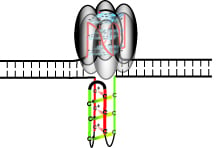
Role of Hfq in Genome Evolution: Instability of G-Quadruplex Sequences in E. coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Media
2.2. Cloning Quadruplex-Forming Repeats
2.3. Measurement of Cmr or Tetr Mutation Rates and Analysis of Revertants
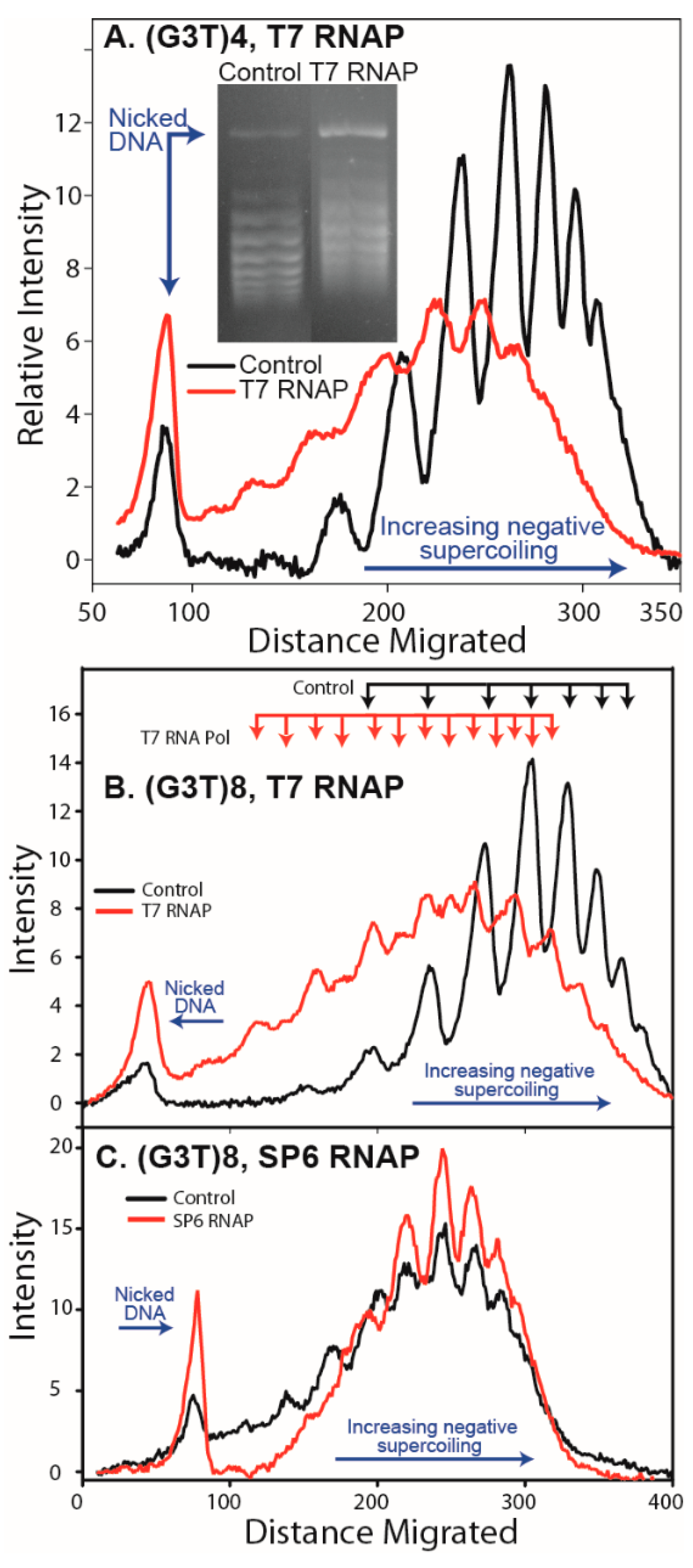
2.4. Analysis of G-Quadruplex Structure Formation in Supercoiled DNA
2.5. Binding Assays of Hfq to dG7
3. Results
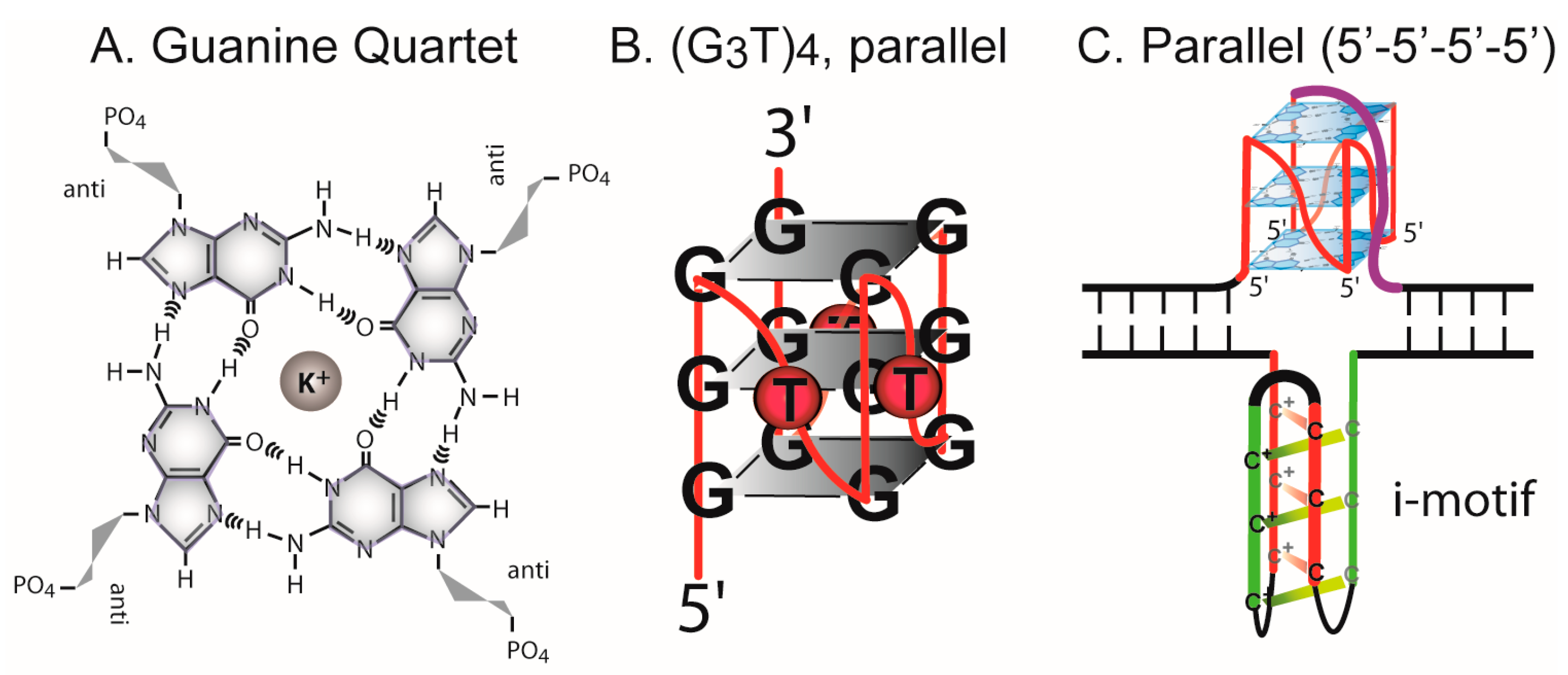
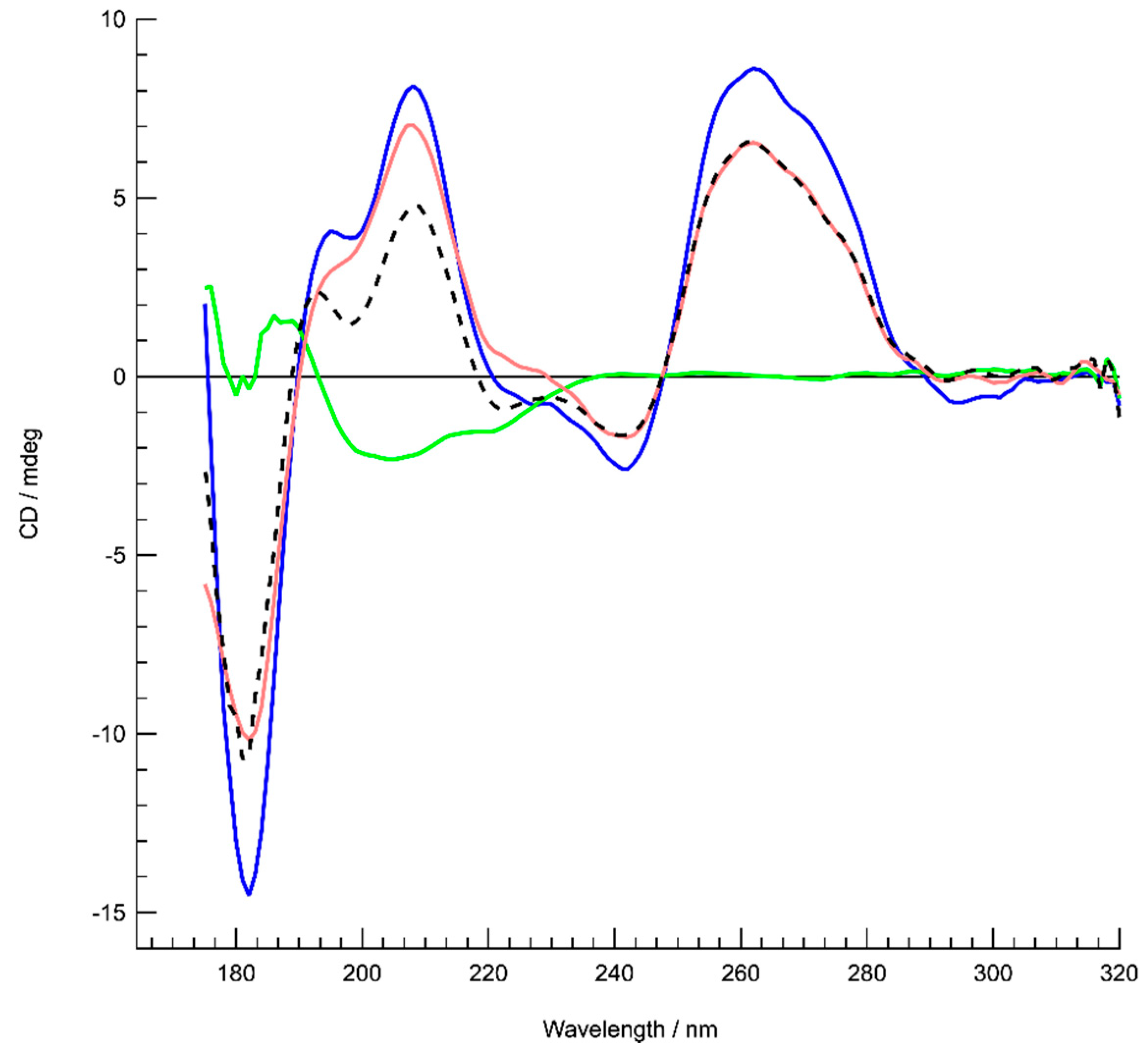
3.1. Formation of G-Quadruplex DNA in Supercoiled Plasmid
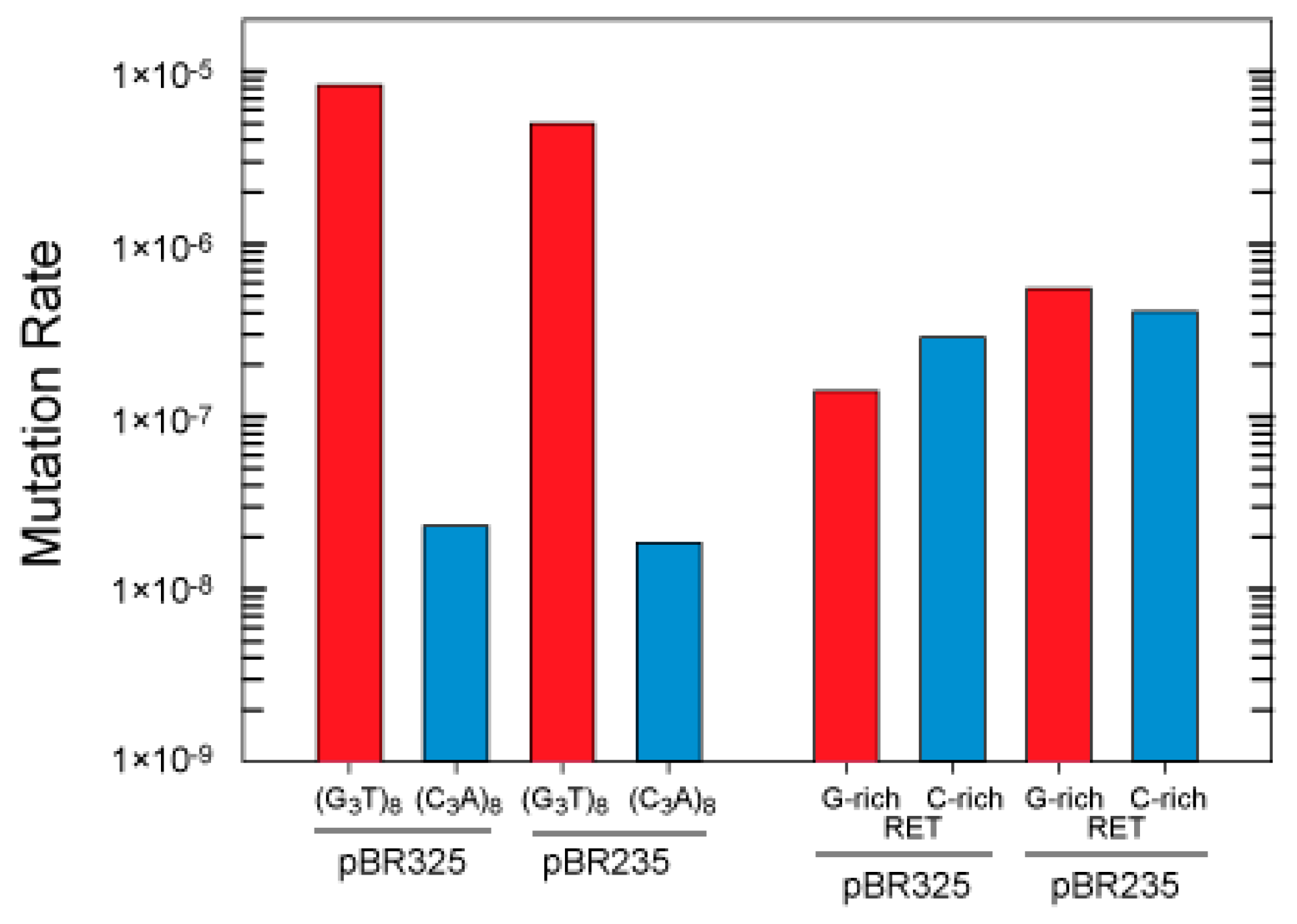
3.2. G-Quadruplex Structures Promote Genetic Instability in E. coli
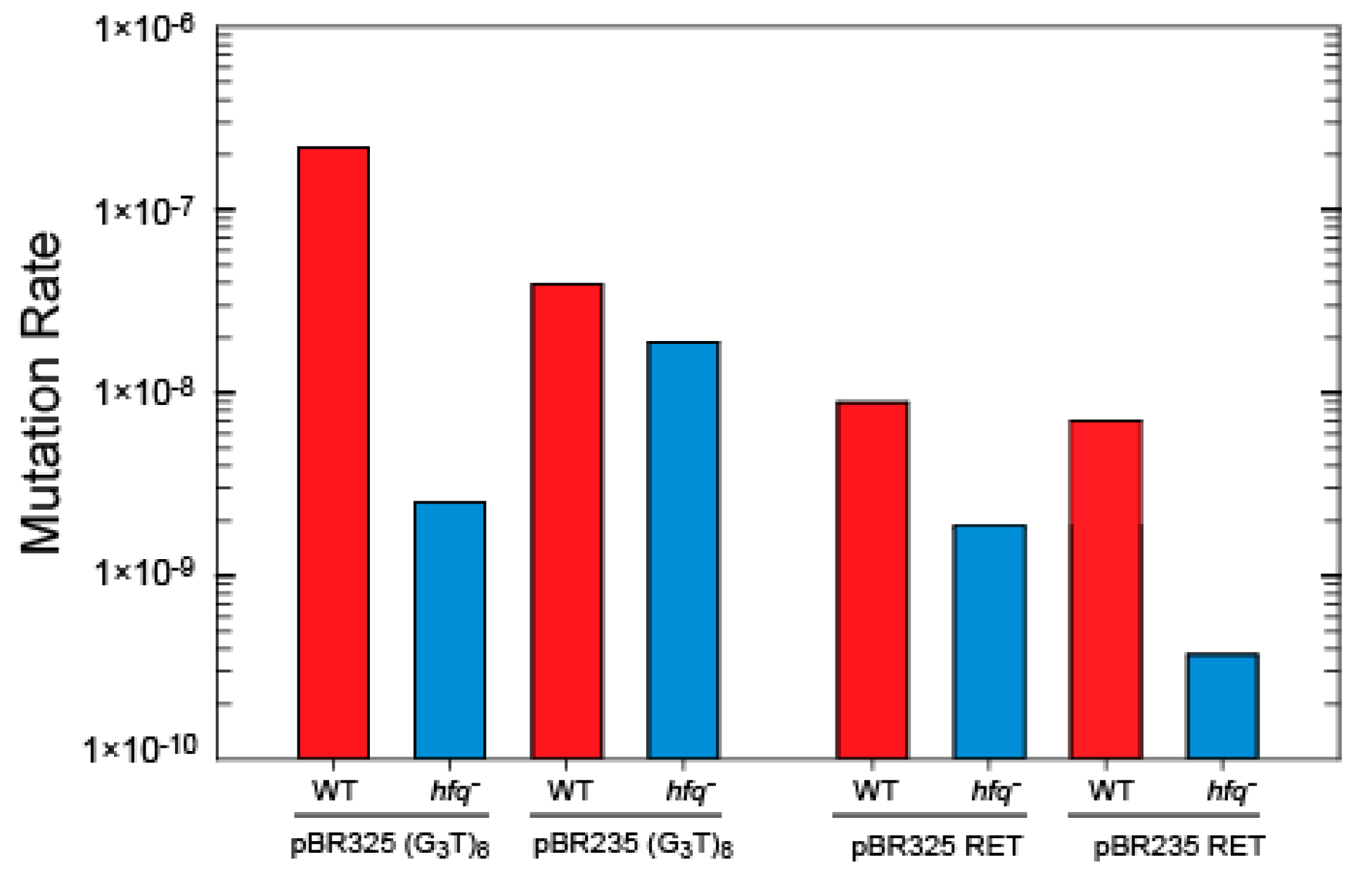
3.3. Influence of Hfq on the Instability of Quadruplex-Forming Repeats
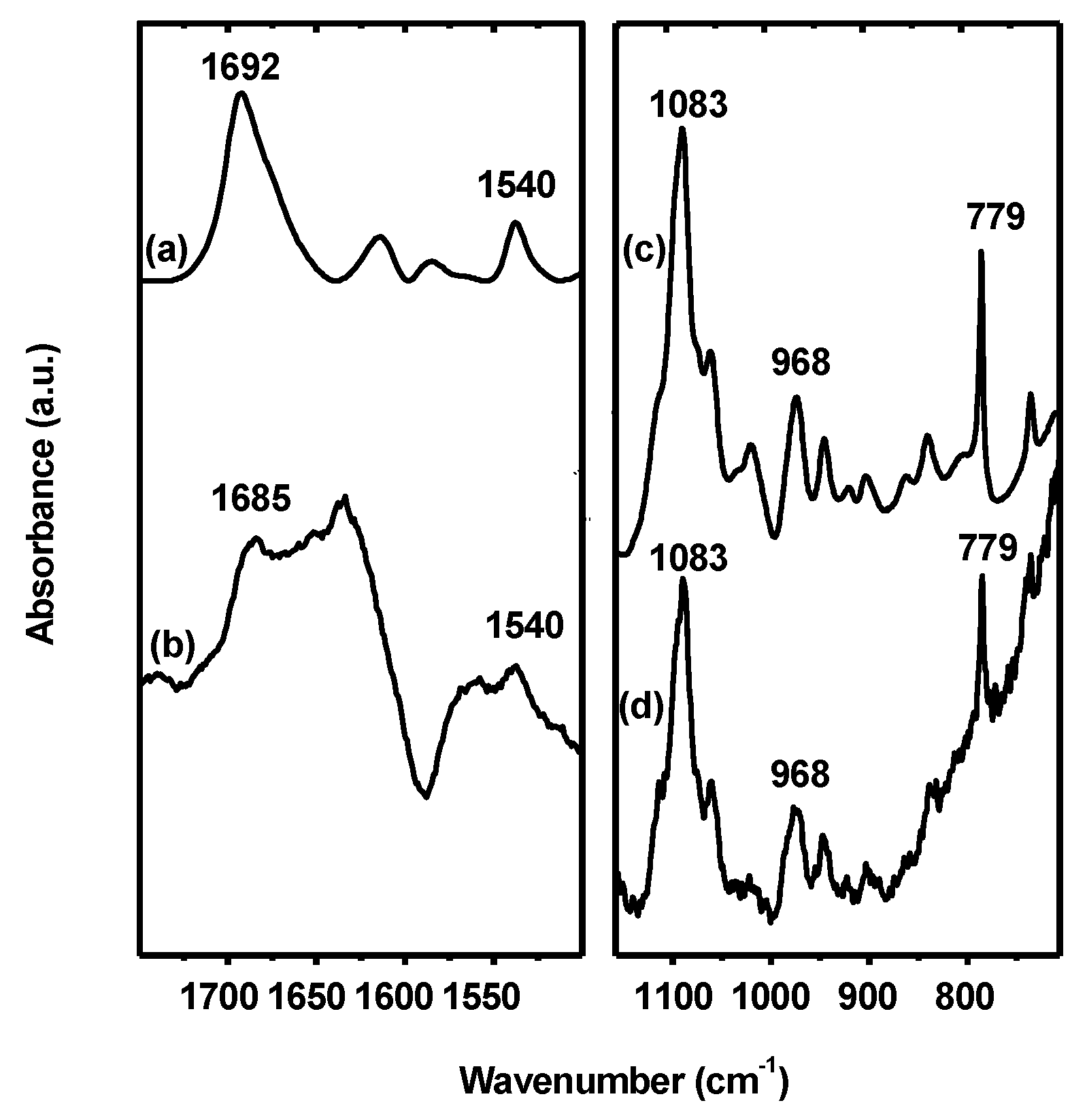
3.4. Interaction of Hfq with G-Quadruplex DNA
4. Discussion
4.1. G-Quadruplex Structures and Pathways to Formation in Cells
4.2. Deletion of G-Quadruplex-Forming Repeats
4.3. A Mutation in hfq Increases Stability of G-Quadruplex Repeats
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sinden, R.R. DNA Structure and Function; Academiic Press: Cambridge, MA, USA, 1994. [Google Scholar]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [Green Version]
- Kypr, J.; Kejnovska, I.; Renciuk, D.; Vorlickova, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [Green Version]
- Patel, D.J.; Phan, A.T.; Kuryavyi, V. Human telomere, oncogenic promoter and 5’-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007, 35, 7429–7455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Y.; Hurley, L.H. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie 2008, 90, 1149–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phan, A.T. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. Febs J. 2010, 277, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Mergny, J.L. Human telomeric DNA: G-quadruplex, i-motif and Watson-Crick double helix. Nucleic Acids Res. 2002, 30, 4618–4625. [Google Scholar] [CrossRef]
- Gehring, K.; Leroy, J.L.; Gueron, M. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature 1993, 363, 561–565. [Google Scholar] [CrossRef]
- Kang, C.H.; Berger, I.; Lockshin, C.; Ratliff, R.; Moyzis, R.; Rich, A. Crystal structure of intercalated four-stranded d(C3T) at 1.4 A resolution. Proc. Natl. Acad. Sci. USA 1994, 91, 11636–11640. [Google Scholar] [CrossRef] [Green Version]
