Reconstructing Genomes of Carbon Monoxide Oxidisers in Volcanic Deposits Including Members of the Class Ktedonobacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequencing
2.2. Quality Control
2.3. Metagenome Assembly and Binning
2.4. Functional Annotation
2.5. Phylogenomic Analysis
2.6. Accession Number
3. Results
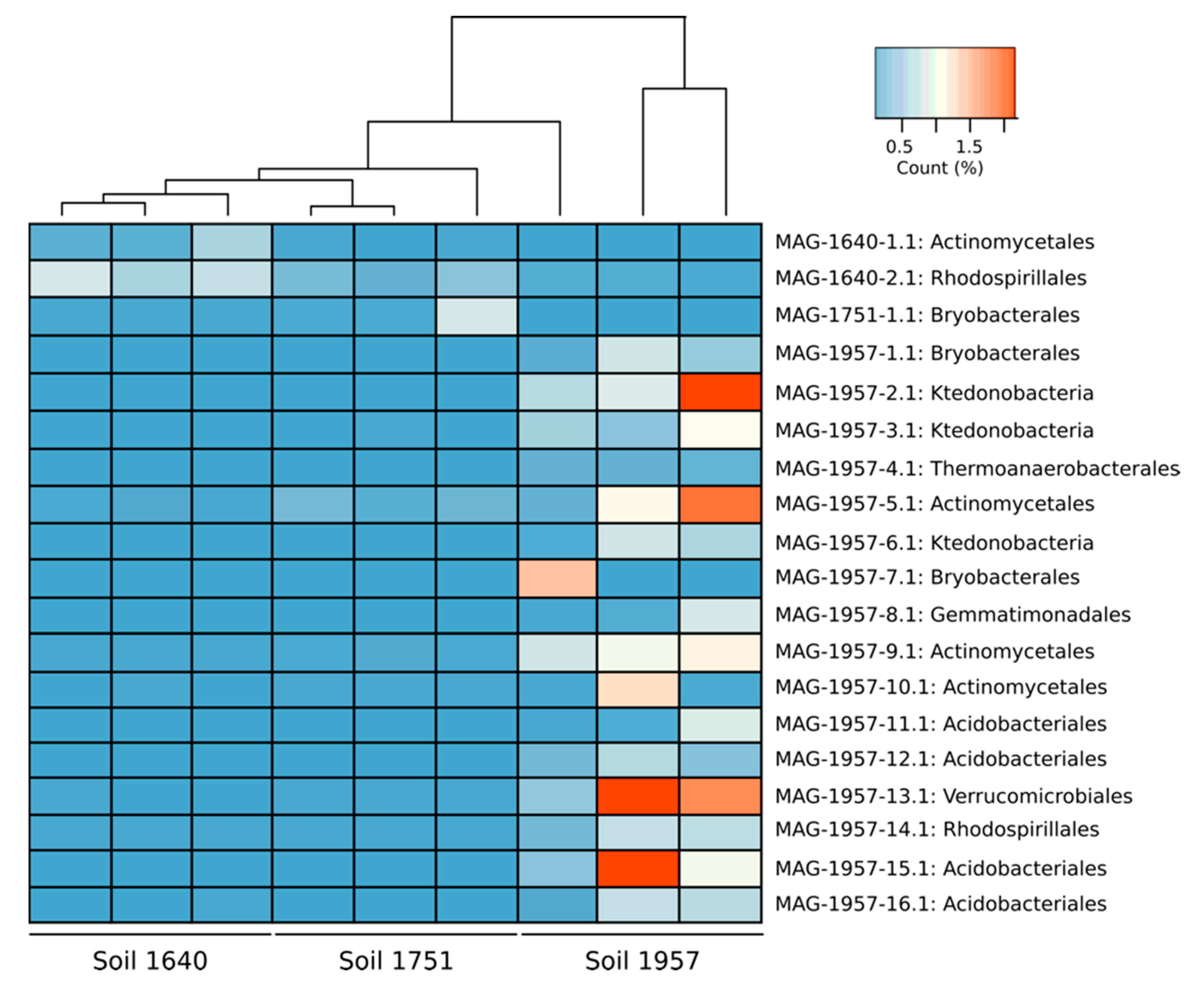
3.1. MAGs Recovery
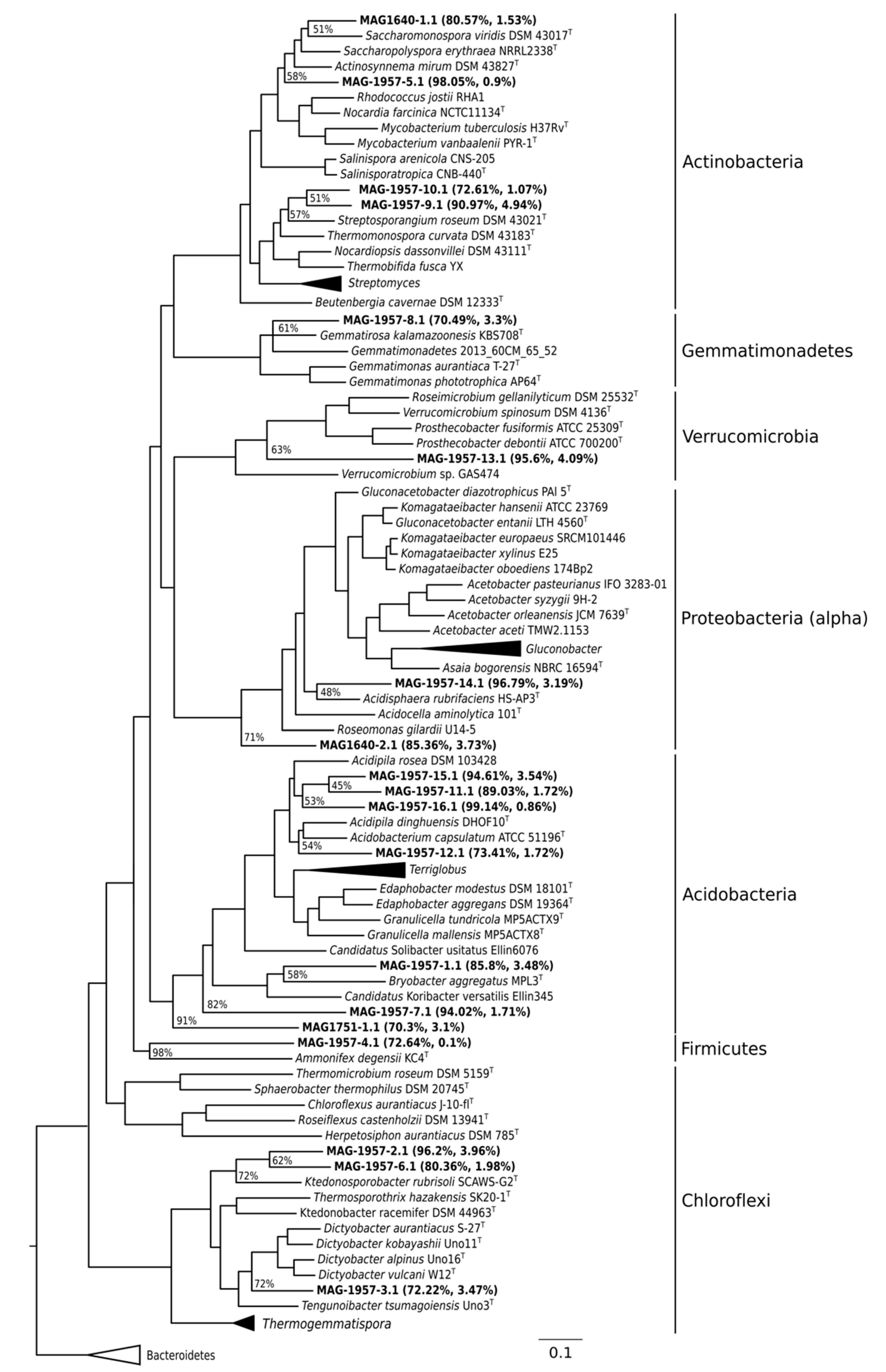
3.2. MAG Identification
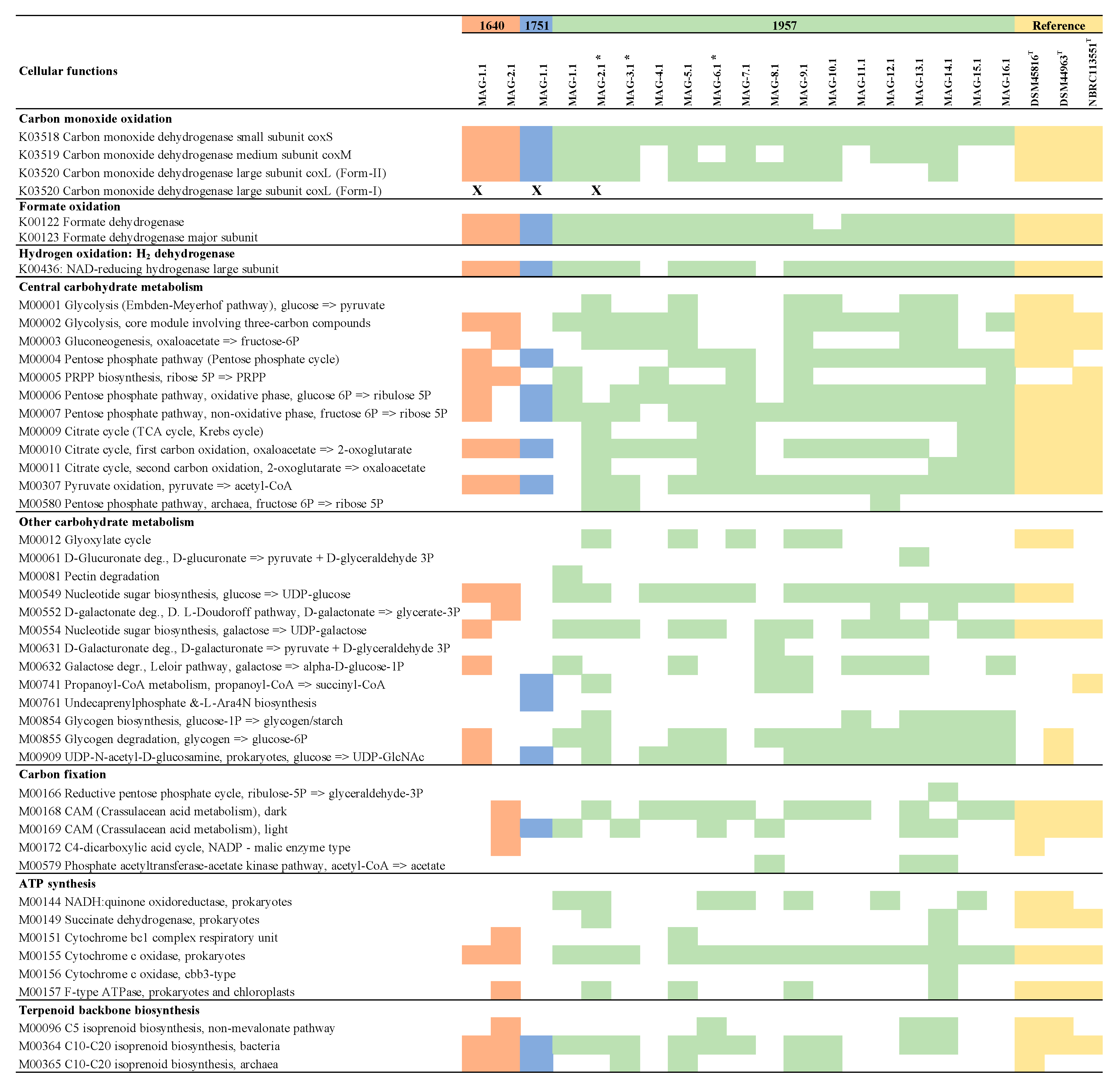
3.3. Metabolic Characterisation of MAGs
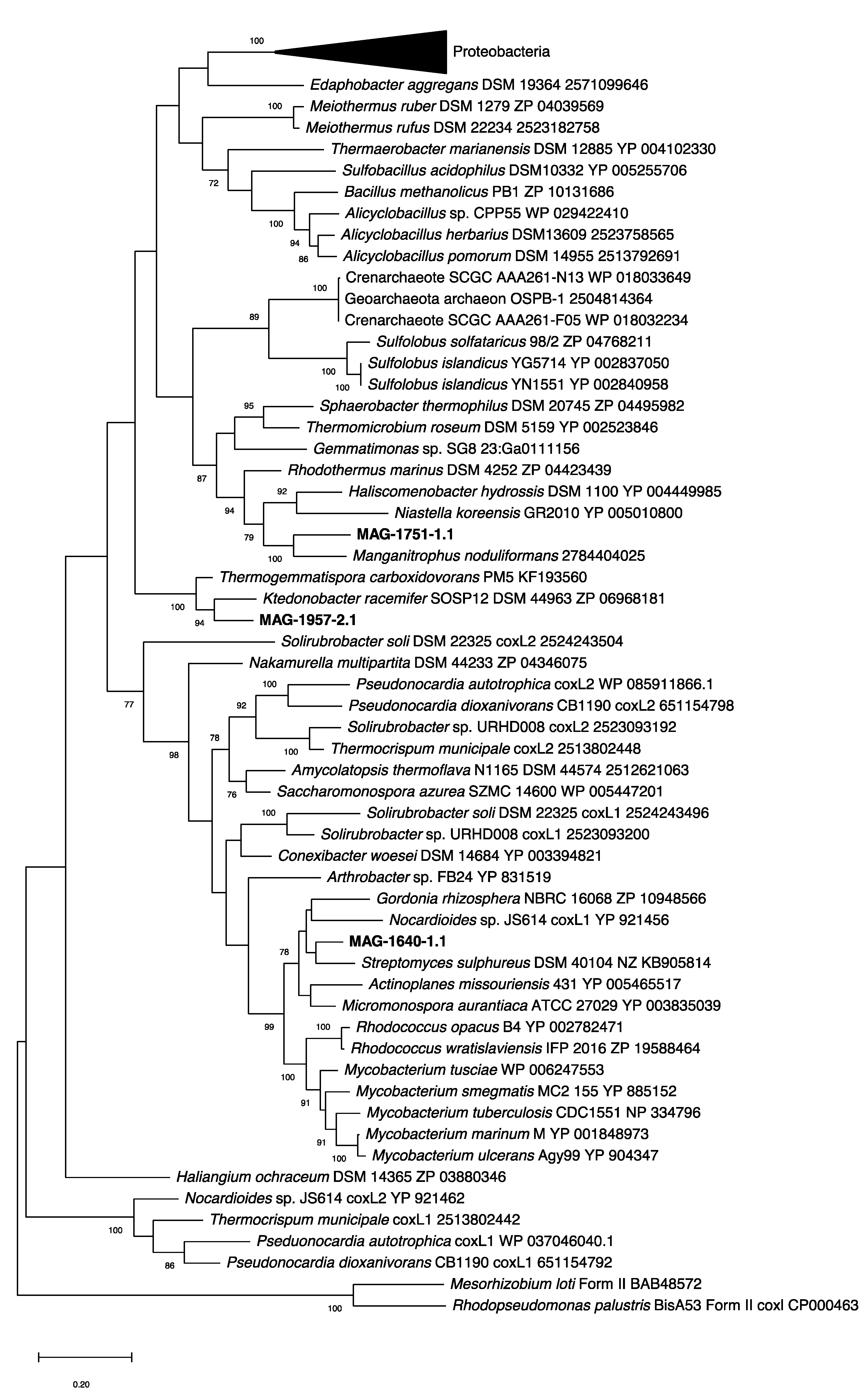
3.3.1. Characterisation of CODH and Hydrogenase Genes in MAGs
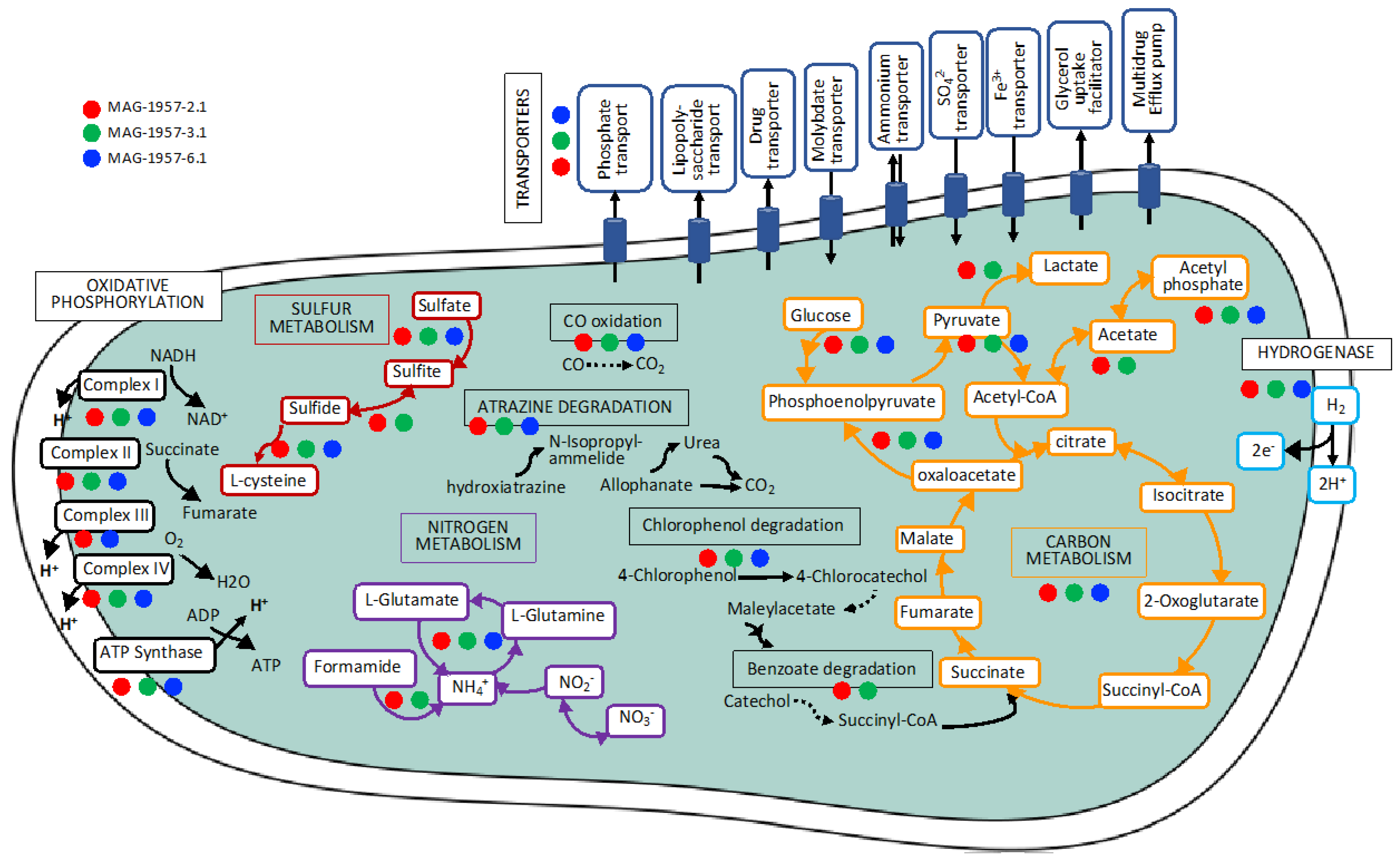
3.3.2. Complete Metabolic Characterisation of Ktedonobacterales MAGs
4. Discussion
4.1. Characterisation of MAGs
4.2. Metabolic Characterisation of MAGs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wubs, E.R.; van der Putten, W.H.; Bosch, M.; Bezemer, T.M. Soil inoculation steers restoration of terrestrial ecosystems. Nat. Plants 2016, 2, 16107. [Google Scholar] [CrossRef] [PubMed]
- Kelly, L.C.; Cockell, C.S.; Thorsteinsson, T.; Marteinsson, V.; Stevenson, J. Pioneer microbial communities of the Fimmvörðuháls Lava Flow, Eyjafjallajökull, Iceland. Microb. Ecol. 2014, 68, 504–518. [Google Scholar] [CrossRef] [PubMed]
