Culturing Ancient Bacteria Carrying Resistance Genes from Permafrost and Comparative Genomics with Modern Isolates
Abstract
1. Introduction
2. Material and Methods
2.1. Specimen Collection
2.2. Specimen Processing to Remove Possible Contaminants
2.3. Permafrost Dating
2.4. Culturomics
2.5. Genomic Study
2.5.1. Choice of Strains and Modern Genomes
2.5.2. DNA Extraction and Genome Sequencing
2.5.3. Genome Assembly
2.5.4. Genome Annotation and Pan-Genome
2.5.5. Phylogeny and Ancestral Single Nucleotide Polymorphism (SNP)
2.6. Antibiotic Resistance
3. Results
3.1. Permafrost Properties
3.2. Microbiology
Microbial Culturomics
3.3. Genomes
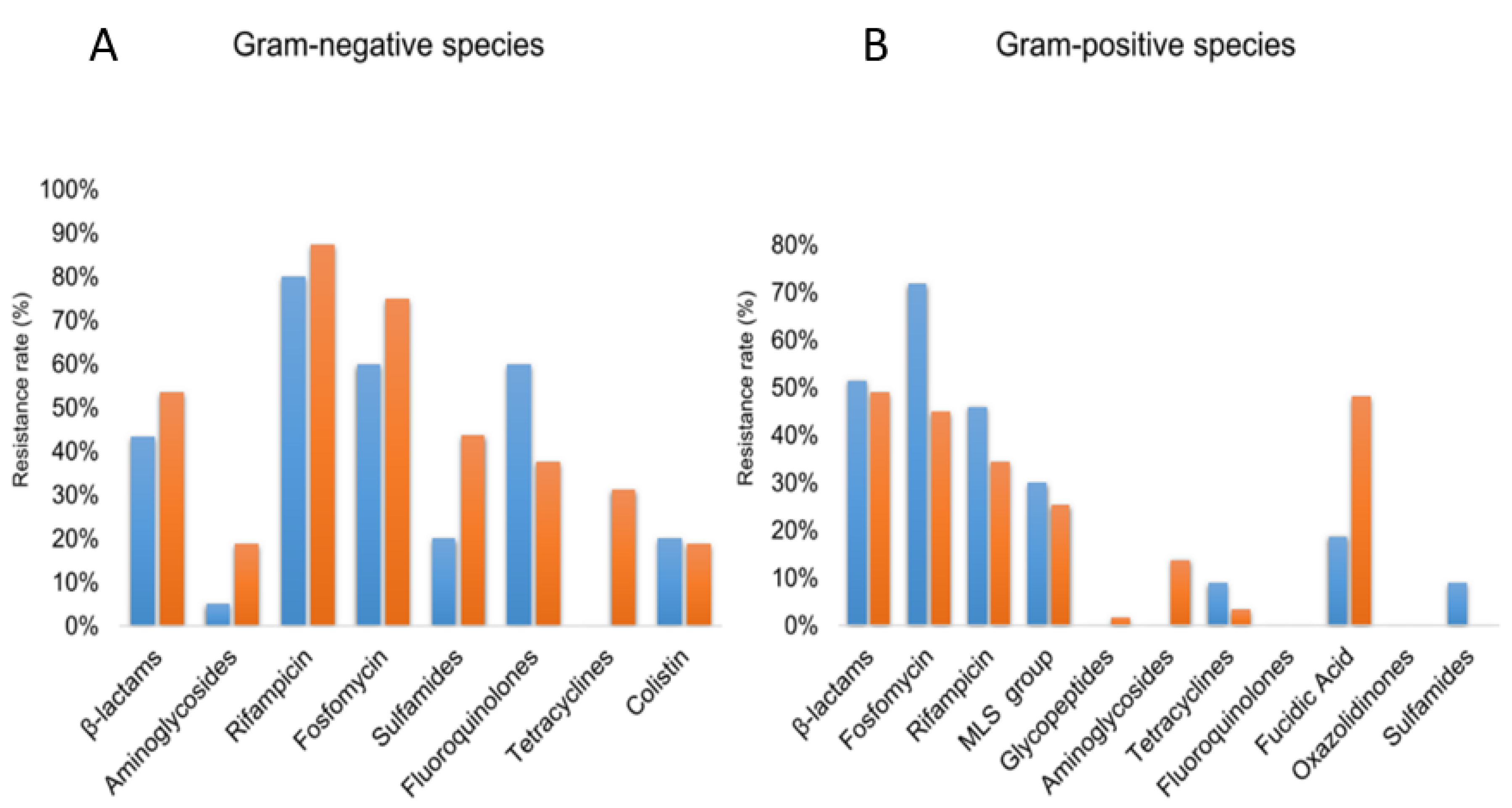
3.4. Antibiotic Resistance
3.4.1. Phenotype of Resistance
3.4.2. Resistome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of Data and Materials
Abbreviations
| CSUR | Collection de Souches de l’Unité des Rickettsies |
| DSMZ | Deutsche Sammlung von Mikroorganismen und Zellkulturen |
| MALDI-TOF MS | Matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry |
| SNP | Single nucleotide polymorphism |
References
- Curtis, T.P.; Sloan, W.T.; Scannell, J.W. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 2002, 99, 10494–10499. [Google Scholar] [CrossRef]
- D’Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the antibiotic resistome. Science 2006, 311, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Mindlin, S.Z.; Soina, V.S.; Petrova, M.A.; Gorlenko, Z.M. Isolation of antibiotic resistance bacterial strains from Eastern Siberia permafrost sediments. Russ. J. Genet. 2008, 44, 27–34. [Google Scholar] [CrossRef]
- Wayne, R.K.; Leonard, J.A.; Cooper, A. Full of Sound and Fury: History of Ancient DNA. Annu. Rev. Ecol. Syst. 1999, 30, 457–477. [Google Scholar] [CrossRef]
- Cano, R.J.; Tiefenbrunner, F.; Ubaldi, M.; Cueto, C.D.; Luciani, S.; Cox, T.; Orkand, P.; Künzel, K.H.; Rollo, F. Sequence analysis of bacterial DNA in the colon and stomach of the Tyrolean Iceman. Am. J. Phys. Anthropol. 2000, 112, 297–309. [Google Scholar] [CrossRef]
- Steven, B.; Léveillé, R.; Pollard, W.H.; Whyte, L.G. Microbial ecology and biodiversity in permafrost. Extremophiles 2006, 10, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Panikov, N.S.; Sizova, M.V. Growth kinetics of microorganisms isolated from Alaskan soil and permafrost in solid media frozen down to -35 degrees C. FEMS Microbiol. Ecol. 2007, 59, 500–512. [Google Scholar] [CrossRef]
