Listeria monocytogenes in Export-approved Beef from Mato Grosso, Brazil: Prevalence, Molecular Characterization and Resistance to Antibiotics and Disinfectants
Abstract
1. Introduction
2. Materials and Methods
2.1. General Procedures
2.2. Listeria sp. Isolation and Species Confirmation
2.3. Real-time PCR Confirmatory Assay
2.4. Molecular Serotyping
2.5. Genotypic Similarity
2.6. Antibiotic Resistance
2.7. Disinfectant Susceptibility
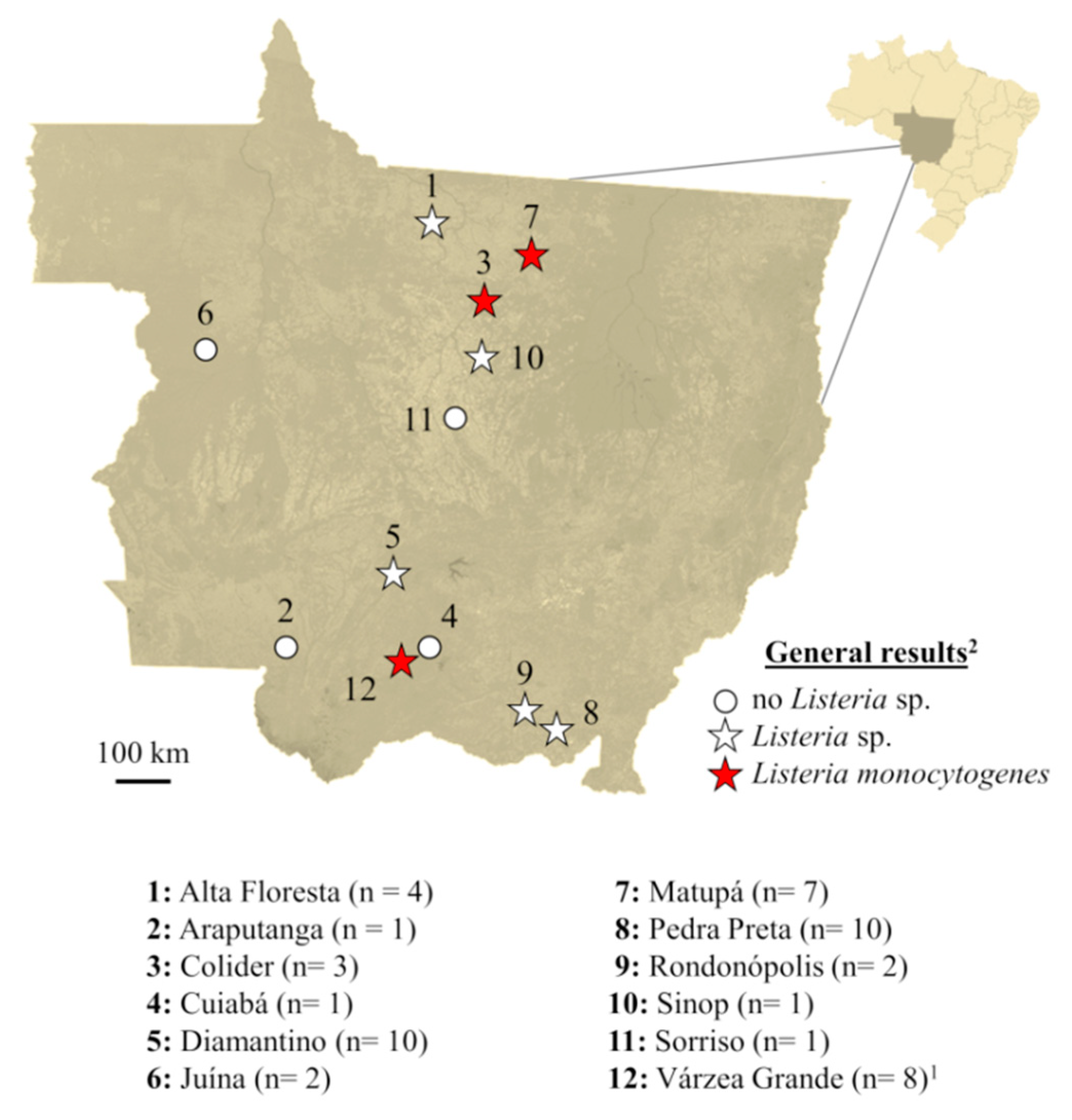
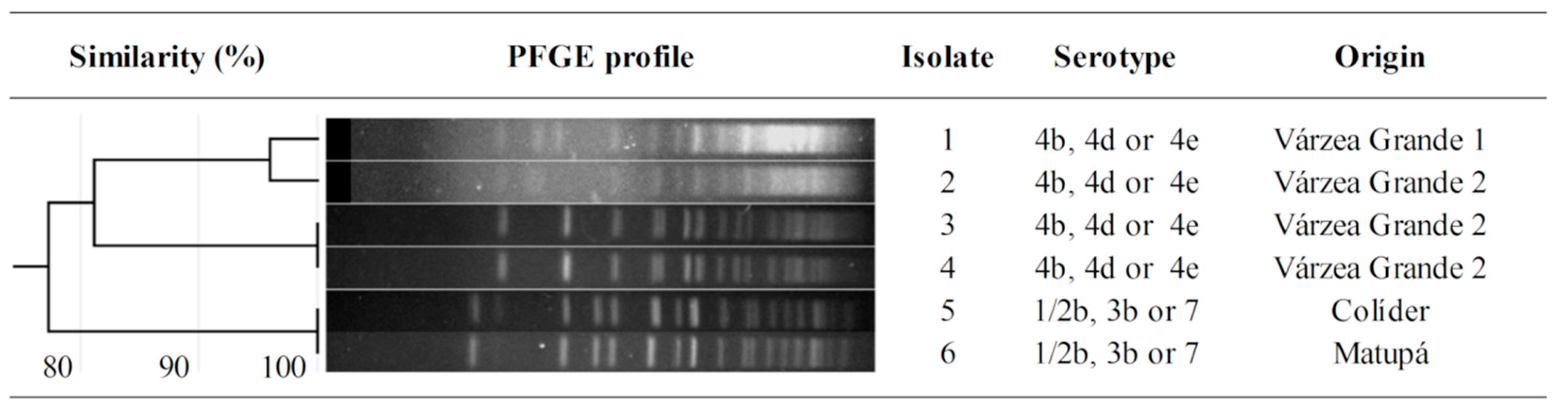
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Buchanan, R.L.; Gorris, L.G.M.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017, 75, 1–13. [Google Scholar] [CrossRef]
- Hernandez-Milian, A.; Payeras-Cifre, A. What is new in listeriosis? Biomed Res. Int. 2014, 2014, 1–7. [Google Scholar] [CrossRef]
- Saludes, M.; Troncoso, M.; Figueroa, G. Presence of Listeria monocytogenes in Chilean food matrices. Food Control 2015, 50, 331–335. [Google Scholar] [CrossRef]
- Dhama, K.; Karthik, K.; Tiwari, R.; Shabbir, M.Z.; Barbuddhe, S.; Malik, S.V.S.; Singh, R.K. Listeriosis in animals, its public health significance (food-borne zoonosis) and advances in diagnosis and control: A comprehensive review. Vet. Q. 2015, 35, 211–235. [Google Scholar] [CrossRef] [PubMed]
- Lakicevic, B.; Nastasijevic, I.; Raseta, M. Sources of Listeria monocytogenes contamination in retail establishments. Procedia Food Sci. 2015, 5, 160–163. [Google Scholar] [CrossRef]
- Carpentier, B.; Cerf, O. Review—Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Langsrud, S.; Sidhu, M.S.; Heir, E.; Holck, A.L. Bacterial disinfectant resistance—A challenge for the food industry. Int. Biodeterior. Biodegrad. 2003, 51, 283–290. [Google Scholar] [CrossRef]
