Abstract
Dengue virus (DENV) is an arboviral human pathogen transmitted through mosquito bite that infects an estimated ~400 million humans (~5% of the global population) annually. To date, no specific therapeutics have been developed that can prevent or treat infections resulting from this pathogen. DENV utilizes numerous host molecules and factors for transcribing the single-stranded ~11 kb positive-sense RNA genome. For example, the glycosylation machinery of the host is required for viral particles to assemble in the endoplasmic reticulum. Since a variety of host factors seem to be utilized by the pathogens, targeting these factors may result in DENV inhibitors, and will play an important role in attenuating the rapid emergence of other flaviviruses. Many experimental studies have yielded findings indicating that host factors facilitate infection, indicating that the focus should be given to targeting the processes contributing to pathogenesis along with many other immune responses. Here, we provide an extensive literature review in order to elucidate the progress made in the development of host-based approaches for DENV viral infections, focusing on host cellular mechanisms and factors responsible for viral replication, aiming to aid the potential development of host-dependent antiviral therapeutics.
1. Introduction
Dengue is an important arthropod-borne viral infectious disease caused by any one of the four-dengue virus (DENV-1 to -4) viral serotypes. The antigenically distinct but closely related serotypes of DENV belong to the genus Flavivirus, family Flaviviridae. Its rapid and intense spread is noted in most of the world’s tropical and subtropical regions, which has led to its categorization as an emerging infectious disease [1,2]. The genus includes more than 70 small-enveloped viruses related to Japanese encephalitis (JEV), Zika viruses (ZIKV), West Nile (WNV), yellow fever virus (YFV), DENV, or tick-borne encephalitis (TBEV), and other medically-important arboviruses [3]. Most importantly, DENV is endemic in 112 countries, and incidence of infection has risen 30-fold over the last five decades [4]. More than one-third of the world population is at risk of infection, and it is estimated that ~400 million individuals suffer annually because of DENV infection [5]. DENV infection results in varying degrees of clinical signs and symptoms (asymptomatic or only mildly symptomatic). In the case of dengue fever (DF), patients may experience headache, myalgia, rash, leukopenia, arthralgia, retro-orbital pain, and hemorrhagic manifestations. Thereafter, patients suffering from dengue hemorrhagic fever (DHF) may develop petechiae, bruising, thrombocytopenia, and shock. Ultimate disease signs include rapid or weak pulse, manifestations of hypotension and clammy skin, and finally cold and restlessness in dengue shock syndrome (DSS) [6].
However, specific therapeutics are presently not available for DENV infections [7], necessitating an improved approach for re-mediating this global burden. In this review, we will explore the host factors or targets that influence DENV replication, focusing on the factors that can potentially be utilized in a new process that may help alleviate this global burden.
1.1. History
As one of the major public health concerns, the term “dengue” is believed to originate from the Swahili word “ki-dinga pepo”, which means “cramp-like pains, produced through the agency of an evil spirit” [8,9]. Joint fever, a type of dengue-like disease, was first recorded during the epidemics in Batavia (Jakarta) and Cairo in 1779. Subsequent epidemics (1780 to 1901) were reported in Philadelphia, Zanzibar, Calcutta, the West Indies, and Hong Kong. Where the relevant factors considered for all these epidemics were thought to be DENV, many others believed that it was not only dengue, but that the Chikungunya virus was also responsible for the epidemics [10,11]. The epidemic capability of dengue acquired a broad geographic distribution before the 18th century; however, the actual causes of dengue infection remained unknown until 1906 [12]. In 1907, Ashburn and Craig provided the first data indicating the filterable, ultramicroscopic character of an etiological agent, namely dengue, which is transmitted by a true vector Aedes aegypti [13]. Cleland et al. (1919) and Siler et al. (1926) subsequently confirmed that the DENV transmission process was akin to the “jungle cycle” of the yellow fever virus [14,15,16]. From 1922 to 1945, many outbreaks were recorded in the United States, Australia, Greece, and Japan. In 1922, it was estimated that 1 to 2 million individuals in the southern United States were affected. In the three decades after World War II, dengue epidemics occurred sporadically in Central America and the Caribbean basin [17]. Socio-economic conditions after the war resulted in poor immunity in most populations and contributed to an increased dengue incidence throughout the world, especially in Southeast Asia [18]. From 1943 to 1944, a major breakthrough occurred in the treatment of dengue fever, when Dr. R. Kimura (Japan) and Dr. S. Hotta (Hawaii) isolated and identified the DENV strains DENV-1 and DENV-2 [19]. In 1953, the DENV-3 and DENV-4 strains were first isolated from infected individuals in the Philippines and Thailand [20].