- Hinsa-Leasure, S.M.; Bhavaraju, L.; Rodrigues, J.L.M.; Bakermans, C.; Gilichinsky, D.A.; Tiedje, J.M. Characterization of a bacterial community from a Northeast Siberian seacoast permafrost sample. FEMS Microbiol. Ecol. 2010, 74, 103–113. [Google Scholar] [CrossRef]
- Dmitriev, V.V.; Suzina, N.E.; Rusakova, T.G.; Gilichinskii, D.A.; Duda, V.I. Ultrastructural Characteristics of Natural Forms of Microorganisms Isolated from Permafrost Grounds of Eastern Siberia by the Method of Low-Temperature Fractionation. Dokl. Biol. Sci. 2001, 378, 304–306. [Google Scholar] [CrossRef]
- Frey, B.; Rime, T.; Phillips, M.; Stierli, B.; Hajdas, I.; Widmer, F.; Hartmann, M. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 2016, 92, fiw018. [Google Scholar] [CrossRef]
- Hu, W.; Zhang, Q.; Li, D.; Cheng, G.; Mu, J.; Wu, Q.; Niu, F.; An, L.; Feng, H. Diversity and community structure of fungi through a permafrost core profile from the Qinghai-Tibet Plateau of China. J. Basic Microbiol. 2014, 54, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.E.; Wallenstein, M.D.; Vishnivetskaya, T.A.; Waldrop, M.P.; Phelps, T.J.; Pfiffner, S.M.; Onstott, T.C.; Whyte, L.G.; Rivkina, E.M.; Gilichinsky, D.A.; et al. Microbes in thawing permafrost: The unknown variable in the climate change equation. ISME J. 2012, 6, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-C.; Brouchkov, A.; Griva, G.; Schinner, F.; Margesin, R. Isolation and characterization of bacteria from ancient siberian permafrost sediment. Biology 2013, 2, 85–106. [Google Scholar] [CrossRef]
- Steven, B.; Briggs, G.; McKay, C.P.; Pollard, W.H.; Greer, C.W.; Whyte, L.G. Characterization of the microbial diversity in a permafrost sample from the Canadian high Arctic using culture-dependent and culture-independent methods. FEMS Microbiol. Ecol. 2007, 59, 513–523. [Google Scholar] [CrossRef]
- Gilichinsky, D.A.; Vorobyova, E.A.; Erokhina, L.G.; Fyordorov-Davydov, D.G.; Chaikovskaya, N.R.; Fyordorov-Dayvdov, D.G. Long-term preservation of microbial ecosystems in permafrost. Adv. Space Res. 1992, 12, 255–263. [Google Scholar] [CrossRef]
- Legendre, M.; Lartigue, A.; Bertaux, L.; Jeudy, S.; Bartoli, J.; Lescot, M.; Alempic, J.-M.; Ramus, C.; Bruley, C.; Labadie, K.; et al. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. USA 2015, 112, E5327–E5335. [Google Scholar] [CrossRef]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef]
- Mindlin, S.; Petrenko, A.; Kurakov, A.; Beletsky, A.; Mardanov, A.; Petrova, M. Resistance of Permafrost and Modern Acinetobacter lwoffii Strains to Heavy Metals and Arsenic Revealed by Genome Analysis. BioMed Res. Int. 2016, 2016. [Google Scholar] [CrossRef]