- Witt, V.; Ayris, P.M.; Damby, D.E.; Cimarelli, C.; Kueppers, U.; Dingwell, D.B.; Wörheide, G. Volcanic ash supports a diverse bacterial community in a marine mesocosm. Geobiology 2017, 15, 453–463. [Google Scholar] [CrossRef]
- Smith, H.G. The colonization of volcanic tephra on deception island by protozoa. Br. Antarct. Surv. Bull. 1974, 38, 49–58. [Google Scholar]
- Chadwick, O.A.; Derry, L.; Vitousek, P.M.; Huebert, B.J.; Hedin, L.O. Changing sources of nutrients during fourmillion years of ecosystem development. Nature 1999, 397, 491–497. [Google Scholar] [CrossRef]
- Fujimura, R.; Sato, Y.; Nishizawa, T.; Nanba, K.; Oshima, K.; Hattori, M.; Kamijo, T.; Ohta, H. Analysis of early bacterial communities on volcanic deposits on the island of Miyake (Miyake-jima), Japan: A 6-year study at a fixed site. Microbes Environ. 2012, 27, 19–29. [Google Scholar] [CrossRef] [PubMed]
- King, G.M. Contributions of atmospheric CO and hydrogen uptake to microbial dynamics on recent Hawaiian volcanic deposits. Appl. Environ. Microbiol. 2003, 69, 4067–4075. [Google Scholar] [CrossRef]
