Comparison of Productivity and Fecal Microbiotas of Sows in Commercial Farms
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sampling
2.2. Analysis of the Fecal Microbiota by 16S rRNA Metagenomics
2.3. Measurement of the Concentrations of Organic Acids in Fecal Samples
2.4. Measurement of the Concentrations of Putrefactive Metabolites in Fecal Samples
2.5. Statistical Analysis
3. Results
3.1. Reproductive Parameters
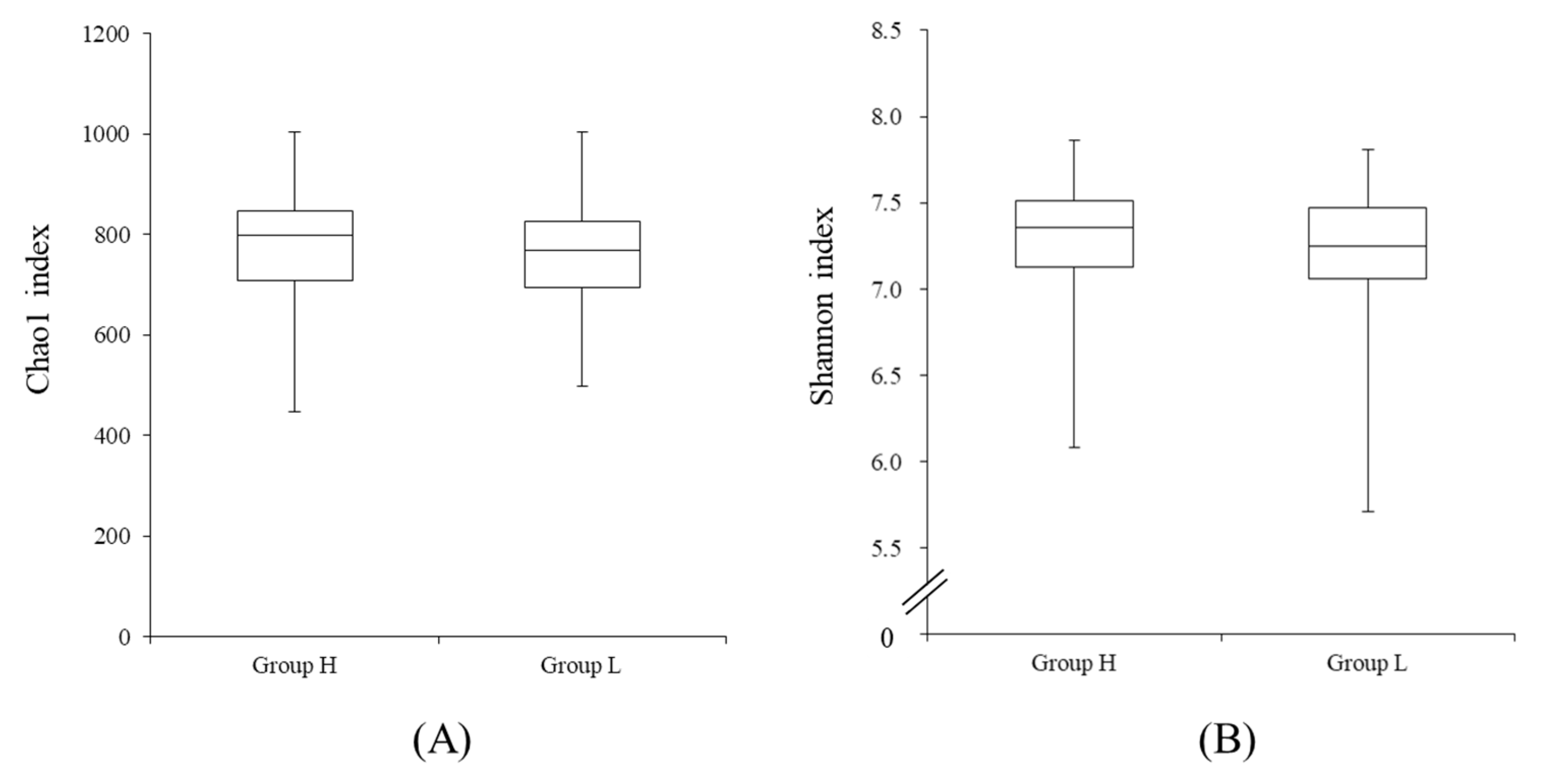
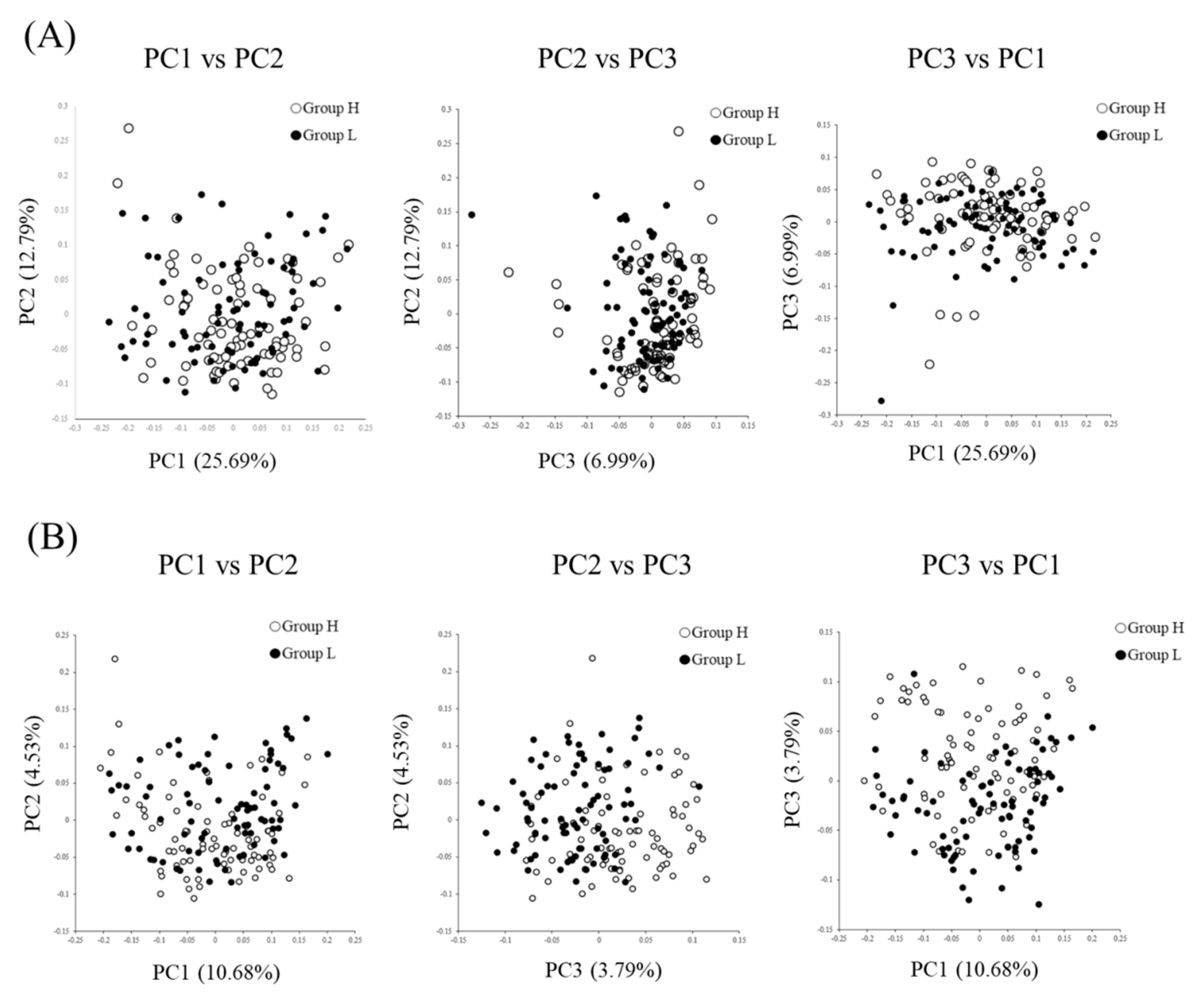
3.2. Comparison of the Fecal Microbiotas of High- and Low-Reproductive Performance Groups
3.3. Metabolites of the Microbiota in Feces
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Britt, J.H. Improving sow productivity through management during gestation, lactation and after weaning. J. Anim. Sci. 1986, 63, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Koketsu, Y.; Tani, S.; Iida, R. Factors for improving reproductive performance of sows and herd productivity in commercial breeding herds. Porcine Health Manag. 2017, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Quesnel, H. Nutritional and lactational effects on follicular development in the pig. In Control of Pig Reproduction VIII; Rodriguez-Martinez, H., Vallet, J.L., Ziecik, A.J., Eds.; Nottingham Univ. Press: Nottingham, UK, 2009; Volume 66, pp. 121–134. ISBN 978-190-476-139-6. [Google Scholar]
- Yun, J.; Valros, A. Benefits of prepartum nest-building behaviour on parturition and lactation in sows—A review. Asian-Australas J. Anim. Sci. 2015, 28, 1519–1524. [Google Scholar] [CrossRef] [PubMed]
- Luckey, T.D. Introduction to intestinal microecology. Am. J. Clin. Nutr. 1972, 25, 1292–1294. [Google Scholar] [CrossRef]
- Savage, D.C. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 1977, 31, 107–133. [Google Scholar] [CrossRef]
- Guarner, F.; Malagelada, J.R. Gut flora in health and disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef]
- Sakata, T. Pitfalls in short-chain fatty acid research: A methodological review. Anim. Sci. J. 2019, 90, 3–13. [Google Scholar] [CrossRef]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- Tiwari, U.P.; Singh, A.K.; Jha, R. Fermentation characteristics of resistant starch, arabinoxylan, and β-glucan and their effects on the gut microbial ecology of pigs: A review. Anim. Nutr. 2019, 5, 217–226. [Google Scholar] [CrossRef]