- Filippova, S.; Surgucheva, N.; Kolganova, T.; Cherbunina, M.Y.; Brushkov, A.; Mulyukin, A.; Gal’chenko, V. Isolation and Identification of Bacteria from an Ice Wedge of the Mamontova Gora Glacial Complex (Central Yakutia). Biol. Bull. 2019, 46, 234–241. [Google Scholar] [CrossRef]
- Perron, G.G.; Whyte, L.; Turnbaugh, P.J.; Goordial, J.; Hanage, W.P.; Dantas, G.; Desai, M.M. Functional characterization of bacteria isolated from ancient arctic soil exposes diverse resistance mechanisms to modern antibiotics. PLoS ONE 2015, 10, e0069533. [Google Scholar] [CrossRef]
- Gilichinsky, D.A.; Nolte, E.; Basilyan, A.E.; Beer, J.; Blinov, A.V.; Lazarev, V.E.; Kholodov, A.L.; Meyer, H.; Nikolskiy, P.A.; Schirrmeister, L.; et al. Dating of syngenetic ice wedges in permafrost with 36Cl. Quat. Sci. Rev. 2007, 26, 1547–1556. [Google Scholar] [CrossRef]
- Tikhomirov, D.A.; Blinov, A.V. Cosmogenic 36Cl as a tool for dating permafrost ice. Bull. Russ. Acad. Sci. Phys. 2009, 73, 384–386. [Google Scholar] [CrossRef]
- Dione, N.; Khelaifia, S.; La Scola, B.; Lagier, J.C.; Raoult, D. A quasi-universal medium to break the aerobic/anaerobic bacterial culture dichotomy in clinical microbiology. Clin. Microbiol. Infect. 2016, 22, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Seck, E.H.; Sankar, S.A.; Khelaifia, S.; Croce, O.; Robert, C.; Couderc, C.; Di Pinto, F.; Sokhna, C.; Fournier, P.-E.; Raoult, D.; et al. Noncontiguous finished genome sequence and description of Planococcus massiliensis sp. nov., a moderately halophilic bacterium isolated from the human gut. New Microbes New Infect. 2016, 10, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinforma. Oxf. Engl. 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0.b5; Citeseer: University Park, PA, USA, 2001. [Google Scholar]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Cadoret, F.; Alou, M.T.; Afouda, P.; Traore, I.S.; Bréchard, L.; Michelle, C.; Di Pinto, F.; Andrieu, C.; Delerce, J.; Levasseur, A.; et al. Noncontiguous finished genome sequences and description of Bacillus massiliglaciei, Bacillus mediterraneensis, Bacillus massilinigeriensis, Bacillus phocaeensis and Bacillus tuaregi, five new species identified by culturomics. New Microbes New Infect. 2017, 19, 45–59. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef]
- Marcolefas, E.; Leung, T.; Okshevsky, M.; McKay, G.; Hignett, E.; Hamel, J.; Aguirre, G.; Blenner-Hasset, O.; Boyle, B.; Lévesque, R.C. Culture-dependent bioprospecting of bacterial isolates from the Canadian high Arctic displaying antibacterial activity. Front. Microbiol. 2019, 10, 1836. [Google Scholar] [CrossRef]