- Ohta, H.; Ogiwara, K.; Murakami, E.; Takahashi, H.; Sekiguchi, M.; Koshida, K.; Someya, T.; Morishima, W.; Rondal, J.D.; Concepcion, R.N.; et al. Quinone profiling of bacterial populations developed in the surface layer of volcanic mudflow deposits from Mt. Pinatubo (the Philippines). Soil Biol. Biochem. 2003, 35, 1155–1158. [Google Scholar] [CrossRef]
- Sato, Y.; Nishihara, H.; Yoshida, M.; Watanabe, M.; Rondal, J.D.; Ohta, H. Occurrence of hydrogen-oxidizing Ralstonia species as primary microorganisms in the Mt. Pinatubo volcanic mudflow deposits. Soil Sci. Plant Nutr. 2004, 50, 855–861. [Google Scholar] [CrossRef]
- Gomez-Alvarez, V.; King, G.M.; Nüsslein, K. Comparative bacterial diversity in recent Hawaiian volcanic deposits of different ages. FEMS Microbiol. Ecol. 2007, 60, 60–73. [Google Scholar] [CrossRef]
- Guo, Y.; Fujimura, R.; Sato, Y.; Suda, W.; Kim, S.W.; Oshima, K.; Hattori, M.; Kamijo, T.; Narisawa, K.; Ohta, H. Characterization of early microbial communities on volcanic deposits along a vegetation gradient on the island of Miyake, Japan. Microbes Environ. 2014, 29, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kim, D.S.; Lee, K.C.; Lee, J.S.; King, G.M.; Kang, S. Microbial community structure and functional potential of lava-formed Gotjawal soils in Jeju, Korea. PLoS ONE 2018, 13, e0204761. [Google Scholar] [CrossRef] [PubMed]