- Tran, T.H.T.; Everaert, N.; Bindelle, J. Review on the effects of potential prebiotics on controlling intestinal enteropathogens Salmonella and Escherichia coli in pig production. J. Anim. Physiol. Anim. Nutr. 2018, 102, 17–32. [Google Scholar] [CrossRef]
- Fouhse, J.M.; Zijlstra, R.T.; Willing, B.P. The role of gut microbiota in the health and disease of pigs. Anim. Front. 2016, 6, 30–36. [Google Scholar] [CrossRef]
- Yang, H.; Huang, X.; Fang, S.; He, M.; Zhao, Y.; Wu, Z.; Yang, M.; Zhang, Z.; Chen, C.; Huang, L. Unraveling the fecal microbiota and metagenomic functional capacity associated with feed efficiency in pigs. Front. Microbiol. 2017, 8, 1555. [Google Scholar] [CrossRef]
- Tan, Z.; Wang, Y.; Yang, T.; Ao, H.; Chen, S.; Xing, K.; Zhang, F.; Zhao, X.; Liu, J.; Wang, C. Differences in gut microbiota composition in finishing Landrace pigs with low and high feed conversion ratios. Antonie Van Leeuwenhoek 2018, 111, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- McCormack, U.M.; Curião, T.; Buzoianu, S.G.; Prieto, M.L.; Ryan, T.; Varley, P.; Crispie, F.; Magowan, E.; Metzler-Zebeli, B.U.; Berry, D.; et al. Exploring a possible link between the intestinal microbiota and feed efficiency in pigs. Appl Environ. Microbiol. 2017, 83, e00380-17. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Zhou, J.; Xiong, X.; Zou, L.; Kong, X.; Tan, B.; Yin, Y. Differences in Gut Microbial and Serum Biochemical Indices Between Sows With Different Productive Capacities During Perinatal Period. Front. Microbiol. 2020, 10, 3047. [Google Scholar] [CrossRef]
- Xu, K.; Bai, M.; Liu, H.; Duan, Y.; Zhou, X.; Wu, X.; Liao, P.; Li, T.; Yin, Y. Gut microbiota and blood metabolomics in weaning multiparous sows: Associations with oestrous. J. Anim. Physiol. Anim. Nutr. 2020, 104, 1155–1168. [Google Scholar] [CrossRef]
- Koketsu, Y. Sangyodobutsujuiryonihitsuyonaseisanshisutemunokangaekata. J. Japan Vet. Med. Assoc. 2006, 59, 567–572. [Google Scholar] [CrossRef]
- Kure, K. tonshashisetsu kanrikizai tonshakankyonadonikansurukadai (The Subjects about Institutions, Managed Equipments and Environment of Pig House). Jpn. J. Swine Sci. 2004, 41, 29–35. [Google Scholar] [CrossRef]
- Yamane, I. How to Handle Data in Epidemiological Research―An Example from Swine Benchmarking System. J. Vet. Epidemiol. 2013, 17, 1–4. [Google Scholar] [CrossRef]