- Romanova, N.; Favrin, S.; Griffiths, M.W. Sensitivity of Listeria monocytogenes to sanitizers used in the meat processing industry. Appl. Environ. Microbiol. 2002, 68, 6405–6409. [Google Scholar] [CrossRef] [PubMed]
- Cetinkaya, F.; Elal Mus, T.; Yibar, A.; Guclu, N.; Tavsanli, H.; Cibik, R. Prevalence, serotype identification by multiplex polymerase chain reaction and antimicrobial resistance patterns of Listeria monocytogenes isolated from retail foods: L. monocytogenes in raw and ready-to-eat foods. J. Food Saf. 2014, 34, 42–49. [Google Scholar] [CrossRef]
- Kevenk, T.O.; Terzi Gulel, G. Prevalence, antimicrobial resistance and serotype distribution of Listeria monocytogenes isolated from raw milk and dairy products: Prevalence, antimicrobial resistance. J. Food Saf. 2016, 36, 11–18. [Google Scholar] [CrossRef]
- Butaye, P.; Devriese, L.A.; Haesebrouck, F. Antimicrobial growth promoters used in animal feed: Effects of less well known antibiotics on Gram-positive bacteria. Clin. Microbiol. Rev. 2003, 16, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, M.K. Use of antibiotics as feed additives: A burning question. Front. Microbiol. 2014, 5, 334. [Google Scholar] [CrossRef] [PubMed]
- IBGE Brazilian Institute of Geography and Statistics. IBGE Indicators: Statistics of Animal Production; IBGE: Brasília, Brazil, 2017. [Google Scholar]
- BRASIL Resolution ANVISA/RDC #12. In Technical Regulation on Food Microbiological Standards; Diário Oficial da União: Brasília, Brazil, 2001.