- King, G.M. Molecular and culture-based analyses of aerobic carbon monoxide oxidizer diversity. Appl. Environ. Microbiol. 2003, 69, 7257–7265. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.F.; King, G.M. Distribution and diversity of carbon monoxide-oxidizing bacteria and bulk bacterial communities across a succession gradient on a Hawaiian volcanic deposit. Environ. Microbiol. 2010, 12, 1855–1867. [Google Scholar] [CrossRef]
- Cordero, P.R.F.; Bayly, K.; Leung, P.M.; Huang, C.; Islam, Z.F.; Schittenhelm, R.B.; King, G.M.; Greening, C. Atmospheric carbon monoxide oxidation is a widespread mechanism supporting microbial survival. ISME J. 2019, 13, 2868–2881. [Google Scholar] [CrossRef]
- Patrauchan, M.A.; Miyazawa, D.; LeBlanc, J.C.; Aiga, C.; Florizone, C.; Dosanjh, M.; Davies, J.; Eltis, L.D.; Mohn, W.W. Proteomic analysis of survival of Rhodococcus jostii RHA1 during carbon starvation. Appl. Environ. Microbiol. 2012, 78, 6714–6725. [Google Scholar] [CrossRef]
- Berney, M.; Cook, G.M. Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS ONE 2010, 5, e8614. [Google Scholar] [CrossRef]