- Inoue, R.; Sakaue, Y.; Sawai, C.; Sawai, T.; Ozeki, M.; Romero-Pérez, G.A.; Tsukahara, T. A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders. Biosci. Biotechnol. Biochem. 2016, 80, 2450–2458. [Google Scholar] [CrossRef]
- Tsukahara, T.; Matsukawa, N.; Tomonaga, S.; Inoue, R.; Ushida, K.; Ochiai, K. High-sensitivity detection of short-chain fatty acids in porcine ileal, cecal, portal and abdominal blood by gas chromatography-mass spectrometry. Anim. Sci. J. 2014, 85, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Morishima, S.; Aoi, W.; Kawamura, A.; Kawase, T.; Takagi, T.; Naito, Y.; Tsukahara, T.; Inoue, R. Intensive, prolonged exercise seemingly causes gut dysbiosis in female endurance runners. J. Clin. Biochem. Nutr. 2020, in press (accepted at 23rd August 2020). [Google Scholar]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed]
- Qi, K.; Men, X.; Wu, J.; Xu, Z. Rearing pattern alters porcine myofiber type, fat deposition, associated microbial communities and functional capacity. BMC Microbiol. 2019, 19, 181. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Bai, Y.; Jiang, L.; Wang, W.; Li, T.; Liu, P.; Tao, S.; Zhao, J.; Han, D.; Wang, J. Effects of oat bran on nutrient digestibility, intestinal microbiota, and inflammatory responses in the hindgut of growing pigs. Int. J. Mol. Sci. 2018, 19, 2407. [Google Scholar] [CrossRef]
- Zhu, J.J.; Gao, M.X.; Song, X.J.; Zhao, L.; Li, Y.W.; Hao, Z.H. Changes in bacterial diversity and composition in the faeces and colon of weaned piglets after feeding fermented soybean meal. J. Med. Microbiol. 2018, 67, 1181–1190. [Google Scholar] [CrossRef]
- Onarman Umu, Ö.C.; Fauske, A.K.; Åkesson, C.P.; Pérez de Nanclares, M.; Sørby, R.; Press, C.M.; Øverland, M.; Sørum, H. Gut microbiota profiling in Norwegian weaner pigs reveals potentially beneficial effects of a high-fiber rapeseed diet. PLoS ONE 2018, 13, e0209439. [Google Scholar] [CrossRef]
- Dehority, B.A. Microbial ecology of cell wall fermentation. In Forage Cell Wall Structure and Digestibility; Jung, H.G., Buxton, D.R., Hatfield, R.D., Ralph, J., Eds.; The American Society of Agronomy, Inc.; Crop Science Society of America, Inc.; Soil Science Society of America, Inc.: Madison, WI, USA, 1993; Volume 17, pp. 425–453. ISBN 978-089-118-115-6. [Google Scholar]