- BRASIL Normative Instruction MAPA #9. In Procedures to Control Listeria monocytogenes Ready to Eat Animal-Source Products; Diário Oficial da União: Brasília, Brazil, 2009.
- Camargo, A.C.; Woodward, J.J.; Call, D.R.; Nero, L.A. Listeria monocytogenes in food-processing facilities, food contamination, and human listeriosis: The Brazilian scenario. Foodborne Pathog. Dis. 2017, 14, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Jay, J. Prevalence of Listeria spp. in meat and poultry products. Food Control 1996, 7, 209–214. [Google Scholar] [CrossRef]
- USDA United States Department of Agriculture. Microbiology Laboratory Guidebook. Isolation and Identification of Listeria monocytogenes from Red Meat, Poultry, Ready-to-Eat Siluriformes (Fish) and Egg Products, and Environmental Samples; USDA: Athens, GA, USA, 2017. [Google Scholar]
- McKellar, R.C. Use of the CAMP test for identification of Listeria monocytogenes. Appl. Environ. Microbiol. 1994, 60, 4219–4225. [Google Scholar]
- Traunsek, U.; Toplak, N.; Jeršek, B.; Lapanje, A.; Majstorović, T.; Kovač, M. Novel cost-efficient real-time PCR assays for detection and quantitation of Listeria monocytogenes. J. Microbiol. Methods 2011, 85, 40–46. [Google Scholar] [CrossRef]
- Moura, G.F.; Tomborelli, P.M.; Carvalho, R.C.T.; Sigarini, C.O.; Carvalho, F.T.; Vieira, B.S.; Figueiredo, E.E.S. Listeria monocytogenes and other species as persistent contaminants in the processing of chicken meat. J. Appl. Poult. Res. 2019, 28, 470–478. [Google Scholar] [CrossRef]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef]
- Graves, L.M.; Swaminathan, B. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresisq. Int. J. Food Microbiol. 2001, 65, 55–62. [Google Scholar] [CrossRef]
- Hunter, S.B.; Vauterin, P.; Lambert-Fair, M.A.; Van Duyne, M.S.; Kubota, K.; Graves, L.; Wrigley, D.; Barrett, T.; Ribot, E. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: Converting the national databases to the new size standard. J. Clin. Microbiol. 2005, 43, 1045–1050. [Google Scholar] [CrossRef]
- Sneath, P.; Sokal, R. Numerical Taxonomy. The Principles and Practice of Numerical Classification; W.H. Freeman and Company: San Francisco, CA, USA, 1973; ISBN 0-7167-0697-0. [Google Scholar]
- CLSI Clinical and Laboratory Standards Institute. CLSI Supplement M02: Performance Standards for Antimicrobial Disk Susceptibility Tests, 12th ed.; CLSI: Wayne, NJ, USA, 2015. [Google Scholar]
- CLSI Clinical and Laboratory Standards Institute. CLSI supplement M100: Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI: Wayne, NJ, USA, 2017. [Google Scholar]
- CLSI Clinical and Laboratory Standards Institute. CLSI Supplement M07: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 10th ed.; CLSI: Wayne, NJ, USA, 2015. [Google Scholar]
- Palma, J.M.; Lisboa, R.C.; Rodrigues, D.P.; Santos, A.F.M.; Hofer, E.; Santana, A.P. Molecular characterization of Listeria monocytogenes from beef samples and cattle slaughterhouses located in the Federal District, Brazil. Pesqui. Veterinária Bras. 2016, 36, 957–964. [Google Scholar] [CrossRef]
- Pesavento, G.; Ducci, B.; Nieri, D.; Comodo, N.; Lo Nostro, A. Prevalence and antibiotic susceptibility of Listeria spp. isolated from raw meat and retail foods. Food Control 2010, 21, 708–713. [Google Scholar] [CrossRef]
