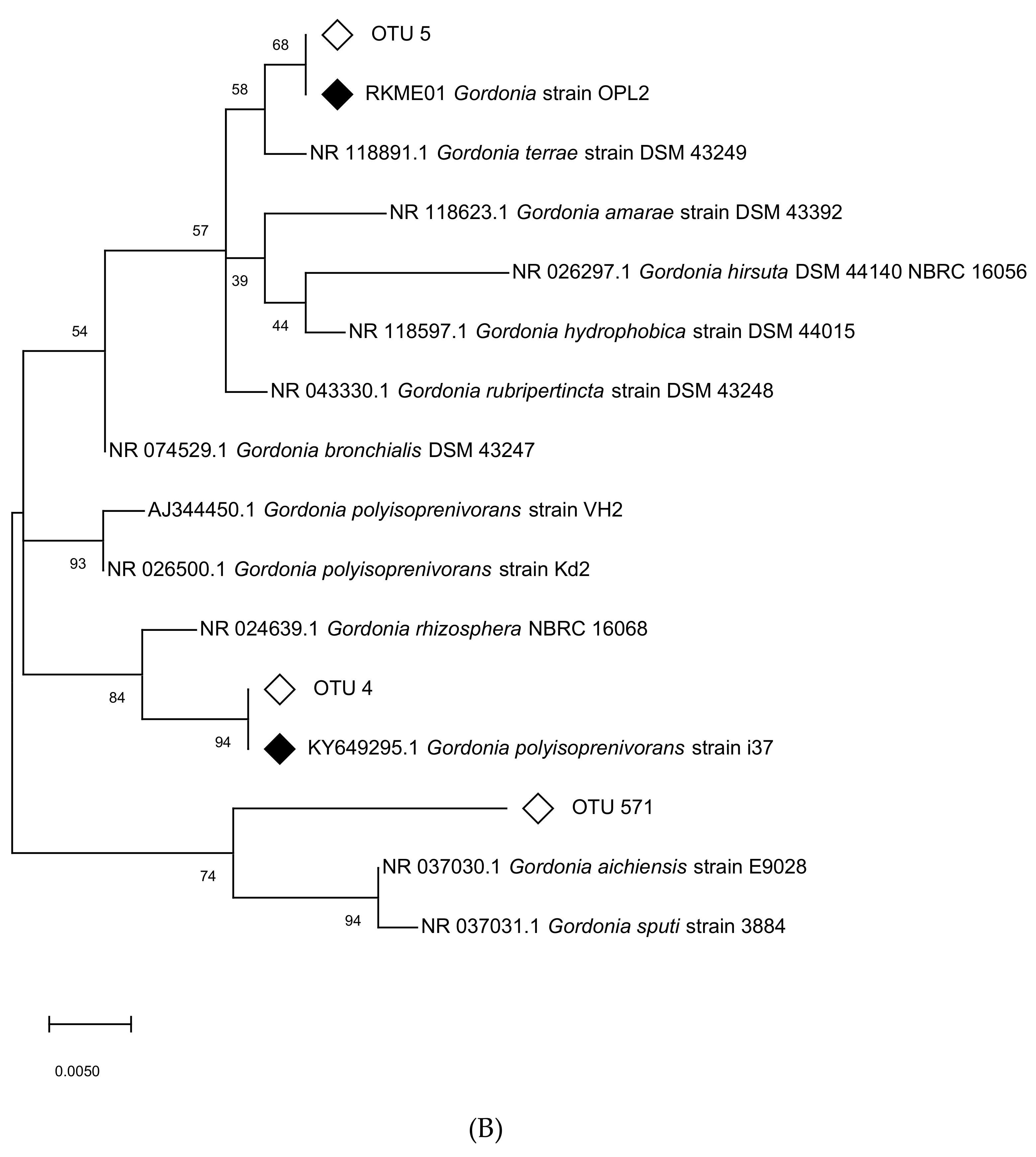
3.6. Targeted Isolation of Isoprene Degraders from Oil Palm
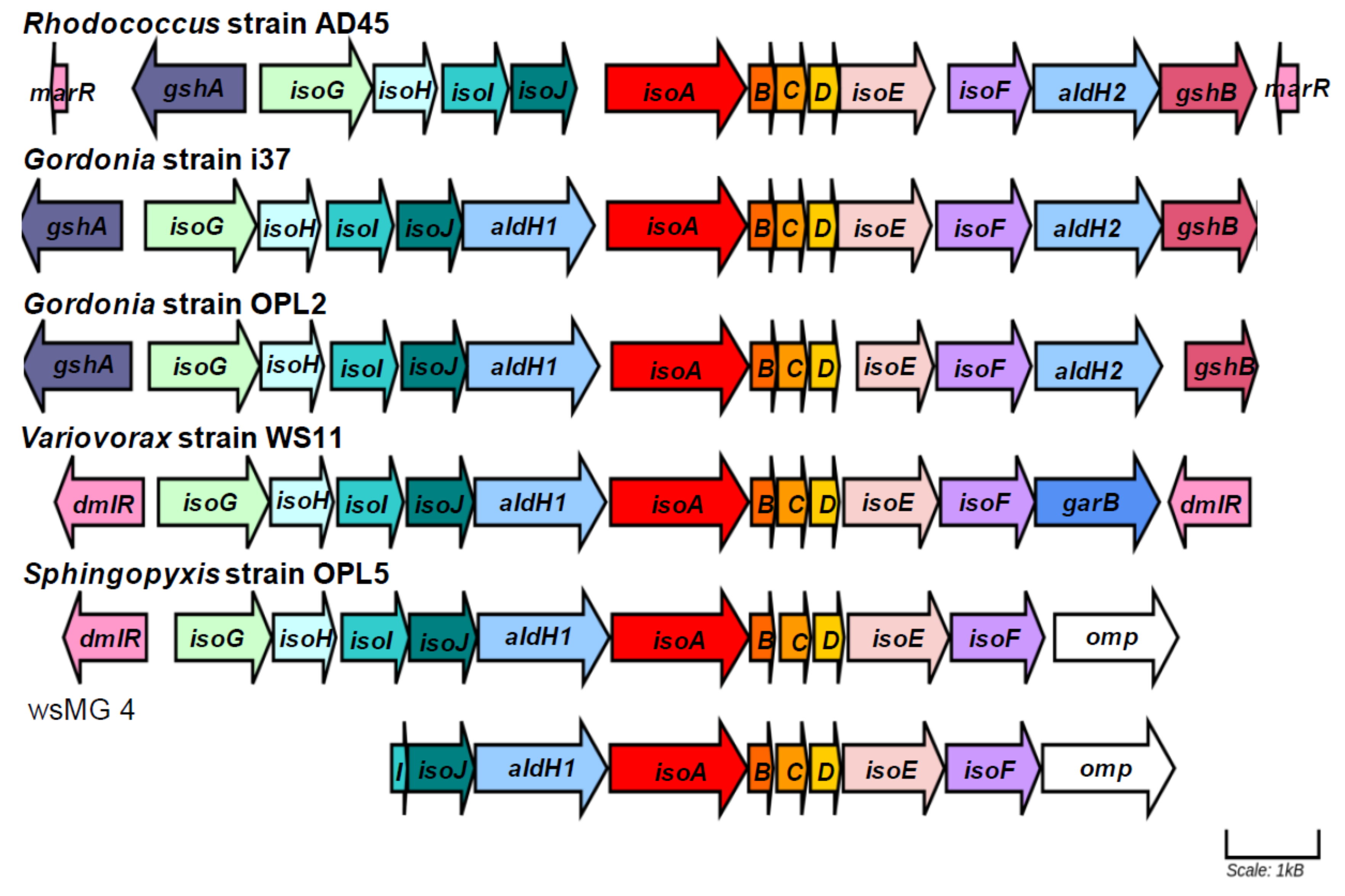
3.7. Genome Sequencing and Characterization of Isoprene Gene Clusters
3.8. Sphingopyxis Strain OPL5 Growth and Affinity for Isoprene
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arneth, A.; Monson, R.K.; Schurgers, G.; Niinemets, Ü.; Palmer, P.I. Why are estimates of global terrestrial isoprene emissions so similar (and why is this not so for monoterpenes)? Atmos. Chem. Phys. 2008, 8, 4605–4620. [Google Scholar] [CrossRef]
- Guenther, A.B.; Jiang, X.; Heald, C.L.; Sakulyanontvittaya, T.; Duhl, T.; Emmons, L.K.; Wang, X. The model of emissions of gases and aerosols from nature version 2.1 (MEGAN2.1): An extended and updated framework for modeling biogenic emissions. Geosci. Model Dev. 2012, 5, 1471–1492. [Google Scholar] [CrossRef]
- Guenther, A. A global model of natural volatile organic compound emissions. J. Geophys. Res. 1995, 100, 8873–8892. [Google Scholar] [CrossRef]
- Atkinson, R.; Arey, J. Atmospheric degradation of volatile organic compounds. Chem. Rev. 2003, 103, 4605–4638. [Google Scholar] [CrossRef]
- Carlton, A.G.; Wiedinmyer, C.; Kroll, J.H. A review of secondary organic aerosol (SOA) formation from isoprene. Atmos. Chem. Phys. 2009, 9, 4987–5005. [Google Scholar] [CrossRef]
- Guenther, A.; Karl, T.; Harley, P.; Wiedinmyer, C.; Palmer, P.I.; Geron, C. Estimates of global terrestrial isoprene emissions using MEGAN. Atmos. Chem. Phys. Discuss. 2006, 6, 107–173. [Google Scholar] [CrossRef]
- Loreto, F.; Sharkey, T.D. On the relationship between isoprene emission and photosynthetic metabolites under different environmental conditions. Planta 1993, 189, 420–424. [Google Scholar] [CrossRef]
- Lantz, A.T.; Allman, J.; Weraduwage, S.M.; Sharkey, T.D. Isoprene: New insights into the control of emission and mediation of stress tolerance by gene expression. Plant Cell Environ. 2019, 42, 2808–2826. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Monson, R.K. Isoprene research – 60 years later, the biology is still enigmatic. Plant Cell Environ. 2017, 40, 1671–1678. [Google Scholar] [CrossRef]