- Chassard, C.; Delmas, E.; Robert, C.; Lawson, P.A.; Bernalier-Donadille, A. Ruminococcus champanellensis sp. nov., a cellulose-degrading bacterium from human gut microbiota. Int. J. Syst. Evol. Microbiol. 2012, 62, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Lu, N.; Xue, Y.; Liu, S.; Lei, H.; Tu, W.; Lu, Y.; Xia, D. Crude fiber modulates the fecal microbiome and steroid hormones in pregnant Meishan sows. Gen. Comp. Endocrinol. 2019, 277, 141–147. [Google Scholar] [CrossRef]
- Engelhardt, W.V. Absorption of short-chain fatty acids from the large intestine. In Physiological and Clinical Aspects of Short-Chain Fatty Acids; Cummings, J.H., Rombeau, J.L., Sakata, T., Eds.; Cambridge University Press: Cambridge, UK, 1995; Volume 10, pp. 149–170. ISBN 978-052-144-048-6. [Google Scholar]
- Umesaki, Y.; Yajima, T.; Yokokuwa, T.; Mutai, M. Effect of organic acid absorption or bicarbonate transport in rat colon. Pflügers Archiv. 1979, 379, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Shimotoyodome, A.; Meguro, S.; Hase, T.; Tokimitsu, I.; Sakata, T. Short-chain fatty acids, but not lactate or succinate, stimulate mucus release in the rat colon. Comp. Biochem. Physiol. 2020, 125A, 525–531. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Kimura-Todani, T.; Hata, T.; Miyata, N.; Takakura, S.; Yoshihara, K.; Zhang, X.T.; Asano, Y.; Altaisaikhan, A.; Tsukahara, T.; Sudo, N. Dietary delivery of acetate to the colon using acylated starches as a carrier exerts anxiolytic effects in mice. Physiol. Behav. 2020, 223, 113004. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.; Aragon, V.; Brockmeier, S.L.; Gebhart, C.; Greeff, A.D.; Kerdsin, A.; O’Dea, M.A.; Okura, M.; Saléry, M. Update on Streptococcus suis Research and Prevention in the Era of Antimicrobial Restriction: 4th International Workshop on S. suis. Pathogens 2020, 9, 374. [Google Scholar] [CrossRef]
- Gottschalk, M.; Segura, M.; Xu, J. Streptococcus suis infections in humans: The Chinese experience and the situation in North America. Anim. Health Res. Rev. 2017, 8, 29–45. [Google Scholar] [CrossRef]
- Vigors, S.; O’Doherty, J.V.; Sweeney, T. Colonic microbiome profiles for improved feed efficiency can be identified despite major effects of farm of origin and contemporary group in pigs. Animal 2020, 1–9. [Google Scholar] [CrossRef]
- Oh, J.K.; Chae, J.P.; Pajarillo, E.A.B.; Kim, S.H.; Kwak, M.J.; Eun, J.S.; Chee, S.W.; Whang, K.; Kim, A.; Kang, D.K. Association between the body weight of growing pigs and the functional capacity of their gut microbiota. Anim. Sci. J. 2020, 91, e13418. [Google Scholar] [CrossRef]