- Hassell, J.M.; Ward, M.J.; Muloi, D.; Bettridge, J.M.; Robinson, T.P.; Kariuki, S.; Ogendo, A.; Kiiru, J.; Imboma, T.; Kang’ethe, E.K. Clinically relevant antimicrobial resistance at the wildlife–livestock–human interface in Nairobi: An epidemiological study. Lancet Planet. Health 2019, 3, e259–e269. [Google Scholar] [CrossRef]
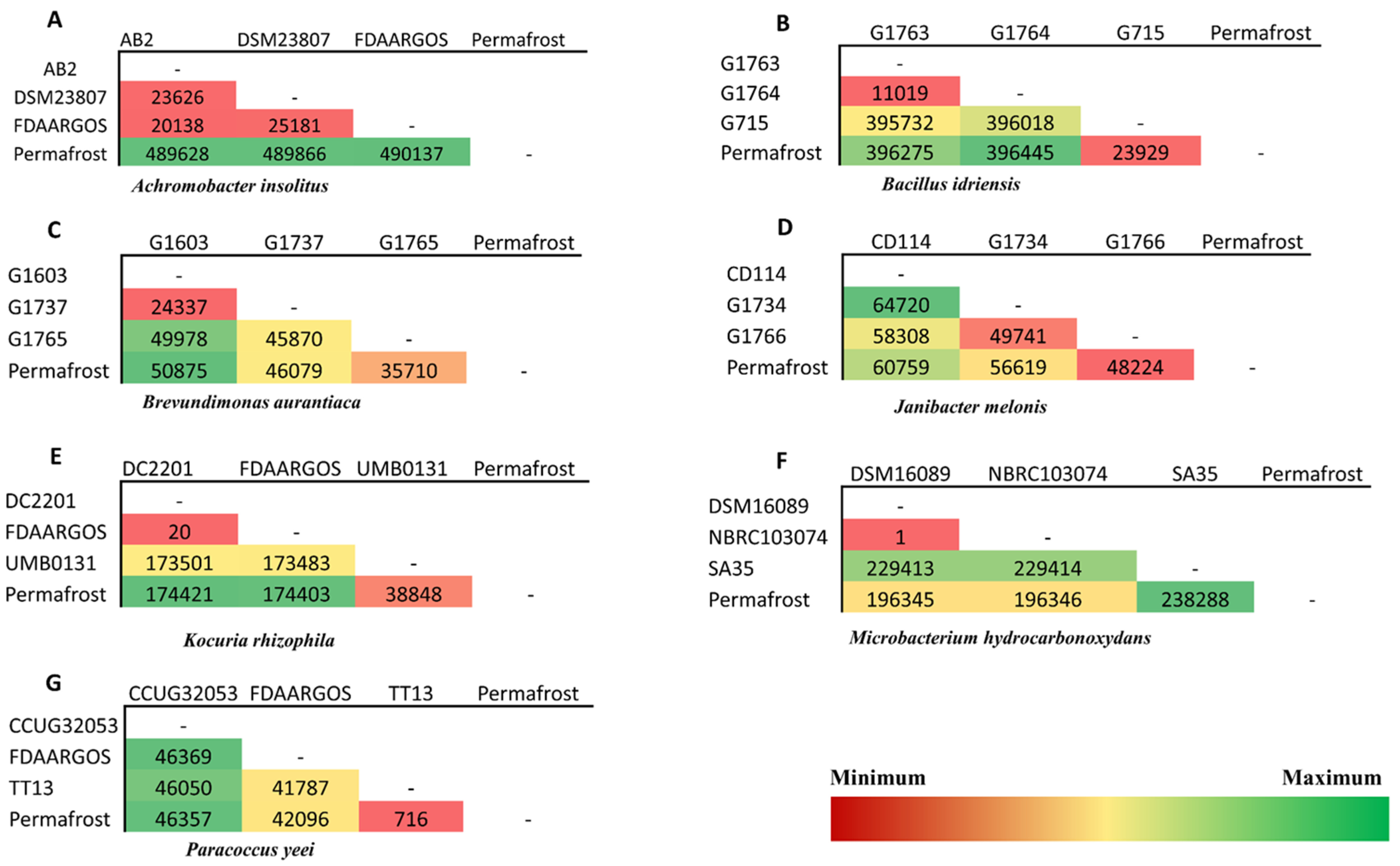
| Nr. | Bacterial Species | Minimal Medium | Room Temperature | Minimal Medium | 30°C | R-Medium | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| COS | R-Medium | COS | CSUR Number | % GC Content | Genome Size (bp) | |||||
| 1 | Achromobacter denitrificans | CSURP2856 | 64.21 | 3,568,352 | ||||||
| 2 | Achromobacter insolitus | CSURP2857 | 65.60 | 3,505,659 | ||||||
| 3 | Achromobacter pulmonis | CSURP5020 | 65.27 | 6,225,599 | ||||||
| 4 | Achromobacter spanius | CSURP2943 | 62.98 | 3,795,947 | ||||||
| 5 | Acinetobacter baumannii | CSURP2941 | 39.4 | 3,547,553 | ||||||
| 6 | Agrococcus baldri | CSURP2731 | 71.77 | 3,021,020 | ||||||
| 7 | Bacillus idriensis | CSURP2855 | 38.67 | 5,097,893 | ||||||
| 8 | Bacillus massiliglaciei | CSURP2600 | 43.77 | 4,145,500 | ||||||
| 9 | Bacillus megaterium | CSURP2736 | 38.30 | 4,914,860 | ||||||
| 10 | Bacillus simplex | CSURP2675 | 41.60 | 3,985,190 | ||||||
| 11 | Brevundimonas aurantiaca | CSURP3291/CSURP3513 | 66.09 | 3,046,902 | ||||||
| 12 | Enterobacter cloacae | CSURP2676 | 55.3 | 4,119,770 | ||||||
| 13 | Janibacter melonis | CSURP2733 | 70.46 | 2,989,541 | ||||||
| 14 | Kocuria rhizophila | CSURP2672 | 70.7 | 2,175,710 | ||||||
| 15 | Microbacterium hydrocarbonoxydans | CSURP2596 | 59.87 | 1,927,472 | ||||||
| 16 | Micrococcus luteus | CSURP2671 | 73.5 | 2,169,972 | ||||||
| 17 | Paenibacillus provencensis | CSURP2737 | 45.98 | 5,620,058 | ||||||
| 18 | Paenibacillus urinalis | CSURP2673/CSURP3512 | 47.2 | 5,466,760 | ||||||
| 19 | Pantoea massiliensis | CSURP5021 | 54.85 | 4,839,004 | ||||||
| 20 | Pantoea septica | CSURP2571 | 58.76 | 3,942,569 | ||||||
| 21 | Paracoccus yeei | CSURP2668 | 68.1 | 2,979,396 | ||||||
| 22 | Pedobacter quisquiliarum | CSURP2597 | 42.6 | 520,513 | ||||||
| 23 | Planomicrobium glaciei | CSURP2599 | 35 | 4,176,240 | ||||||
| 24 | Sphingomonas paucimobilis | CSURP3292/CSURP3510 | 55.68 | 2,442,583 | ||||||
| 25 | Staphylococcus capitis | CSURP5018 | 33.3 | 3,786,748 | ||||||
| 26 | Staphylococcus epidermidis | CSURP3290/CSURP3514 | 32.3 | 2,318,162 | ||||||
| 27 | Staphylococcus pasteuri | CSURP2670 | 37.35 | 2,414,997 | ||||||