- Klinger, L.F.; Greenburg, J.; Guenther, A.; Tyndall, G.; Zimmerman, P.; M’Bangui, M.; Moutsamboté, J.-M.; Kenfack, D. Patterns in volatile organic compound emissions along a savanna-rainforest gradient in central Africa. J. Geophys. Res. Atmos. 1998, 103, 1443–1454. [Google Scholar] [CrossRef]
- Hewitt, C.N.N.; Street, R.A. A qualitative assessment of the emission of non-methane hydrocarbon compounds from the biosphere to the atmosphere in the U.K.: Present knowledge and uncertainties. Atmos. Environ. Part A Gen. Top. 1992, 26, 3069–3077. [Google Scholar] [CrossRef]
- Cheng, Y.; Yu, L.; Xu, Y.; Lu, H.; Cracknell, A.P.; Kanniah, K.; Gong, P. Mapping oil palm plantation expansion in Malaysia over the past decade (2007–2016) using ALOS-1/2 PALSAR-1/2 data. Int. J. Remote Sens. 2019, 40, 7389–7408. [Google Scholar] [CrossRef]
- Hewitt, C.N.; MacKenzie, A.R.; Di Carlo, P.; Di Marco, C.F.; Dorsey, J.R.; Evans, M.; Fowler, D.; Gallagher, M.W.; Hopkins, J.R.; Jones, C.E.; et al. Nitrogen management is essential to prevent tropical oil palm plantations from causing ground-level ozone pollution. Proc. Natl. Acad. Sci. USA 2009, 106, 18447–18451. [Google Scholar] [CrossRef] [PubMed]
- Van Ginkel, C.G.; De Jong, E.; Tilanus, J.W.R.; De Bont, J.A.M. Microbial oxidation of isoprene, a biogenic foliage volatile and of 1,3-butadiene, an anthropogenic gas. FEMS Microbiol. Lett. 1987, 45, 275–279. [Google Scholar] [CrossRef]
- Fall, R.; Copley, S.D. Bacterial sources and sinks of isoprene, a reactive atmospheric hydrocarbon. Environ. Microbiol. 2000, 2, 123–130. [Google Scholar] [CrossRef]
- Murrell, J.C.; McGenity, T.J.; Crombie, A.T. Microbial metabolism of isoprene: A much-neglected climate-active gas. Microbiology 2020, 1–25. [Google Scholar] [CrossRef]
- Carrión, O.; McGenity, T.J.; Murrell, J.C. Molecular ecology of isoprene-degrading bacteria. Microorganisms 2020, 8, 967. [Google Scholar] [CrossRef]
- McGenity, T.J.; Crombie, A.T.; Murrell, J.C. Microbial cycling of isoprene, the most abundantly produced biological volatile organic compound on Earth. ISME J. 2018, 12, 931–941. [Google Scholar] [CrossRef]
- El Khawand, M.; Crombie, A.T.; Johnston, A.; Vavlline, D.V.; McAuliffe, J.C.; Latone, J.A.; Primak, Y.A.; Lee, S.K.; Whited, G.M.; McGenity, T.J.; et al. Isolation of isoprene degrading bacteria from soils, development of isoA gene probes and identification of the active isoprene-degrading soil community using DNA-stable isotope probing. Environ. Microbiol. 2016, 18, 2743–2753. [Google Scholar] [CrossRef]