- Lin, C.; Wan, J.; Su, Y.; Zhu, W. Effects of early intervention with maternal fecal microbiota and antibiotics on the gut microbiota and metabolite profiles of piglets. Metabolites 2018, 8, 89. [Google Scholar] [CrossRef]
- Yang, Q.; Huang, X.; Zhao, S.; Sun, W.; Yan, Z.; Wang, P.; Li, S.; Huang, W.; Zhang, S.; Liu, L.; et al. Structure and function of the fecal microbiota in diarrheic neonatal piglets. Front. Microbiol. 2017, 8, 502. [Google Scholar] [CrossRef]
- Modolo, J.R.; Margato, L.F.F.; Gottschalk, A.F.; Lopes, C.A.D.M. Incidence of Campylobacter in pigs with and without diarrhea. Rev. Microbiol. 1999, 30, 19–21. [Google Scholar] [CrossRef]
- De Rodas, B.; Youmans, B.P.; Danzeisen, J.L.; Tran, H.; Johnson, T.J. Microbiome profiling of commercial pigs from farrow to finish. J. Anim. Sci. 2018, 96, 1778–1794. [Google Scholar] [CrossRef]
- Jha, R.; Berrocoso, J.F. Dietary fiber and protein fermentation in the intestine of swine and their interactive effects on gut health and on the environment: A review. Anim. Feed Sci. Technol. 2016, 212, 18–26. [Google Scholar] [CrossRef]
- Isaacson, R.; Kim, H.B. The intestinal microbiome of the pig. Anim. Health Res. Rev. 2012, 13, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Patil, Y.; Gooneratne, R.; Ju, X.H. Interactions between host and gut microbiota in domestic pigs: A review. Gut Microbes 2020, 11, 310–334. [Google Scholar] [CrossRef] [PubMed]
- Kubasova, T.; Davidova-Gerzova, L.; Merlot, E.; Medvecky, M.; Polansky, O.; Gardan-Salmon, D.; Quesnel, H.; Rychlik, I. Housing systems influence gut microbiota composition of sows but not of their piglets. PLoS ONE 2017, 12, e0170051. [Google Scholar] [PubMed]
- Arruda, A.G.; Deblais, L.; Hale, V.; Pairis-Garcia, M.; Srivastava, V.; Kathayat, D.; Kumar, A.; Rajashekara, G. Nasal and gut microbiota for sows of different health status within six commercial swine farms from one swine production system. BioRxiv 2019, 596130. [Google Scholar] [CrossRef]
| Parameters of Reproductive Performance | Group H (n = 9) | Group L (n = 9) | Welch’s t-Test p Values |
|---|---|---|---|
| Number of piglets weaned/sow/year | 27.69 ± 0.33 | 19.33 ± 0.62 | 5.3 × 10−8 |
| Farrowing rate | 0.89 ± 0.01 | 0.80 ± 0.02 | 2.7 × 10−3 |
| Farrowing/sow/year | 2.41 ± 0.02 | 2.21 ± 0.02 | 4.3 × 10−6 |