1.2. Vector and Non-Vector Transmission of Dengue
In most cases, DENV transmission is accomplished by the primary vectors, whereby A. aegypti and Aedes albopictus have long been recognized as hosts for the viruses [21,22]. A. aegypti is the most common primary epidemic-causing and predominant vector of dengue [6]. Other species that act as secondary vectors for carrying DENV include Aedes scutellaris, Aedes africanus (subgenus Stegomyia) and Aedes niveus (subgenus Finlaya), which are considered as sylvatic vectors. Meanwhile, other species such as Aedes taylorior, Aedes furcifer (subgenus Diceromyia), as well as Aedes mediovittatus (Gymnometopa) and Aedes triseriatus (Protomacleaya) also play key roles as secondary dengue vectors for carrying dengue infectious virus [22,23].
The non-mosquito DENV transmission route is infrequent and accidental, but it has great importance for physicians. Several authors have reported cases of transmission without a mosquito vector, which occurred in different ways, including through needles, congenitally, through mucocutaneous exposure, and through bone marrow transplants [24]. DENV infection of healthcare workers usually occurs as a result of needle injuries [25,26,27,28,29], while vertical transmission is responsible for dengue fever cases in infants born to a DHF-diagnosed mother [30]. A child from Puerto Rico attained DENV-4 via bone marrow transplant and subsequently died [31], and DENV-3 was transmitted to a healthcare worker through blood (mucocutaneous exposure) [32]. These are only a few examples of non-vector dengue transmission.
1.3. Transmission Process
A few hundred years ago, dengue was primarily a sylvatic disease. The sylvatic cycle is ecologically and evolutionarily-distinct from the human transmission cycle, causing sporadic outbreaks in humans [33,34]. A sylvatic cycle that serves as an enzootic cycle involving canopy-dwelling Aedes mosquitoes and lower primates for dengue transmission has been well-documented in the rainforests of western Africa and Southeast Asia, including Peninsular Malaysia and eastern Senegal [6,33]. Aedes luteocephalus, A. furcifer, and A. taylori were the principal transmission vectors in Africa [35,36,37,38,39], and the primatophilic canopy-dwelling mosquitoes of the A. niveus s.l. complex—a group that includes Aedes pseudoniveus, Aedes subniveus, Aedes vanus, Aedes albolateralis, Aedes niveoidesan, and Aedes novoniveus—were the principal vectors in Asia (Figure 1) [40]. In other locations in tropical Africa and Asia, probable rates of sylvatic DENV transmission to humans are unknown, but appeared to be minimal [41,42]. In North America, several species of new-world non-human primates were found to be resistant to DENV infection, and no cases of sylvatic DENV transmission were recorded. Similarly, no evidence of enzootic circulation through lower primates have been found in Panama [43]. However, seroconversions among indigenous Ayoreo Indians living in an isolated forested region of Bolivia, where A. aegypti is absent, recommend that sylvatic DENV transmission may happen in the region [44]. Cross-species transmission of DENV in rural areas of Africa and Asia from non-human primates to humans tends to occur when great numbers of the enzootic vector(s) are present [45]. The sylvan environments in rural areas of Africa and Asia can be a source of an abundant amount of primary vector A. albopictus, which is responsible for the transfer of the virus into human habitats [46]. Endemic/epidemic cycles involved the human host, and viruses are transmitted mainly by A. aegypti, A. albopictus, and other mosquitos that serve as secondary vectors, such as A. mediovittatus, Aedes polynesiensis and other members of Aedes scutellarin [47,48].
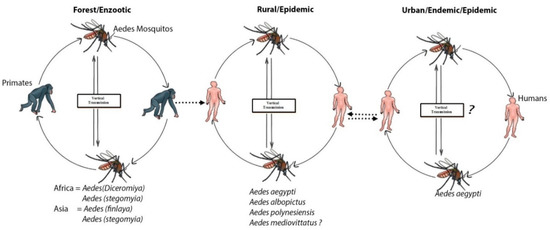
Figure 1.