- Crombie, A.T.; Emery, H.; McGenity, T.J.; Murrell, J.C. Draft genome sequences of three terrestrial isoprene-degrading Rhodococcus strains. Genome Announc. 2017, 5, e01256-17. [Google Scholar] [CrossRef]
- Johnston, A.; Crombie, A.T.; El Khawand, M.; Sims, L.; Whited, G.M.; McGenity, T.J.; Murrell, J.C. Identification and characterisation of isoprene-degrading bacteria in an estuarine environment. Environ. Microbiol. 2017, 19, 3526–3537. [Google Scholar] [CrossRef] [PubMed]
- Gibson, L.; Larke-Mejía, N.L.; Murrell, J.C. Complete genome of isoprene degrading Nocardioides sp. WS12. Microorganisms 2020, 8, 889. [Google Scholar] [CrossRef] [PubMed]
- Van Hylckama Vlieg, J.E.T.T.; Leemhuis, H.; Lutje Spelberg, J.H.; Janssen, D.B.; Jeffrey, H.; Spelberg, L.; Janssen, D.B.; Ad, S.; Vlieg, J.E.T.V.H.; Leemhuis, H.; et al. Characterization of the gene cluster involved in isoprene metabolism in Rhodococcus sp. strain AD45. J. Bacteriol. 2000, 182, 1956–1963. [Google Scholar] [CrossRef] [PubMed]
- Crombie, A.T.; Khawand, M.E.; Rhodius, V.A.; Fengler, K.A.; Miller, M.C.; Whited, G.M.; McGenity, T.J.; Murrell, J.C. Regulation of plasmid-encoded isoprene metabolism in Rhodococcus, a representative of an important link in the global isoprene cycle. Environ. Microbiol. 2015, 17, 3314–3329. [Google Scholar] [CrossRef] [PubMed]
- Larke-Mejía, N.L.; Crombie, A.T.; Pratscher, J.; McGenity, T.J.; Murrell, J.C. Novel isoprene-degrading Proteobacteria from soil and leaves identified by cultivation and metagenomics analysis of stable isotope probing experiments. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Dawson, R.A.; Larke-Mejía, N.L.; Crombie, A.T.; Ul Haque, M.F.; Murrell, J.C. Isoprene oxidation by the Gram-negative model bacterium Variovorax sp. WS11. Microorganisms 2020, 8, 349. [Google Scholar] [CrossRef] [PubMed]
- Crombie, A.T.; Larke-Mejia, N.L.; Emery, H.; Dawson, R.; Pratscher, J.; Murphy, G.P.; McGenity, T.J.; Murrell, J.C. Poplar phyllosphere harbors disparate isoprene-degrading bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, 13081–13086. [Google Scholar] [CrossRef]