- Gebretsadik, S.; Kassa, T.; Alemayehu, H.; Huruy, K.; Kebede, N. Isolation and characterization of Listeria monocytogenes and other Listeria species in foods of animal origin in Addis Ababa, Ethiopia. J. Infect. Public Health 2011, 4, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, K.; Dmowska, K.; Osek, J. Prevalence, characterization, and antimicrobial resistance of Listeria monocytogenes isolates from bovine hides and carcasses. Appl. Environ. Microbiol. 2012, 78, 2043–2045. [Google Scholar] [CrossRef] [PubMed]
- Kalender, H. Prevalence of Listeria species in ground beef and chicken meat sold in eastern Turkey. Pak. Vet. J. 2012, 32, 456–458. [Google Scholar]
- Sauders, B.D.; Wiedmann, M. Ecology of Listeria species and L. monocytogenes in the natural environment. In Listeria, Listeriosis and Food Safety; CRC Press: Boca Raton, FL, USA, 2007; pp. 21–44. [Google Scholar]
- Borucki, M.K.; Call, D.R. Listeria monocytogenes serotype identification by PCR. J. Clin. Microbiol. 2003, 41, 5537–5540. [Google Scholar] [CrossRef]
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef]
- Montero, D.; Bodero, M.; Riveros, G.; Lapierre, L.; Gaggero, A.; Vidal, R.M.; Vidal, M. Molecular epidemiology and genetic diversity of Listeria monocytogenes isolates from a wide variety of ready-to-eat foods and their relationship to clinical strains from listeriosis outbreaks in Chile. Front. Microbiol. 2015, 6, 384. [Google Scholar] [CrossRef]
- Bertrand, S.; Ceyssens, P.J.; Yde, M.; Dierick, K.; Boyen, F.; Vanderpas, J.; Vanhoof, R.; Mattheus, W. Diversity of Listeria monocytogenes strains of clinical and food chain origins in Belgium between 1985 and 2014. PLoS ONE 2016, 11, e0164283. [Google Scholar] [CrossRef]
- Tappero, J.W. Reduction in the incidence of human listeriosis in the United States: Effectiveness of prevention efforts? JAMA 1995, 273, 1118–1122. [Google Scholar] [CrossRef]
- Gilot, P.; Genicot, A.; Andre, P. Serotyping and esterase typing for analysis of Listeria monocytogenes populations recovered from foodstuffs and from human patients with listeriosis in Belgium. J. Clin. Microbiol. 1996, 34, 1007–1010. [Google Scholar] [PubMed]
- Carvalho, F.T.; Vieira, B.S.; Vallim, D.C.; Carvalho, L.A.; Carvalho, R.C.T.; Pereira, R.C.L.; Figueiredo, E.E.S. Genetic similarity, antibiotic resistance and disinfectant susceptibility of Listeria monocytogenes isolated from chicken meat and chicken-meat processing environment in Mato Grosso, Brazil. LWT 2019, 109, 77–82. [Google Scholar] [CrossRef]
- Jacquet, C.; Doumith, M.; Gordon, J.I.; Martin, P.M.V.; Cossart, P.; Lecuit, M. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 2004, 189, 2094–2100. [Google Scholar] [CrossRef] [PubMed]
- Nightingale, K.K.; Ivy, R.A.; Ho, A.J.; Fortes, E.D.; Njaa, B.L.; Peters, R.M.; Wiedmann, M. InlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl. Environ. Microbiol. 2008, 74, 6570–6583. [Google Scholar] [CrossRef] [PubMed]
- Toledo, V.; den Bakker, H.; Hormazábal, J.; González-Rocha, G.; Bello-Toledo, H.; Toro, M.; Moreno-Switt, A. Genomic diversity of Listeria monocytogenes isolated from clinical and non-clinical samples in Chile. Genes 2018, 9, 396. [Google Scholar] [CrossRef]