- Guo, K.; Pourpak, A.; Beetz-Rogers, K.; Gokhale, V.; Sun, D.; Hurley, L.H. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007, 129, 10220–10228. [Google Scholar] [CrossRef] [Green Version]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef]
- Eddy, J.; Maizels, N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006, 34, 3887–3896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplan, O.I.; Berber, B.; Hekim, N.; Doluca, O. G-quadruplex prediction in E. coli genome reveals a conserved putative G-quadruplex-Hairpin-Duplex switch. Nucleic Acids Res. 2016, 44, 9083–9095. [Google Scholar] [PubMed] [Green Version]
- Du, X.; Wojtowicz, D.; Bowers, A.A.; Levens, D.; Benham, C.J.; Przytycka, T.M. The genome-wide distribution of non-B DNA motifs is shaped by operon structure and suggests the transcriptional importance of non-B DNA structures in Escherichia coli. Nucleic Acids Res. 2013, 41, 5965–5977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartas, M.; Cutova, M.; Brazda, V.; Kaura, P.; Stastny, J.; Kolomaznik, J.; Coufal, J.; Goswami, P.; Cerven, J.; Pecinka, P. The presence and localization of G-quadruplex forming sequences in the domain of bacteria. Molecules 2019, 24, 1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Salvo, M.; Pinatel, E.; Tala, A.; Fondi, M.; Peano, C.; Alifano, P. G4PromFinder: An algorithm for predicting transcription promoters in GC-rich bacterial genomes based on AT-rich elements and G-quadruplex motifs. BMC Bioinform. 2018, 19, 36. [Google Scholar] [CrossRef] [Green Version]
- Saranathan, N.; Vivekanandan, P. G-Quadruplexes: More Than Just a Kink in Microbial Genomes. Trends Microbiol. 2019, 27, 148–163. [Google Scholar] [CrossRef] [Green Version]
- Rawal, P.; Kummarasetti, V.B.; Ravindran, J.; Kumar, N.; Halder, K.; Sharma, R.; Mukerji, M.; Das, S.K.; Chowdhury, S. Genome-wide prediction of G4 DNA as regulatory motifs: Role in Escherichia coli global regulation. Genome Res. 2006, 16, 644–655. [Google Scholar] [CrossRef] [Green Version]
- Beaume, N.; Pathak, R.; Yadav, V.K.; Kota, S.; Misra, H.S.; Gautam, H.K.; Chowdhury, S. Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: Radioresistance of D. radiodurans involves G4 DNA-mediated regulation. Nucleic Acids Res. 2013, 41, 76–89. [Google Scholar] [CrossRef] [Green Version]