- Coleman, N.V.; Bui, N.B.; Holmes, A.J. Soluble di-iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environ. Microbiol. 2006, 8, 1228–1239. [Google Scholar] [CrossRef]
- Carrión, O.; Larke-Mejía, N.L.; Gibson, L.; Farhan Ul Haque, M.; Ramiro-García, J.; McGenity, T.J.; Murrell, J.C. Gene probing reveals the widespread distribution, diversity and abundance of isoprene-degrading bacteria in the environment. Microbiome 2018, 6, 219. [Google Scholar] [CrossRef]
- Carrión, O.; Gibson, L.; Elias, D.M.O.; McNamara, N.P.; van Alen, T.A.; Op den Camp, H.J.M.; Supramaniam, C.V.; McGenity, T.J.; Murrell, J.C. Diversity of isoprene-degrading bacteria in phyllosphere and soil communities from a high isoprene-emitting environment: a Malaysian oil palm plantation. Microbiome 2020, 8, 81. [Google Scholar] [CrossRef]
- Hedin, G.; Rynbäck, J.; Loré, B. New technique to take samples from environmental surfaces using flocked nylon swabs. J. Hosp. Infect. 2010, 75, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Ewers, J.; Freier-Schröder, D.; Knackmuss, H.J. Selection of trichloroethene (TCE) degrading bacteria that resist inactivation by TCE. Arch. Microbiol. 1990, 154, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, 1–11. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic acid techniques in bacterial systematics.; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons, Inc.: New York, NY, USA, 1991; ISBN 0471929069. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 1–15. [Google Scholar] [CrossRef]
- Vallenet, D.; Engelen, S.; Mornico, D.; Cruveiller, S.; Fleury, L.; Lajus, A.; Rouy, Z.; Roche, D.; Salvignol, G.; Scarpelli, C.; et al. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford). 2009, 2009, bap021. [Google Scholar] [CrossRef]
- Altschul, S. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Gunturu, S.; Harvey, W.T.; Rosselló-Mora, R.; Tiedje, J.M.; Cole, J.R.; Konstantinidis, K.T. The Microbial Genomes Atlas (MiGA) webserver: Taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res. 2018, 46, W282–W288. [Google Scholar] [CrossRef]
- Durinck, S.; Bullard, J.; Spellman, P.T.; Dudoit, S. GenomeGraphs: Integrated genomic data visualization with R. BMC Bioinform. 2009, 10, 2. [Google Scholar] [CrossRef]