- Krawczyk-Balska, A.; Markiewicz, Z. The intrinsic cephalosporin resistome of Listeria monocytogenes in the context of stress response, gene regulation, pathogenesis and therapeutics. J. Appl. Microbiol. 2016, 120, 251–265. [Google Scholar] [CrossRef]
- Haubert, L.; Mendonça, M.; Lopes, G.V.; de Itapema Cardoso, M.R.; da Silva, W.P. Listeria monocytogenes isolates from food and food environment harbouring tetM and ermB resistance genes. Lett. Appl. Microbiol. 2016, 62, 23–29. [Google Scholar] [CrossRef]
- Stoller, A.; Stevens, M.; Stephan, R.; Guldimann, C. Characteristics of Listeria monocytogenes strains persisting in a meat processing facility over a 4-year period. Pathogens 2019, 8, 32. [Google Scholar] [CrossRef]
- Carandina, D. Evaluation of Biofilms Formed by Listeria Monocytogenes Isolated from Dairy Plants and Resistance to Sanitizing Agents. Master’s Thesis, University of Sao Paulo, Pirassununga, São Paulo, Brazil, 2013. [Google Scholar]
- Kocot, A.M.; Olszewska, M.A. Biofilm formation and microscopic analysis of biofilms formed by Listeria monocytogenes in a food processing context. Food Sci. Technol. 2017, 84, 47–57. [Google Scholar] [CrossRef]
| Code | Sequence (5′–3′) | Specificity |
|---|---|---|
| hlyA | F: AGAAGTNATTAGTTTTAAACAAATTTACTATAACG | Listeria monocytogenes |
| R: AACTGCTCTTTAGTNACAGCTTTGC | ||
| hlyA Probe | FAM –TGAACCTACANGACCTTCC– MGB | Listeria monocytogenes |
| prs | F: GCTGAAGAGATTGCGAAAGAAG | Listeria sp. |
| R: CAAAGAAACCTTGGATTTGCGG | ||
| lmo0737 | F: AGGGCTTCAAGGACTTACCC | Listeria monocytogenes 1/2a, 1/2c, 3a, 3c |
| R: ACGATTTCTGCTTGCCATTC | ||
| lmo1118 | F: AGGGGTCTTAAATCCTGGAA | Listeria monocytogenes 1/2c, 3c |
| R: CGGCTTGTTCGGCATACTTA | ||
| ORF2819 | F: AGCAAAATGCCAAAACTCGT | Listeria monocytogenes 1/2b, 3b, 4b, 4d, 4e |
| R: CATCACTAAAGCCTCCCATTG | ||
| ORF2110 | F: AGTGGACAATTGATTGGTGAA | Listeria monocytogenes 4b, 4d, 4e |
| R: CATCCATCCCTTACTTTGGAC |
| Antibiotic | Code | Class | Disk Content (µg) | Zone Diameter Breakpoints (mm) for Each Phenotype | ||
|---|---|---|---|---|---|---|
| S | I | R | ||||
| Ciprofloxacin | CIP | Fluoroquinolone | 5 | ≥21 | 16–20 | ≤15 |
| Enrofloxacin | ENR | Fluoroquinolone | 5 | ≥18 | 15–17 | ≤14 |
| Sulfonamides | SSS | Folate pathway inhibitor | 300 | ≥17 | 13–16 | ≤12 |
| Trimethoprim | TMP | Folate pathway inhibitor | 5 | ≥16 | 11–15 | ≤10 |
| Trimethoprim + sulfamethoxazole | SXT | Folate pathway inhibitor | 23.75 | ≥16 | 11–15 | ≤10 |
| Ampicillin | AMP | Penicillin | 10 | ≥29 | - | ≤28 |
| Nitrofurantoin | NIT | Nitrofuran | 300 | ≥17 | 15–16 | ≤14 |
| Gentamicin | GEN | Aminoglycoside | 10 | ≥15 | 13–14 | ≤12 |
| Rifampin | RIF | Ansamycin | 5 | ≥20 | 17–19 | ≤16 |
| Chloramphenicol | CHL | Phenicol | 30 | ≥18 | 13–17 | ≤12 |
| Florfenicol | FLF | Phenicol | 30 | ≥18 | 13–17 | ≤12 |
| Erythromycin | ERY | Macrolide | 15 | ≥23 | 14–22 | ≤13 |
| Azithromycin | AZI | Macrolide | 15 | ≥18 | 14–17 | ≤13 |
| Imipenem | IPM | Carbapenem | 10 | ≥22 | - | ≤21 |
| Tetracycline | TET | Tetracycline | 30 | ≥19 | 15–18 | ≤14 |
| Cefoxitin | FOX | Cephem | 30 | ≥22 | - | ≤21 |