- Duquette, M.L.; Handa, P.; Vincent, J.A.; Taylor, A.F.; Maizels, N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004, 18, 1618–1629. [Google Scholar] [CrossRef] [Green Version]
- Holder, I.T.; Hartig, J.S. A matter of location: Influence of G-quadruplexes on Escherichia coli gene expression. Chem. Biol. 2014, 21, 1511–1521. [Google Scholar] [CrossRef] [Green Version]
- Wu, R.Y.; Zheng, K.W.; Zhang, J.Y.; Hao, Y.H.; Tan, Z. Formation of DNA:RNA hybrid G-quadruplex in bacterial cells and its dominance over the intramolecular DNA G-quadruplex in mediating transcription termination. Angew. Chem. 2015, 54, 2447–2451. [Google Scholar] [CrossRef] [PubMed]
- Endoh, T.; Kawasaki, Y.; Sugimoto, N. Suppression of gene expression by G-quadruplexes in open reading frames depends on G-quadruplex stability. Angew. Chem. 2013, 52, 5522–5526. [Google Scholar] [CrossRef] [PubMed]
- Weitzmann, M.N.; Woodford, K.J.; Usdin, K. The development and use of a DNA polymerase arrest assay for the evaluation of parameters affecting intrastrand tetraplex formation. J. Biol. Chem. 1996, 271, 20958–20964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, H.; Hurley, L.H.; Salazar, M. A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res. 1999, 27, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.; Piazza, A.; Bermejo, R.; Kriegsman, B.; Colosio, A.; Teulade-Fichou, M.P.; Foiani, M.; Nicolas, A. G-quadruplex-induced instability during leading-strand replication. Embo J. 2011, 30, 4033–4046. [Google Scholar] [CrossRef]
- Ribeyre, C.; Lopes, J.; Boule, J.B.; Piazza, A.; Guedin, A.; Zakian, V.A.; Mergny, J.L.; Nicolas, A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. Plos Genet. 2009, 5, e1000475. [Google Scholar] [CrossRef] [Green Version]
- Oliver, A.W.; Bogdarina, I.; Schroeder, E.; Taylor, I.A.; Kneale, G.G. Preferential binding of fd gene 5 protein to tetraplex nucleic acid structures. J. Mol. Biol. 2000, 301, 575–584. [Google Scholar] [CrossRef]
- Teng, F.Y.; Hou, X.M.; Fan, S.H.; Rety, S.; Dou, S.X.; Xi, X.G. Escherichia coli DNA polymerase I can disrupt G-quadruplex structures during DNA replication. Febs J. 2017, 284, 4051–4065. [Google Scholar] [CrossRef] [Green Version]
- Ehrat, E.A.; Johnson, B.R.; Williams, J.D.; Borchert, G.M.; Larson, E.D. G-quadruplex recognition activities of E. coli MutS. BMC Mol. Biol. 2012, 13, 23. [Google Scholar] [CrossRef] [Green Version]