- Clark, L.C.; Wolf, R.; Granger, D.; Taylor, Z. Continuous recording of blood oxygen tensions by polarography. J. Appl. Physiol. 1953, 6, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, J.D.; Vohra, J.; Dumont, M.G.; Lueders, T.; Manefield, M.; Friedrich, M.W.; Murrell, C.J. DNA stable-isotope probing. Nat. Protoc. 2007, 2, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, F.; Muzica, L.; Schuster, J.; Treuter, N.; Rosell, M.; Harms, H.; Müller, R.H.; Rohwerder, T. Alkene formation from tertiary alkyl ether and alcohol degradation by Aquincola tertiaricarbonis L108 and Methylibium spp. Am. Soc. Microbiol. 2011, 77, 5981–5987. [Google Scholar] [CrossRef][Green Version]
- Schuster, J.; Schäfer, F.; Hübler, N.; Brandt, A.; Rosell, M.; Härtig, C.; Harms, H.; Müller, R.H.; Rohwerder, T. Bacterial degradation of tert-amyl alcohol proceeds via hemiterpene 2-methyl-3-buten-2-ol by employing the tertiary alcohol desaturase function of the Rieske nonheme mononuclear iron oxygenase MdpJ. J. Bacteriol. 2012, 194, 972–981. [Google Scholar] [CrossRef]
- Sy, A.; Timmers, A.C.J.; Knief, C.; Vorholt, J.A. Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl. Environ. Microbiol. 2005, 71, 7245–7252. [Google Scholar] [CrossRef]
- Srivastva, N.; Vishwakarma, P.; Bhardwaj, Y.; Singh, A.; Manjunath, K.; Dubey, S.K. Kinetic and molecular analyses reveal isoprene degradation potential of Methylobacterium sp. Bioresour. Technol. 2017, 242, 87–91. [Google Scholar] [CrossRef]
- Murphy, G.P. Isoprene degradation in the terrestrial environment. Ph.D Thesis, University of Essex, Colchester, UK, 2016. [Google Scholar]
- Posman, K.M.; DeRito, C.M.; Madsen, E.L. Benzene degradation by a Variovorax species within a coal tar-contaminated groundwater microbial community. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Lin, Z.; Marett, L.; Hughen, R.W.; Flores, M.; Forteza, I.; Ammon, M.A.; Concepcion, G.P.; Espino, S.; Olivera, B.M.; Rosenberg, G.; et al. Neuroactive diol and acyloin metabolites from cone snail-associated bacteria. Bioorganic Med. Chem. Lett. 2013, 23, 4867–4869. [Google Scholar] [CrossRef]
- Lee, H.W.; Ten, I.L.; Jung, H.M.; Liu, Q.M.; Im, W.T.; Lee, S.T. Sphingopyxis panaciterrae sp. nov., isolated from soil of ginseng field. J. Microbiol. Biotechnol. 2008, 18, 1011–1015. [Google Scholar]