Sylvatic transmission of dengue virus (DENV) from lower primates to humans: Serotypes of DENV-1–4 emerged independently via the sylvatic cycle and subsequently disseminated among human populations. The forests of Southeast Asia and West Africa maintained the sylvatic cycle and supported contact with human populations, leading to an urban endemic/epidemic cycle between Aedes mosquitoes and human repository hosts.
DENV-infected small A. aegypti bite humans and thus initiate DENV transmission. These mosquitos lay eggs and rest indoors in coconut shells, artificial containers, vases with wastewater, and old automobile tires found in and around homes. At dawn, 2–3 h after sunrise, as well as during sunset (in daylight hours), adult mosquitoes prefer to feed on humans, to which these periods are known as the “two peaks of biting activity”. A. aegypti females very often feed on several persons, thus rapidly accelerating the transmission of DENV [49]. After transmission into the human host and following a 3 to 14-day incubation period, most affected individuals enter a 2 to 10-day acute febrile period and experience nonspecific signs and symptoms. During this viremic stage, other biting mosquitoes become infected and viruses circulate in the peripheral blood.
3. Targeting Host Metabolic Pathway
Viral infections can modify many physiological as well as metabolic pathways. Metabolic changes include lipid metabolism, in addition to stimulation of glycolytic pathways toward an energetically favorable state, which modifies membrane lipid and other composition for viral replication and virion envelopment. Targeting intracellular metabolic pathways and their pharmacological inhibition can reduce DENV RNA synthesis and infectious virion production [60], which may serve as successful DENV antiviral strategies. For example, lipid and glucose metabolic pathways are necessary for every step in the replication cycle of DENV. Both steps in the replication cycles of DENV can be inhibited by different pharmacological agents, and the agents/inhibitors developed mainly target the host factors that mediate lipid synthesis, lipid and glucose metabolism, and trafficking pathways. Despite this, targeting host glucose and lipid metabolism and trafficking as an antiviral strategy by blockade of entire pathways may be limited because of host toxicity [50]. Knowledge of the molecular details of lipid and glucose metabolic pathways, regulatory enzymes of the pathways and metabolic function in replication, and the mechanisms by which specific glucose and lipids are generated during DENV infection, as well as its trafficking to the relevant factors, will help to enable more targeted antiviral strategies without creating any toxic effects on the host cell.
3.1. Targeting the Host Glycolytic Pathway
The host’s cellular metabolism provides the necessary energy (ATP), biosynthetic building blocks, and other important molecules required for viral replication. In DENV infection, a major change occurs in the central carbon metabolism, especially in glycolysis, whereby the expression of both glucose transporter I (GLUTI) and hexokinase II (HK-II) is up-regulated and glucose consumption is increased in DENV-infected cells. DENV activates the glycolytic pathway for viral metabolic requirements and life cycles, including energy, replication, and biosynthetic building blocks [60]. Glucose and glutamine serve as the main carbon sources in healthy cells and the tricarboxylic acid (TCA) cycle generates ATP using the oxidation of glucose via glycolysis. However, in some cases, glutamine serves as an ATP generator in the TCA cycle instead of glucose, so that it can be utilized for biosynthetic processes (Figure 2), such as in the case cancer cells and human cytomegalovirus (HCMV) cells [3,4,5,6]. As DENV activates the host glycolytic pathway for generating their necessary building blocks, pharmacologic regulation of glycolysis significantly blocks infectious DENV production. Krystal and colleagues reported that glycolysis inhibition through sodium oxamate and 2-deoxy-d-glucose (2DG) treatment can result in a significant reduction in DENV replication [5].