- Saha, T.; Shukla, K.; Thakur, R.S.; Desingu, A.; Nagaraju, G. Mycobacterium tuberculosis UvrD1 and UvrD2 helicases unwind G-quadruplex DNA. Febs J. 2019, 286, 2062–2086. [Google Scholar] [CrossRef]
- Shukla, K.; Thakur, R.S.; Ganguli, D.; Rao, D.N.; Nagaraju, G. Escherichia coli and Neisseria gonorrhoeae UvrD helicase unwinds G4 DNA structures. Biochem. J. 2017, 474, 3579–3597. [Google Scholar] [CrossRef] [PubMed]
- Thakur, R.S.; Desingu, A.; Basavaraju, S.; Subramanya, S.; Rao, D.N.; Nagaraju, G. Mycobacterium tuberculosis DinG is a structure-specific helicase that unwinds G4 DNA: Implications for targeting G4 DNA as a novel therapeutic approach. J. Biol. Chem. 2014, 289, 25112–25136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, S.G.; Henderson, E. Identification of non-telomeric G4-DNA binding proteins in human, E. coli, yeast, and Arabidopsis. Mol. Cells 2002, 14, 404–410. [Google Scholar] [PubMed]
- Malabirade, A.; Jiang, K.; Kubiak, K.; Diaz-Mendoza, A.; Liu, F.; van Kan, J.A.; Berret, J.F.; Arluison, V.; van der Maarel, J.R.C. Compaction and condensation of DNA mediated by the C-terminal domain of Hfq. Nucleic Acids Res. 2017, 45, 7299–7308. [Google Scholar] [CrossRef]
- van der Maarel, J.R.; Guttula, D.; Arluison, V.; Egelhaaf, S.U.; Grillo, I.; Forsyth, V.T. Structure of the H-NS-DNA nucleoprotein complex. Soft Matter 2016, 12, 3636–3642. [Google Scholar] [CrossRef] [Green Version]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef] [Green Version]
- Takada, A.; Wachi, M.; Kaidow, A.; Takamura, M.; Nagai, K. DNA binding properties of the hfq gene product of Escherichia coli. Biochem. Biophys. Res. Commun. 1997, 236, 576–579. [Google Scholar] [CrossRef]
- Jiang, K.; Zhang, C.; Guttula, D.; Liu, F.; van Kan, J.A.; Lavelle, C.; Kubiak, K.; Malabirade, A.; Lapp, A.; Arluison, V.; et al. Effects of Hfq on the conformation and compaction of DNA. Nucleic Acids Res. 2015, 43, 4332–4341. [Google Scholar] [CrossRef] [Green Version]
- Cech, G.M.; Szalewska-Palasz, A.; Kubiak, K.; Malabirade, A.; Grange, W.; Arluison, V.; Wegrzyn, G. The Escherichia Coli Hfq Protein: An Unattended DNA-Transactions Regulator. Front. Mol. Biosci. 2016, 3, 36. [Google Scholar] [CrossRef] [Green Version]
- Malabirade, A.; Partouche, D.; El Hamoui, O.; Turbant, F.; Geinguenaud, F.; Recouvreux, P.; Bizien, T.; Busi, F.; Wien, F.; Arluison, V. Revised role for Hfq bacterial regulator on DNA topology. Sci. Rep. 2018, 8, 16792. [Google Scholar] [CrossRef]