| Cefepime | FEP | Cephem | 30 | ≥24 | 21–23 | ≤20 |
| Antibiotic | Code | Resistance Profile 1 | |||||
|---|---|---|---|---|---|---|---|
| Isolate 1 | Isolate 2 | Isolate 3 | Isolate 4 | Isolate 5 | Isolate 6 | ||
| Ciprofloxacin | CIP | S | S | S | S | S | S |
| Enrofloxacin | ENR | S | S | S | S | S | S |
| Sulfonamides | SSS | I | I | R | R | S | R |
| Trimethoprim | TMP | S | S | S | S | S | S |
| Trimethoprim + sulfamethoxazole | SXT | S | S | S | S | S | S |
| Ampicillin | AMP | S | S | S | S | S | S |
| Nitrofurantoin | NIT | S | S | S | S | S | S |
| Gentamicin | GEN | S | S | S | S | S | S |
| Rifampin | RIF | S | S | S | S | S | S |
| Chloramphenicol | CHL | S | S | S | S | S | S |
| Florfenicol | FLF | S | S | S | S | S | S |
| Erythromycin | ERY | S | S | S | S | S | S |
| Azithromycin | AZI | S | S | S | S | S | S |
| Imipenem | IPM | S | S | S | S | S | S |
| Tetracycline | TET | S | S | S | S | S | S |
| Cefoxitin | FOX | R | R | R | R | R | R |
| Cefepime | FEP | R | S | R | R | R | R |
| Chemical Disinfectant 2 | Recommended Concentration (mg/L) | Minimal Inhibitory Concentration (mg/L) 1 | ||||||
|---|---|---|---|---|---|---|---|---|
| Isolate 1 | Isolate 2 | Isolate 3 | Isolate 4 | Isolate 5 | Isolate 6 | Average | ||
| Benzalkonium chloride | 5000 | 0.6 | 9.8 | 4.9 | 4.9 | 4.9 | 1.2 | 4.4 |
| Chlorhexidine | 20,000 | 0.2 | 0.2 | 0.4 | 0.2 | 1.6 | 0.4 | 0.5 |
| Peracetic acid | 187.5 | 2.9 | 23.4 | 11.7 | 46.9 | 23.4 | 1.5 | 18.3 |
| Quaternary ammonium | 2000 | 2.0 | 31.3 | 7.8 | 31.3 | 15.6 | 3.9 | 15.3 |
| Sodium hypochlorite | 2400 | 7200 | 1800 | 1800 | 1800 | 450 | 7200 | 3375 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teixeira, L.A.C.; Carvalho, F.T.; Vallim, D.C.; Pereira, R.C.L.; Cunha Neto, A.; Vieira, B.S.; Carvalho, R.C.T.; Figueiredo, E.E.S. Listeria monocytogenes in Export-approved Beef from Mato Grosso, Brazil: Prevalence, Molecular Characterization and Resistance to Antibiotics and Disinfectants. Microorganisms 2020, 8, 18. https://doi.org/10.3390/microorganisms8010018
Teixeira LAC, Carvalho FT, Vallim DC, Pereira RCL, Cunha Neto A, Vieira BS, Carvalho RCT, Figueiredo EES. Listeria monocytogenes in Export-approved Beef from Mato Grosso, Brazil: Prevalence, Molecular Characterization and Resistance to Antibiotics and Disinfectants. Microorganisms. 2020; 8(1):18. https://doi.org/10.3390/microorganisms8010018
Chicago/Turabian StyleTeixeira, Larrayane A.C., Fernanda T. Carvalho, Deyse C. Vallim, Rodrigo C.L. Pereira, Adelino Cunha Neto, Bruno S. Vieira, Ricardo C.T. Carvalho, and Eduardo E.S. Figueiredo. 2020. "Listeria monocytogenes in Export-approved Beef from Mato Grosso, Brazil: Prevalence, Molecular Characterization and Resistance to Antibiotics and Disinfectants" Microorganisms 8, no. 1: 18. https://doi.org/10.3390/microorganisms8010018
APA StyleTeixeira, L. A. C., Carvalho, F. T., Vallim, D. C., Pereira, R. C. L., Cunha Neto, A., Vieira, B. S., Carvalho, R. C. T., & Figueiredo, E. E. S. (2020). Listeria monocytogenes in Export-approved Beef from Mato Grosso, Brazil: Prevalence, Molecular Characterization and Resistance to Antibiotics and Disinfectants. Microorganisms, 8(1), 18. https://doi.org/10.3390/microorganisms8010018