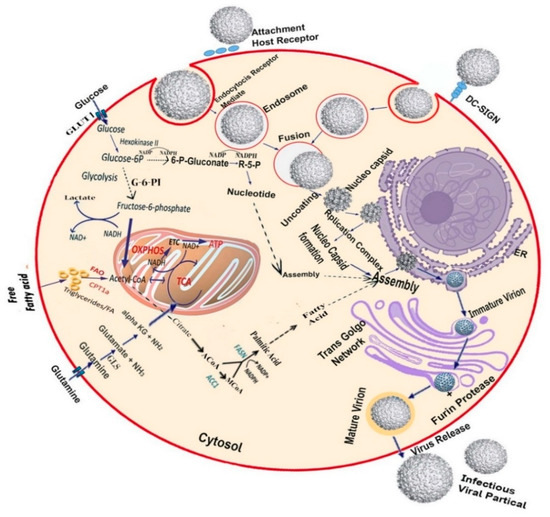
Figure 2.
Host metabolic pathways are necessary for DENV replication: As an enveloped virus, DENV requires fatty acids for replication and virion envelopment, resulting in virus-mediated modifications in the host cellular systems. These modifications lead to alterations in glucose and glutamine metabolism, as well as fatty acid synthesis. At the primary level of infection, lipid droplets and nucleotides are reabsorbed into the endoplasmic reticulum (ER) and subsequently assemble with the DENV virus. Glucose uptake in DENV-infected cells may increase through the induction of the glucose transporter 4 (GLUT-4) or overexpression of glucose transporter 1 (GLUT-1) and hexokinase II (HK-II), the first enzyme of glycolysis. DENV infection alters glucose metabolism allosterically by up-regulation of glycolytic enzymes. Infected cells stimulate glycolysis to produce ATP through the tricarboxylic acid (TCA) cycle. It also generates citrate, which is a precursor of fatty acid biosynthesis. Glucose carbons are diverted and subsequently migrate to the cytoplasm from the TCA cycle through citrate. Exogenous glutamine uptake is increased in DENV-infected cells [5]. The TCA cycle is maintained by glutaminolysis enzymes that are induced by DENV, whereas imported glutamine was converted into α-ketoglutarate. Fatty acid and sterol synthesis are upregulated, so that acetyl-coenzyme A (AcCoA) can be used for fatty acid synthesis. Lipid synthetic enzymes are modified to generate a large amount of distinct membrane lipid [60,61]. Experimentally limiting glucose and fatty acid synthesis during DENV infection, along with limiting glutamine levels, can help prevent infections.
3.2. Targeting the Host Lipid Biosynthesis Pathway
As an enveloped virus, DENV stimulates the lipid biosynthesis pathway for essential membrane formation. Fatty acids, triglycerides, and other lipid compositions of the host are utilized by Flaviviruses for envelope formation. Membrane composition is not only required for the formation of the envelope but is also needed for inducing viral infection in many ways. Replication includes virion egress and assembly requires a great number of fatty acids and their derivatives that generate membrane. In DENV-infected cells, fatty acid synthesis is regulated to utilize acetyl-coenzyme A (AcCoA) for generating most distinct membrane lipids (Figure 2). DENV stimulates fatty acid biosynthesis by the help of the important cofactor fatty acid synthase (FASN), which was first identified through the DENV-2 replicon mediated siRNA screening [60,62]. Lipophagy is known as a type of selective autophagy that transports lipids for β-oxidation. Several studies indicate that DENV infection induces pro-viral autophagy [62,63,64,65,66]. The lipids accumulated in auto-phagosomes next transport to mitochondria, increasing the β-oxidation rate, which generates energy and plays a key role in lipophagy, thus assisting DENV replication. Additionally, NADPH that arises through β-oxidation utilizes a cofactor of FASN and it may stimulate the fatty acid synthesis for DENV replication. In DENV-infected cells, both fatty acid synthesis and lipophagy process take place at the same time. In contrast, both processes do not occur in natural cells at the same time [67]. Extant research has shown that pharmacological inhibition of FASN [68] and mevalonate diphosphate decarboxylase, an enzyme required for cholesterol biosynthesis [60], can decrease DENV production in host cells.