- Geinguenaud, F.; Calandrini, V.; Teixeira, J.; Mayer, C.; Liquier, J.; Lavelle, C.; Arluison, V. Conformational transition of DNA bound to Hfq probed by infrared spectroscopy. Phys. Chem. Chem. Phys. PCCP 2011, 13, 1222–1229. [Google Scholar] [CrossRef] [PubMed]
- Tsui, H.C.; Leung, H.C.; Winkler, M.E. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994, 13, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Hashem, V.I.; Rosche, W.A.; Sinden, R.R. Genetic assays for measuring rates of (CAG).(CTG) repeat instability in Escherichia coli. Mutat. Res. 2002, 502, 25–37. [Google Scholar] [CrossRef]
- Hazel, P.; Huppert, J.; Balasubramanian, S.; Neidle, S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004, 126, 16405–16415. [Google Scholar] [CrossRef] [PubMed]
- Guedin, A.; Alberti, P.; Mergny, J.L. Stability of intramolecular quadruplexes: Sequence effects in the central loop. Nucleic Acids Res. 2009, 37, 5559–5567. [Google Scholar] [CrossRef] [Green Version]
- Rachwal, P.A.; Findlow, I.S.; Werner, J.M.; Brown, T.; Fox, K.R. Intramolecular DNA quadruplexes with different arrangements of short and long loops. Nucleic Acids Res. 2007, 35, 4214–4222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trinh, T.Q.; Sinden, R.R. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature 1991, 352, 544–547. [Google Scholar] [CrossRef]
- Sinden, R.R.; Zheng, G.X.; Brankamp, R.G.; Allen, K.N. On the deletion of inverted repeated DNA in Escherichia coli: Effects of length, thermal stability, and cruciform formation in vivo. Genetics 1991, 129, 991–1005. [Google Scholar]
- Edwards, S.F.; Hashem, V.I.; Klysik, E.A.; Sinden, R.R. Genetic instabilities of (CCTG).(CAGG) and (ATTCT).(AGAAT) disease-associated repeats reveal multiple pathways for repeat deletion. Mol. Carcinog. 2009, 48, 336–349. [Google Scholar] [CrossRef]
- Hashem, V.I.; Sinden, R.R. Duplications between direct repeats stabilized by DNA secondary structure occur preferentially in the leading strand during DNA replication. Mutat. Res. 2005, 570, 215–226. [Google Scholar] [CrossRef]
- Luria, S.E.; Delbruck, M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics 1943, 28, 491–511. [Google Scholar] [PubMed]