- Islam, Z.F.; Cordero, P.R.F.; Feng, J.; Chen, Y.J.; Bay, S.K.; Jirapanjawat, T.; Gleadow, R.M.; Carere, C.R.; Stott, M.B.; Chiri, E.; et al. Two Chloroflexi classes independently evolved the ability to persist on atmospheric hydrogen and carbon monoxide. ISME J. 2019, 13, 1801–1813. [Google Scholar] [CrossRef]
- Hernández, M.; Calabi, M.; Conrad, R.; Dumont, M.G. Analysis of the microbial communities in soils of different ages following volcanic eruptions. Pedosphere 2020, 30, 126–134. [Google Scholar] [CrossRef]
- Hernández, M.; Dumont, M.G.; Calabi, M.; Basualto, D.; Conrad, R. Ammonia oxidizers are pioneer microorganisms in the colonization of new acidic volcanic soils from South of Chile. Environ. Microbiol. Rep. 2014, 6, 70–79. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High throughput Sequence Data 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 31 December 2019).
- Bushnell, B.; Rood, J.; Singer, E. BBMerge—Accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Mikheenko, A.; Saveliev, V.; Gurevich, A. MetaQUAST: Evaluation of metagenome assemblies. Bioinformatics 2016, 32, 1088–1090. [Google Scholar] [CrossRef]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef]
- Uritskiy, G.V.; DiRuggiero, J.; Taylor, J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef]
- Wu, Y.W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Laczny, C.C.; Sternal, T.; Plugaru, V.; Gawron, P.; Atashpendar, A.; Margossian, H.H.; Coronado, S.; van der Maaten, L.; Vlassis, N.; Wilmes, P. VizBin—An application for reference-independent visualization and human-augmented binning of metagenomic data. Microbiome 2015, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Olm, M.R.; Brown, C.T.; Brooks, B.; Banfield, J.F. dRep: A tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017, 11, 2864–2868. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, C.M.; Rodriguez-Valera, F.; Kimes, N.E.; Ghai, R. Expanding the marine virosphere using metagenomics. PLoS Genet. 2013, 9, e1003987. [Google Scholar] [CrossRef]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, W.H.A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. Gplots: Various R Programming Tools for Plotting Data. The Comprehensive R Archive Network. 2009. Available online: https://cran.r-project.org/web/packages/gplots/index.html (accessed on 31 May 2020).