- Verma, H.; Dhingra, G.G.; Sharma, M.; Gupta, V.; Negi, R.K.; Singh, Y.; Lal, R. Comparative genomics of Sphingopyxis spp. unravelled functional attributes. Genomics 2020, 112, 1956–1969. [Google Scholar] [CrossRef]
- Van Hylckama Vlieg, J.E.; Kingma, J.; van den Wijngaard, A.J.; Janssen, D.B. A glutathione S-transferase with activity towards cis-1, 2-dichloroepoxyethane is involved in isoprene utilization by Rhodococcus sp. strain AD45. Appl. Environ. Microbiol. 1998, 64, 2800–2805. [Google Scholar] [CrossRef] [PubMed]
| Gordonia OPL2 | Sphingopyxis OPL5 | |
|---|---|---|
| NCBI Tax ID | 2486274 | 2486273 |
| Length (bp) | 5,759,526 | 4,676,975 |
| GC (%) | 67.3 | 65.89 |
| Contigs | 132 | 1 |
| N50 | 80,039 | 4,676,975 |
| CDS (total) | 5313 | 4403 |
| Genes (coding) | 5200 | 4392 |
| Genes (RNA) | 55 | 51 |
| rRNAs (5S, 16S, 23S) | 3, 3, 1 | 1, 1, 1 |
| tRNAs | 45 | 55 |
| Pseudogenes (total) | 113 | 69 |
| Coding Density (%) | 91.2 | 91.9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larke-Mejía, N.L.; Carrión, O.; Crombie, A.T.; McGenity, T.J.; Murrell, J.C. Sphingopyxis sp. Strain OPL5, an Isoprene-Degrading Bacterium from the Sphingomonadaceae Family Isolated from Oil Palm Leaves. Microorganisms 2020, 8, 1557. https://doi.org/10.3390/microorganisms8101557
Larke-Mejía NL, Carrión O, Crombie AT, McGenity TJ, Murrell JC. Sphingopyxis sp. Strain OPL5, an Isoprene-Degrading Bacterium from the Sphingomonadaceae Family Isolated from Oil Palm Leaves. Microorganisms. 2020; 8(10):1557. https://doi.org/10.3390/microorganisms8101557
Chicago/Turabian StyleLarke-Mejía, Nasmille L., Ornella Carrión, Andrew T. Crombie, Terry J. McGenity, and J. Colin Murrell. 2020. "Sphingopyxis sp. Strain OPL5, an Isoprene-Degrading Bacterium from the Sphingomonadaceae Family Isolated from Oil Palm Leaves" Microorganisms 8, no. 10: 1557. https://doi.org/10.3390/microorganisms8101557
APA StyleLarke-Mejía, N. L., Carrión, O., Crombie, A. T., McGenity, T. J., & Murrell, J. C. (2020). Sphingopyxis sp. Strain OPL5, an Isoprene-Degrading Bacterium from the Sphingomonadaceae Family Isolated from Oil Palm Leaves. Microorganisms, 8(10), 1557. https://doi.org/10.3390/microorganisms8101557