- Sarkar, S.; Ma, W.T.; Sandri, G.H. On fluctuation analysis: A new, simple and efficient method for computing the expected number of mutants. Genetica 1992, 85, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.M.; Ma, C.X.; Liang, P.; Singh, K.K. Fluctuation analysis CalculatOR: A web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 2009, 25, 1564–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosche, W.A.; Foster, P.L. Determining mutation rates in bacterial populations. Methods 2000, 20, 4–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, P.L. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006, 409, 195–213. [Google Scholar] [PubMed] [Green Version]
- Hwang, W.; Arluison, V.; Hohng, S. Dynamic competition of DsrA and rpoS fragments for the proximal binding site of Hfq as a means for efficient annealing. Nucleic Acids Res. 2011, 39, 5131–5139. [Google Scholar] [CrossRef] [Green Version]
- Refregiers, M.; Wien, F.; Ta, H.P.; Premvardhan, L.; Bac, S.; Jamme, F.; Rouam, V.; Lagarde, B.; Polack, F.; Giorgetta, J.L.; et al. DISCO synchrotron-radiation circular-dichroism endstation at SOLEIL. J. Synchrotron. Radiat. 2012, 19, 831–835. [Google Scholar] [CrossRef] [Green Version]
- Wien, F.; Wallace, B.A. Calcium fluoride micro cells for synchrotron radiation circular dichroism spectroscopy. Appl. Spectrosc. 2005, 59, 1109–1113. [Google Scholar] [CrossRef]
- Lees, J.G.; Smith, B.R.; Wien, F.; Miles, A.J.; Wallace, B.A. CDtool-an integrated software package for circular dichroism spectroscopic data processing, analysis, and archiving. Anal. Biochem. 2004, 332, 285–289. [Google Scholar] [CrossRef] [Green Version]
- Potaman, V.N.; Bissler, J.J.; Hashem, V.I.; Oussatcheva, E.A.; Lu, L.; Shlyakhtenko, L.S.; Lyubchenko, Y.L.; Matsuura, T.; Ashizawa, T.; Leffak, M.; et al. Unpaired structures in SCA10 (ATTCT)n.(AGAAT)n repeats. J. Mol. Biol. 2003, 326, 1095–1111. [Google Scholar] [CrossRef]
- Sun, D.; Hurley, L.H. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: Implications for drug targeting and control of gene expression. J. Med. Chem. 2009, 52, 2863–2874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekibo, D.A.T.; Fox, K.R. The effects of DNA supercoiling on G-quadruplex formation. Nucleic Acids Res. 2017, 45, 12069–12079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, K.; Chedin, F.; Hsieh, C.L.; Wilson, T.E.; Lieber, M.R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003, 4, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Duquette, M.L.; Huber, M.D.; Maizels, N. G-rich proto-oncogenes are targeted for genomic instability in B-cell lymphomas. Cancer Res. 2007, 67, 2586–2594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arluison, V.; Hohng, S.; Roy, R.; Pellegrini, O.; Regnier, P.; Ha, T. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic Acids Res. 2007, 35, 999–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taillandier, E.; Liquier, J. Vibrational spectroscopy of nucleic acids. In Handbook of Vibrational Spectroscopy; Chalmers, J.M., Griffiths, P.R., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2002; Volume 5. [Google Scholar]
- Guzman, M.R.; Liquier, J.; Brahmachari, S.K.; Taillandier, E. Characterization of parallel and antiparallel G-tetraplex structures by vibrational spectroscopy. Spectrochim. Acta. Part A Mol. Biomol. Spectrosc. 2006, 64, 495–503. [Google Scholar] [CrossRef] [Green Version]
- Tsuboi, M. Application of infrared spectroscopy to structure studies of nucleic acids. In Applied Spectroscopy Reviews; Brame, E.G.J., Ed.; Dekker: New York, NY, USA, 1969; pp. 45–90. [Google Scholar]
- Holm, A.I.S.; Nielsen, L.M.; Hoffmann, S.V.; Nielsen, S.B. Vacuum-ultraviolet circular dichroism spectroscopy of DNA: A valuable tool to elucidate topology and electronic coupling in DNA. Phys. Chem. Chem. Phys. PCCP 2010, 12, 9581–9596. [Google Scholar] [CrossRef]
- Nambiar, M.; Goldsmith, G.; Moorthy, B.T.; Lieber, M.R.; Joshi, M.V.; Choudhary, B.; Hosur, R.V.; Raghavan, S.C. Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res. 2011, 39, 936–948. [Google Scholar] [CrossRef] [Green Version]
- Belotserkovskii, B.P.; Liu, R.; Tornaletti, S.; Krasilnikova, M.M.; Mirkin, S.M.; Hanawalt, P.C. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl. Acad. Sci. USA 2010, 107, 12816–12821. [Google Scholar] [CrossRef] [Green Version]
- Hashem, V.I.; Rosche, W.A.; Sinden, R.R. Genetic recombination destabilizes (CTG)n.(CAG)n repeats in E. coli. Mutat. Res. 2004, 554, 95–109. [Google Scholar] [CrossRef]
- Trinh, T.Q.; Sinden, R.R. The influence of primary and secondary DNA structure in deletion and duplication between direct repeats in Escherichia coli. Genetics 1993, 134, 409–422. [Google Scholar] [PubMed]
- Kim, S.H.; Pytlos, M.J.; Sinden, R.R. Replication restart: A pathway for (CTG).(CAG) repeat deletion in Escherichia coli. Mutat. Res. 2006, 595, 5–22. [Google Scholar] [CrossRef]