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Bose, T.; Haque, M.M.; Reddy, C.; Mande, S.S. COGNIZER: A framework for functional annotation of metagenomic datasets. PLoS ONE 2015, 10, e0142102. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef]
- King, G.M.; Weber, C.F. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 2007, 5, 107–118. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206D214. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016, 4, e1900v1. [Google Scholar]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Jospin, G.; Lowe, E.; Matsen, F.A.; Bik, H.M.; Eisen, J.A. PhyloSift: Phylogenetic analysis of genomes and metagenomes. PeerJ 2014, 2, e243. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comp. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- King, C.E.; King, G.M. Description of Thermogemmatispora carboxidivorans sp. Nov., a carbon-monoxide-oxidizing member of the class Ktedonobacteria isolated from a geothermally heated biofilm, and analysis of carbon monoxide oxidation by members of the class Ktedonobacteria. Int. J. Syst. Evol. Microbiol. 2014, 64, 1244–1251. [Google Scholar] [CrossRef]
- Chang, Y.J.; Land, M.; Hauser, L.; Chertkov, O.; Del Rio, T.G.; Nolan, M.; Copeland, A.; Tice, H.; Cheng, J.F.; Lucas, S.; et al. Non-contiguous finished genome sequence and contextual data of the filamentous soil bacterium Ktedonobacter racemifer type strain (SOSP1-21). Stand. Genomic. Sci. 2011, 5, 97–111. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, C.M.; Sakai, Y.; Abe, K.; Yokota, A.; Yabe, S. Dictyobacter vulcani sp. nov., belonging to the class Ktedonobacteria, isolated from soil of the Mt Zao volcano. Int. J. Syst. Evol. Microbiol. 2020, 70, 1805–1813. [Google Scholar] [CrossRef]
- Qin, Q.L.; Xie, B.B.; Zhang, X.Y.; Chen, X.L.; Zhou, B.C.; Zhou, J.; Oren, A.; Zhang, Y.Z. A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 2014, 196, 2210–2215. [Google Scholar] [CrossRef] [PubMed]
- King, G.M.; Weber, C.F.; Nanba, K.; Sato, Y.; Ohta, H. Atmospheric CO and hydrogen uptake and CO oxidizer phylogeny for Miyake-jima, Japan volcanic deposits. Microbes Environ. 2008, 23, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Hernandez, F.; Fornas, O.; Lluesma Gomez, M.; Bolduc, B.; de la Cruz Peña, M.J.; Martínez, J.M.; Anton, J.; Gasol, J.M.; Rosselli, R.; Valera, F.R.; et al. Single-virus genomics reveals hidden cosmopolitan and abundant viruses. Nat. Commun. 2017, 8, 15892. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Barbero, M.D.; Martin-Cuadrado, A.B.; Viver, T.; Santos, F.; Martinez-Garcia, M.; Anton, J. Recovering microbial genomes from metagenomes in hypersaline environments: The good, the bad and the ugly. Syst. Appl. Microbiol. 2019, 42, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.F.; King, G.M. Quantification of Burkholderia coxL genes in Hawaiian volcanic deposits. Appl. Environ. Microbiol. 2010, 76, 2212–2217. [Google Scholar] [CrossRef]
- Dunfield, K.E.; King, G.M. Molecular analysis of carbon monoxide-oxidizing bacteria associated with recent Hawaiian volcanic deposits. Appl. Environ. Microbiol. 2004, 70, 4242–4248. [Google Scholar] [CrossRef][Green Version]
- Cavaletti, L.; Monciardini, P.; Bamonte, R.; Schumann, P.; Rohde, M.; Sosio, M.; Donadio, S. New lineage of filamentous, spore-forming, Gram-positive bacteria from Soil. Appl. Environ. Microbiol. 2006, 72, 4360–4369. [Google Scholar] [CrossRef]
- Yabe, S.; Sakai, Y.; Abe, K.; Yokota, A.; Také, A.; Matsumoto, A.; Sugiharto, A.; Susilowati, D.; Hamada, M.; Nara, K.; et al. Dictyobacter aurantiacus gen. nov., sp. nov., a member of the family Ktedonobacteraceae, isolated from soil, and emended description of the genus Thermosporothrix. Int. J. Syst. Evol. Microbiol. 2017, 67, 2615–2621. [Google Scholar] [CrossRef]
- Yabe, S.; Aiba, Y.; Sakai, Y.; Hazaka, M.; Yokota, A. Thermosporothrix hazakensis gen. nov., sp. nov., isolated from compost and description of Thermosporotrichaceae fam. nov. within the class Ktedonobacteria. Int. J. Syst. Evol. Microbiol. 2010, 60, 1794–1801. [Google Scholar] [CrossRef]
- Yabe, S.; Sakai, Y.; Yokota, A. Thermosporothrix narukonensis sp. nov., belonging to the class Ktedonobacteria, isolated from fallen leaves on geothermal soil, and emended description of the genus Thermosporothrix. Int. J. Syst. Evol. Microbiol. 2016, 66, 2152–2157. [Google Scholar] [CrossRef]