3.3. Targeting the Host Nucleoside Biosynthesis Pathway
All viruses require host nucleosides for replication. Host proteins associated with nucleoside biosynthesis can thus be targeted as anti-dengue therapeutics. Nucleotide guanosine 5′-triphosphate (GTP) pool depletion has emerged as a significant system for repressing Flaviviruses. Guanine biosynthesis can be inhibited through antiviral ribavirin [69]. Dihydroorotate dehydrogenase (DHODH) is a mitochondrial protein that catalyzes the oxidation of dihydroorotate to orotate. It is an essential enzyme in the de novo pyrimidine biosynthesis pathway. Available evidence indicates that using brequinar (a known DHODH inhibitor), an anti-metabolite in cancer and immune-suppression, can inhibit DENV type 1,2,3 (DENV-1,2,3) serotypes. However, it cannot inhibit DENV type 2 (DENV-2) variants because of resistance against brequinar, which was also cross-resistant to compound NITD-982 [70,71]. The NITD-982 analogue directly bound to the DHODH protein. A study also suggests that compound NITD-982 is also capable of inhibiting host DHODH [69]. An in vitro study of the compound shows a great potency against DENV serotypes, but the compound did not show any efficacy because of the exogenous uptake of pyrimidine from the diet in the DENV-AG129 mouse model (deficient in interferon alpha/beta and gamma receptor signalling). Targeting the enzymes that play a key role in supplying DENV nucleoside can therefore be effective for antiviral therapeutics.
4. Targeting Host Cellular Receptors and Attachment Factors
Several host factors at the cellular level play a key role in the DENV virus entry process, but attachment factors and receptors are deemed the most important factors. Molecules in mammalian cells can act as attachment factors and receptors. Dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) [72,73] and glycosaminoglycans (GAGs) [74] are the first line of attachment factors and receptors, while second-line molecular factors include the GRP-78—also known as binding immunoglobulin protein (BiP), the laminin receptor [75] and the T-cell immunoglobulin and mucin domain (TIM), and Tyro3, Axl, and Mer (TAM) receptors [76]. Glycosphingolipids (GSLs), chaperone-proteins, and undefined proteins have been reported as potential treatment candidates. Targeting host factors involved in DENV attachment can thus have a beneficial antiviral potential.
4.1. Heparin and Heparan Sulfate (Glycosaminoglycans)
Heparan sulfate (HS) belongs to the family of glycosaminoglycans (GAGs). It initiates interactions between the DENV envelope protein and the host cell (Figure 3). HS also exhibits anti-DENV like molecular properties. For example, HS that was extracted from shrimp heads exhibits a strong inhibitory effect on infections caused by DENV [74]. Chondroitin sulfate, a curdlan sulfate that inhibits all DENV serotypes, has been found in baby hamster kidney 21 (BHK-21), Vero cells, and the rhesus monkey kidney cell (LLC-MK2) line [77,78]. Suramin [79,80], heparin [81], fucoidans [82], K5 polysaccharide from Escherichia coli [83], the heparan sulfate (HS) mimetic PI88 [80], α-d-glucan [84], GAG [85], and dextran sulfate (DS, MW > 500,000 Da) [85] are also able to inhibit DENV-2, whereas carrageenan [86] and DL-galactan hybrids extracted from red seaweed that lack cytotoxic effects and anticoagulant properties [86] can inhibit both DENV-2 and DENV-3 virus-infected cells (Table 1). Hence, targeting host heparin and heparan sulfate mimetics may act as potential antivirals that can aid in remediating the disease.
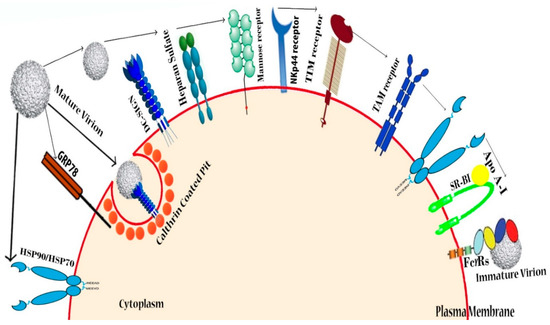
Figure 3.
DENV receptors in human cells: DENV recognizes various types of receptors present in the host cells. Heparan sulfate was identified as the first molecule responsible for DENV entry into mammalian cells, while sulfated glycosaminoglycans—ubiquitous molecules on the cell surface—were found to play a role in mediating DENV attachment. CLEC4L (C-type lectin domain family 4, member L), CLEC4M (C-type lectin domain family 4, member M), and mannose receptor/CLEC13D/CD206 (cluster of differentiation 206) are some conventional C-type lectin receptors (CLRs) that have a high affinity towards high-mannose ligands involved in DENV entry into target cells, where C-type lectin domain family 5 member A (CLEC5A) lectin acts as a signaling receptor for releasing proinflammatory cytokine. Cell surface chaperones HSP-90, HSP-70 and GRP-78, all of which are part of the receptor complex, assist in the DENV binding. Lipid receptors phosphatydil serine (PtdSer), T-cell immunoglobulin and mucin domain (TIM), and Tyro3, Axl, and Mer (TAM) types of putative receptors are relevant for viral entry into the human 293 T-cell line [87]. Another receptor, SR-BI, is also responsible for lipoprotein-associated interaction with DENV and allows the virus to enter the host [76].