| 28 | Staphylococcus saprophyticus | CSURP2669 | 32.9 | 2,291,952 | ||||||
| Nbr of Contigs | Genome Size (Mb) | %GC content | Ac | Ag | Bct | BL | C | E | FF | FA | GP | ML | Mi | MDE | Mup | Nov | Ph | Pol | Q | Rif | Stg | Sul | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Achromobacter insolitus G1433 | 1 | 6.5 | 65.6 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | 40 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 |
| Achromobacter pulmonis G1574 | 94 | 6.23 | 65.27 | 2 | 1 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 30 | 0 | 3 | 0 | 1 | 1 | 0 | 0 | 0 |
| Achromobacter spanius | 98 | 6.47 | 62.98 | 2 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 1 | 0 | 37 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Acinetobacter baumannii | 1 | 4.33 | 39.4 | 0 | 1 | 0 | 5 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 19 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 |
| Agroccocus massilioglaciei G1167 | 3 | 3.02 | 71.77 | 2 | 0 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 6 | 0 | 1 | 0 | 0 | 0 | |
| Bacillus idriensis G1436 | 46 | 5.1 | 38.67 | 3 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 1 |
| Bacillus massilioglaciei | 19 | 4.14 | 43.77 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Bacillus megaterium G1443 | 1 | 5.34 | 38.3 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 2 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| Bacillus simplex G1422 | 1 | 5.34 | 41.6 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 6 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | |
| Brevundimonas aurantiaca G1452 | 94 | 3.12 | 66.09 | 2 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Enterobacter cloacae | 1 | 5.32 | 55.3 | 0 | 1 | 1 | 8 | 1 | 3 | 2 | 0 | 0 | 1 | 1 | 33 | 0 | 0 | 0 | 6 | 14 | 0 | 0 | 0 |
| Janibacter melonis | 7 | 3.2 | 70.46 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 4 | 1 | 6 | 0 | 1 | 2 | 0 | 0 | ||
| Kocuria rhizophila G1424 | 1 | 2.7 | 70.7 | 2 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Microbacterium hydrocarbonoxydans G1438 | 10 | 3.95 | 59.87 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 3 | 1 | 0 | 2 | 3 | 3 | 0 | 0 | 0 | 1 | 0 | 0 |
| Micrococcus luteus G1425 | 1 | 2.5 | 73.5 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Paenibacillus provencensis G1439 | 20 | 5.62 | 45.98 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Paenibacillus urinalis G1453 | 3 | 5.47 | 47.2 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pantoea massiliensis G1572 | 65 | 4.83 | 54.85 | 1 | 0 | 1 | 8 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 32 | 0 | 1 | 5 | 3 | 0 | 2 | 0 | |