- Yan, B.; Guo, X.; Liu, M.; Huang, Y. Ktedonosporobacter rubrisoli gen. nov., sp. nov., a novel representative of the class Ktedonobacteria, isolated from red soil, and proposal of Ktedonosporobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 1015–1025. [Google Scholar] [CrossRef]
- Wang, C.M.; Zheng, Y.; Sakai, Y.; Toyoda, A.; Minakuchi, Y.; Abe, K.; Yokota, A.; Yabe, S. Tengunoibacter tsumagoiensis gen. nov., sp. nov., Dictyobacter kobayashii sp. nov., Dictyobacter alpinus sp. nov., and description of Dictyobacteraceae fam. nov. within the order Ktedonobacterales isolated from Tengu-no-mugimeshi, a soil-like granular mass of microorganisms, and emended descriptions of the genera Ktedonobacter and Dictyobacter. Int. J. Syst. Evol. Microbiol. 2019, 69, 1910–1918. [Google Scholar] [PubMed]
- Zheng, Y.; Wang, C.M.; Sakai, Y.; Abe, K.; Yokota, A.; Yabe, S. Thermogemmatispora aurantia sp. nov. and Thermogemmatispora argillosa sp. nov., within the class Ktedonobacteria, and emended description of the genus Thermogemmatispora. Int. J. Syst. Evol. Microbiol. 2019, 69, 1744–1750. [Google Scholar] [CrossRef]
- Yabe, S.; Aiba, Y.; Sakai, Y.; Hazaka, M.; Yokota, A. Thermogemmatispora onikobensis gen. nov., sp. nov. and Thermogemmatispora foliorum sp. nov., isolated from fallen leaves on geothermal soils, and description of Thermogemmatisporaceae fam. nov. and Thermogemmatisporales ord. nov. within the class Ktedonobacteria. Int. J. Syst. Evol. Microbiol. 2011, 61, 903–910. [Google Scholar] [PubMed]
- Greening, C.; Biswas, A.; Carere, C.R.; Jackson, C.J.; Taylor, M.C.; Stott, M.B.; Cook, G.M.; Morales, S.E. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 2016, 10, 761–777. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Nishihara, H.; Yoshida, M.; Watanabe, M.; Rondal, J.D.; Concepcion, R.N.; Ohta, H. Cupriavidus pinatubonensis sp. nov. and Cupriavidus laharis sp. nov., novel hydrogen- oxidizing, facultatively chemolithotrophic bacteria isolated from volcanic mudflow deposits from Mt. Pinatubo in the Philippines. Int. J. Syst. Evol. Microbiol. 2006, 56, 973–978. [Google Scholar] [CrossRef] [PubMed][Green Version]


| Statistics | 1640 | 1751 | 1957 |
| Number of scaffolds (>= 0 bp) | 3,631,380 | 4,047,900 | 3,138,527 |
| Number of scaffolds (>= 500 bp) | 1,488,437 | 1,609,020 | 1,578,887 |
| Number of scaffolds (>= 1000 bp) | 326,980 | 333,885 | 447,418 |
| Number of scaffolds (>= 25,000 bp) | 172 | 195 | 1551 |
| Number of scaffolds (>= 50,000 bp) | 12 | 40 | 499 |
| Total length (>= 0 bp) | 2,320,437,372 | 2,460,323,814 | 2,396,069,091 |
| Total length (>= 500 bp) | 1,385,386,471 | 1,404,094,641 | 1,717,252,425 |
| Total length (>= 1000 bp) | 631,179,105 | 570,404,360 | 964,085,094 |
| Total length (>= 25,000 bp) | 5,697,241 | 8,152,100 | 8,236,5310 |
| Total length (>= 50,000 bp) | 686,375 | 2,983,958 | 46,419,897 |
| N50 | 914 | 850 | 700 |
| L50 | 391,387 | 477,193 | 348,604 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández, M.; Vera-Gargallo, B.; Calabi-Floody, M.; King, G.M.; Conrad, R.; Tebbe, C.C. Reconstructing Genomes of Carbon Monoxide Oxidisers in Volcanic Deposits Including Members of the Class Ktedonobacteria. Microorganisms 2020, 8, 1880. https://doi.org/10.3390/microorganisms8121880
Hernández M, Vera-Gargallo B, Calabi-Floody M, King GM, Conrad R, Tebbe CC. Reconstructing Genomes of Carbon Monoxide Oxidisers in Volcanic Deposits Including Members of the Class Ktedonobacteria. Microorganisms. 2020; 8(12):1880. https://doi.org/10.3390/microorganisms8121880
Chicago/Turabian StyleHernández, Marcela, Blanca Vera-Gargallo, Marcela Calabi-Floody, Gary M. King, Ralf Conrad, and Christoph C. Tebbe. 2020. "Reconstructing Genomes of Carbon Monoxide Oxidisers in Volcanic Deposits Including Members of the Class Ktedonobacteria" Microorganisms 8, no. 12: 1880. https://doi.org/10.3390/microorganisms8121880
APA StyleHernández, M., Vera-Gargallo, B., Calabi-Floody, M., King, G. M., Conrad, R., & Tebbe, C. C. (2020). Reconstructing Genomes of Carbon Monoxide Oxidisers in Volcanic Deposits Including Members of the Class Ktedonobacteria. Microorganisms, 8(12), 1880. https://doi.org/10.3390/microorganisms8121880