| Number of piglets born/litter | 14.68 ± 0.43 | 11.48 ± 0.30 | 2.4 × 10−5 |
| Number of piglets born alive/litter | 13.27 ± 0.33 | 10.24 ± 0.32 | 6.9 × 10−6 |
| Number of stillbirths/litters | 1.42 ± 0.14 | 1.23 ± 0.17 | 0.42 |
| Pre-weaning mortality | 0.13 ± 0.02 | 0.15 ± 0.02 | 0.53 |
| Parameters of Reproductive Performance | Correlation with the Number of Piglets Weaned Per Sow Per Year | |
|---|---|---|
| Correlation Rate | p-Value | |
| Farrowing rate | 0.71 | 3.37 × 10−3 |
| Farrowing/sow/year | 0.75 | 2.04 × 10−3 |
| Number of piglets born/litter | 0.88 | 2.93 × 10−4 |
| Number of piglets born alive/litter | 0.88 | 2.75 × 10−4 |
| Number of stillbirths/litter | 0.18 | 0.47 |
| Pre-weaning mortality | −0.18 | 0.45 |
| Phylum | Class | Order | Family | Genus | Group H | Group L |
|---|---|---|---|---|---|---|
| Firmicutes | Clostridia | Clostridiales | Unclassified | Unclassified | 5.81 ± 0.25 | 6.79 ± 0.33 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | Treponema | 4.79 ± 0.31 | 3.59 ± 0.24 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminococcus | 3.76 ± 0.19 | 3.10 ± 0.16 |
| Planctomycetes | Planctomycetia | Pirellulales | Pirellulaceae | Unclassified | 1.92 ± 0.17 | 3.01 ± 0.35 |
| Verrucomicrobia | Verruco-5 | WCHB1-41 | RFP12 | Unclassified | 0.87 ± 0.07 | 0.62 ± 0.05 |
| Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | Unclassified | 0.76 ± 0.04 | 0.62 ± 0.03 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | 0.68 ± 0.14 | 1.99 ± 0.28 |
| Fibrobacteres | Fibrobacteria | Fibrobacterales | Fibrobacteraceae | Fibrobacter | 0.57 ± 0.07 | 0.37 ± 0.04 |
| Bacteroidetes | Bacteroidia | Bacteroidales | [Paraprevotellaceae] | CF231 | 0.54 ± 0.05 | 0.73 ± 0.07 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Porphyromonadaceae | Parabacteroides | 0.50 ± 0.04 | 0.67 ± 0.04 |
| Firmicutes | Bacilli | Bacillales | Planococcaceae | Rummeliibacillus | 0.49 ± 0.18 | 0.03 ± 0.01 |
| Firmicutes | Clostridia | Clostridiales | Veillonellaceae | Phascolarctobacterium | 0.49 ± 0.03 | 0.36 ± 0.02 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Butyricicoccus | 0.44 ± 0.03 | 0.30 ± 0.03 |
| Firmicutes | Erysipelotrichi | Erysipelotrichales | Erysipelotrichaceae | Bulleidia | 0.39 ± 0.04 | 0.24 ± 0.02 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Oribacterium | 0.32 ± 0.05 | 0.16 ± 0.03 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Blautia | 0.27 ± 0.03 | 0.17 ± 0.01 |