| Pantoea septica G1442 | 37 | 4.55 | 58.76 | 2 | 1 | 1 | 6 | 1 | 2 | 1 | 0 | 0 | 3 | 1 | 35 | 0 | 2 | 1 | 6 | 4 | 1 | 1 | 0 |
| Paracoccus yeei G1426 | 1 | 3.62 | 68.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pedobacter cryoconitis | 1 | 5.95 | 42.6 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Planomicrobium glaciei G1601 | 35 | 4.18 | 35 | 0 | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| Sphingomonas paucimobilis G1448 | 84 | 4.33 | 55.68 | 2 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Staphylococcus saprophyticus G1441 | 1 | 2.52 | 33.3 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 0 | 4 | 0 | 0 | 2 |
| Staphylococcus epidermidis | 1 | 2.5 | 32.3 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 0 | 4 | 0 | 0 | 2 |
| Staphylococcus capitis G1573 | 11 | 2.42 | 37.35 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 0 | 5 | 0 | 0 | 2 |
| Staphylococcus pasteuri G1440 | 1 | 2.56 | 32.9 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 0 | 4 | 0 | 0 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afouda, P.; Dubourg, G.; Levasseur, A.; Fournier, P.-E.; Delerce, J.; Mediannikov, O.; Diene, S.M.; Nahon, D.; Bourlès, D.; Rolain, J.-M.; et al. Culturing Ancient Bacteria Carrying Resistance Genes from Permafrost and Comparative Genomics with Modern Isolates. Microorganisms 2020, 8, 1522. https://doi.org/10.3390/microorganisms8101522
Afouda P, Dubourg G, Levasseur A, Fournier P-E, Delerce J, Mediannikov O, Diene SM, Nahon D, Bourlès D, Rolain J-M, et al. Culturing Ancient Bacteria Carrying Resistance Genes from Permafrost and Comparative Genomics with Modern Isolates. Microorganisms. 2020; 8(10):1522. https://doi.org/10.3390/microorganisms8101522
Chicago/Turabian StyleAfouda, Pamela, Grégory Dubourg, Anthony Levasseur, Pierre-Edouard Fournier, Jeremy Delerce, Oleg Mediannikov, Seydina M. Diene, Daniel Nahon, Didier Bourlès, Jean-Marc Rolain, and et al. 2020. "Culturing Ancient Bacteria Carrying Resistance Genes from Permafrost and Comparative Genomics with Modern Isolates" Microorganisms 8, no. 10: 1522. https://doi.org/10.3390/microorganisms8101522
APA StyleAfouda, P., Dubourg, G., Levasseur, A., Fournier, P.-E., Delerce, J., Mediannikov, O., Diene, S. M., Nahon, D., Bourlès, D., Rolain, J.-M., & Raoult, D. (2020). Culturing Ancient Bacteria Carrying Resistance Genes from Permafrost and Comparative Genomics with Modern Isolates. Microorganisms, 8(10), 1522. https://doi.org/10.3390/microorganisms8101522