- Puig Lombardi, E.; Holmes, A.; Verga, D.; Teulade-Fichou, M.P.; Nicolas, A.; Londono-Vallejo, A. Thermodynamically stable and genetically unstable G-quadruplexes are depleted in genomes across species. Nucleic Acids Res. 2019, 47, 6098–6113. [Google Scholar] [CrossRef] [PubMed]
- van Noort, V.; Worning, P.; Ussery, D.W.; Rosche, W.A.; Sinden, R.R. Strand misalignments lead to quasipalindrome correction. Trends Genet. TIG 2003, 19, 365–369. [Google Scholar] [CrossRef]
- Tsui, H.C.; Feng, G.; Winkler, M.E. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 1997, 179, 7476–7487. [Google Scholar] [CrossRef] [Green Version]
- Updegrove, T.B.; Zhang, A.; Storz, G. Hfq: The flexible RNA matchmaker. Curr. Opin. Microbiol. 2016, 30, 133–138. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Gottesman, S. Hfq links translation repression to stress-induced mutagenesis in E. coli. Genes Dev. 2017, 31, 1382–1395. [Google Scholar] [CrossRef] [Green Version]
- Foster, P.L. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 373–397. [Google Scholar] [CrossRef]
- Larson, E.D.; Duquette, M.L.; Cummings, W.J.; Streiff, R.J.; Maizels, N. MutSalpha binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr. Biol. CB 2005, 15, 470–474. [Google Scholar] [CrossRef] [Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parekh, V.J.; Niccum, B.A.; Shah, R.; Rivera, M.A.; Novak, M.J.; Geinguenaud, F.; Wien, F.; Arluison, V.; Sinden, R.R. Role of Hfq in Genome Evolution: Instability of G-Quadruplex Sequences in E. coli. Microorganisms 2020, 8, 28. https://doi.org/10.3390/microorganisms8010028
Parekh VJ, Niccum BA, Shah R, Rivera MA, Novak MJ, Geinguenaud F, Wien F, Arluison V, Sinden RR. Role of Hfq in Genome Evolution: Instability of G-Quadruplex Sequences in E. coli. Microorganisms. 2020; 8(1):28. https://doi.org/10.3390/microorganisms8010028
Chicago/Turabian StyleParekh, Virali J., Brittany A. Niccum, Rachna Shah, Marisa A. Rivera, Mark J. Novak, Frederic Geinguenaud, Frank Wien, Véronique Arluison, and Richard R. Sinden. 2020. "Role of Hfq in Genome Evolution: Instability of G-Quadruplex Sequences in E. coli" Microorganisms 8, no. 1: 28. https://doi.org/10.3390/microorganisms8010028
APA StyleParekh, V. J., Niccum, B. A., Shah, R., Rivera, M. A., Novak, M. J., Geinguenaud, F., Wien, F., Arluison, V., & Sinden, R. R. (2020). Role of Hfq in Genome Evolution: Instability of G-Quadruplex Sequences in E. coli. Microorganisms, 8(1), 28. https://doi.org/10.3390/microorganisms8010028