| Spirochaetes | Spirochaetes | Sphaerochaetales | Sphaerochaetaceae | Sphaerochaeta | 0.19 ± 0.01 | 0.10 ± 0.01 |
| Bacteroidetes | Bacteroidia | Bacteroidales | RF16 | Unclassified | 0.18 ± 0.02 | 0.08 ± 0.01 |
| Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | Collinsella | 0.17 ± 0.02 | 0.06 ± 0.01 |
| Firmicutes | Clostridia | Clostridiales | Peptococcaceae | Peptococcus | 0.13 ± 0.01 | 0.10 ± 0.01 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | Campylobacter | 0.08 ± 0.01 | 0.16 ± 0.04 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | Oxalobacter | 0.07 ± 0.01 | 0.03 ± 0.00 |
| Lentisphaerae | [Lentisphaeria] | Z20 | R4-45B | Unclassified | 0.07 ± 0.01 | 0.02 ± 0.00 |
| Tenericutes | Mollicutes | Anaeroplasmatales | Anaeroplasmataceae | Anaeroplasma | 0.05 ± 0.01 | 0.03 ± 0.00 |
| Verrucomicrobia | Verruco-5 | WCHB1-41 | Unclassified | Unclassified | 0.05 ± 0.01 | 0.02 ± 0.00 |
| Firmicutes | Erysipelotrichi | Erysipelotrichales | Erysipelotrichaceae | RFN20 | 0.04 ± 0.00 | 0.03 ± 0.00 |
| Synergistetes | Synergistia | Synergistales | Synergistaceae | Synergistes | 0.03 ± 0.01 | 0.01 ± 0.00 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Lachnospira | 0.02 ± 0.01 | 0.01 ± 0.00 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | Enterococcus | 0.02 ± 0.01 | 0.01 ± 0.00 |
| Firmicutes | Clostridia | Clostridiales | Eubacteriaceae | Anaerofustis | 0.02 ± 0.00 | 0.02 ± 0.00 |
| Verrucomicrobia | Verruco-5 | WCHB1-41 | WCHB1-25 | Unclassified | 0.02 ± 0.00 | 0.01 ± 0.00 |
| Proteobacteria | Alphaproteobacteria | RF32 | Unclassified | Unclassified | 0.02 ± 0.00 | 0.01 ± 0.00 |
| Tenericutes | RF3 | ML615J-28 | Unclassified | Unclassified | 0.01 ± 0.00 | 0.01 ± 0.00 |
| Elusimicrobia | Elusimicrobia | Elusimicrobiales | Elusimicrobiaceae | Unclassified | 0.01 ± 0.00 | <0.01 |
| Lentisphaerae | [Lentisphaeria] | Victivallales | Victivallaceae | Unclassified | 0.01 ± 0.00 | <0.01 |
| Firmicutes | Bacilli | Lactobacillales | Leuconostocaceae | Weissella | 0.01 ± 0.00 | 0.03 ± 0.01 |
| Firmicutes | Erysipelotrichi | Erysipelotrichales | Erysipelotrichaceae | Asteroleplasma | 0.01 ± 0.00 | <0.01 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Rikenellaceae | Unclassified | <0.01 | <0.01 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Anaerofilum | <0.01 | 0.02 ± 0.00 |
| Deferribacteres | Deferribacteres | Deferribacterales | Deferribacteraceae | Mucispirillum | <0.01 | 0.01 ± 0.00 |
| Firmicutes | Bacilli | Lactobacillales | Leuconostocaceae | Leuconostoc | <0.01 | 0.02 ± 0.01 |
| Fusobacteria | Fusobacteriia | Fusobacteriales | Fusobacteriaceae | Fusobacterium | <0.01 | 0.06 ± 0.03 |
| Firmicutes | Clostridia | Clostridiales | [Tissierellaceae] | Parvimonas | <0.01 | <0.01 |
| Organic Acids (µmol/g of Wet Feces) | Group H (n = 87) | Group L (n = 88) | Wilcoxon Rank Sum Test p Values |
|---|---|---|---|
| Lactate | 0.13 ± 0.05 | 0.24 ± 0.05 | 0.20 |
| Formate | 0.02 ± 0.02 | 0.01 ± 0.01 | 0.99 |
| Acetate | 86.53 ± 2.43 | 81.00 ± 2.73 | 0.04 |
| Propionate | 37.49 ± 1.27 | 33.28 ± 1.61 | 0.01 |
| iso-Butyrate | 0.13 ± 0.14 | 0.24 ± 0.16 | 0.36 |
| n-Butyrate | 16.11 ± 0.79 | 14.48 ± 0.96 | 0.045 |
| iso-Valerate | 0.13 ± 0.22 | 0.24 ± 0.24 | 0.38 |
| n-Valerate | 0.13 ± 0.17 | 0.24 ± 0.21 | 0.93 |
| Putrefactive Metabolites (µmol/g of Wet Feces) | Group H (n = 76) | Group L (n = 87) | Wilcoxon Rank Sum Test p Values |
|---|---|---|---|
| phenol | 52.67 ± 4.34 | 51.56 ± 2.79 | 0.77 |
| para-cresol | 62.95 ± 5.08 | 61.32 ± 3.32 | 0.81 |
| indole | 2.32 ± 0.21 | 2.30 ± 0.19 | 0.95 |
| para-ethylphenol | 0.19 ± 0.03 | 0.18 ± 0.03 | 0.68 |
| skatole | 9.54 ± 0.88 | 10.97 ± 0.81 | 0.13 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uryu, H.; Tsukahara, T.; Ishikawa, H.; Oi, M.; Otake, S.; Yamane, I.; Inoue, R. Comparison of Productivity and Fecal Microbiotas of Sows in Commercial Farms. Microorganisms 2020, 8, 1469. https://doi.org/10.3390/microorganisms8101469
Uryu H, Tsukahara T, Ishikawa H, Oi M, Otake S, Yamane I, Inoue R. Comparison of Productivity and Fecal Microbiotas of Sows in Commercial Farms. Microorganisms. 2020; 8(10):1469. https://doi.org/10.3390/microorganisms8101469
Chicago/Turabian StyleUryu, Haruka, Takamitsu Tsukahara, Hiromichi Ishikawa, Munetaka Oi, Satoshi Otake, Itsuro Yamane, and Ryo Inoue. 2020. "Comparison of Productivity and Fecal Microbiotas of Sows in Commercial Farms" Microorganisms 8, no. 10: 1469. https://doi.org/10.3390/microorganisms8101469
APA StyleUryu, H., Tsukahara, T., Ishikawa, H., Oi, M., Otake, S., Yamane, I., & Inoue, R. (2020). Comparison of Productivity and Fecal Microbiotas of Sows in Commercial Farms. Microorganisms, 8(10), 1469. https://doi.org/10.3390/microorganisms8101469