Table 1.
List of DENV inhibitors isolated by targeting host process. LLC-MK2 = rhesus monkey kidney cell, BHK = baby hamster kidney, GAG = glycosaminoglycan, DC-SIGN = dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin, MEK = MAPK/extracellular receptor kinase (ERK) kinase, NTRK1 = neurotrophic receptor tyrosine kinase 1, MAPKAPK = MAP kinase-activated protein kinase, RIG-I = retinoic-acid inducible gene I, IMP dehydrogenase = Inosine-5′-monophosphate dehydrogenase.
4.2. DC-SIGN
DC (dendritic cell) is a cell surface attachment factor present in every tissue [88]. It is known as Cluster of Differentiation 209 (CD209) and was found to regulate DC trafficking and T-cell synapse formation expressed by human immature dendritic cells in the plasma membrane capable of recognizing DENV. It not only recognizes DENV, but also allows its surface transport through the process of endocytosis, resulting in cell infectivity (Figure 3) [73,89]. Although the role of DC-SIGN in DENV entry remains controversial [90,91,92], many authors posit that DENV is handed over to another unidentified co-receptor for movement throughout the cells [90]. Findings yielded by a large number of studies indicate that DENV can recognize cell surface DC-SIGN, as well as move into the plasma membrane through various co-receptors. For example, HIV co-receptor CCR5 allows viral attachment, resulting in entry into the cell [89,93]. As DENV can pass to clathrin-coated structures (CCS) through virus receptor complexes [22], receptors that mediate DC-SIGN are potential targets for DENV antiviral treatment. Several compounds, such as glycomimetic DC-SIGN ligand and plant lectins from Hippeastrum hybrid, Galanthus nivalis, and Urtica dioica (Table 1), were found to act as strong inhibitors of DENV infection in DC-transfected cells. These findings can potentially be utilized in novel strategies aimed at enhancing the efficiency of a wide spectrum of antiviral therapies to block DENV virus uptake.
4.3. Other Possible Receptors
Viral attachment to the cell requires many sequential interactions with various receptors. DC-SIGN and glycosaminoglycans (GAGs) are known as the first line of attachment receptors. The second line of higher affinity receptors may then be recruited to permit DENV entry because of the diverse tissue tropism of the virus [94]. DENV uses two pathways to enter DCs, whereby infection can enter immature DCs through DC-SIGN, or through Fc gamma receptors (FcɣRs) in mature DCs. DCs expressed FcɣRs as the second line of a higher affinity receptor in host cells, and activation of FcɣRs in hematopoietic cells serves to remove antibody-opsonized antigens—including DENV—from the body circulation system. However, cross-reactive or sub-neutralizing levels of antibodies grant an alternative entry pathway of DENV, where DENV enters monocytes, macrophages, and dendritic cells through the activating FcɣRs [95]. DENV can enter cells via cellular attachment molecules’ TIM and TAM receptors [76], as both are able to recognize the apoptotic marker phosphatidylserine (PtdSer) and are responsible for the engulfment and removal of apoptotic cells. Since DENV is a virus that exposes PtdSer in its membrane, it naturally enters the cell as a PtdSer through direct binding of the TIM receptor or indirectly via the TAM receptor. Moreover, apoptotic marker PtdSer binds with TIM and TAM by the help of the growth arrest-specific 6 (Gas6) binder molecule (Figure 3) [96,97]. Cell surface chaperones, heat shock protein (HSP-90, HSP-70), and GRP-78 are known as a receptor complex, which allows DENV entry into human cells from hepatic, neural, and monocytic cells [75,98]. The interaction between apolipoprotein A-I and the scavenger receptor class B type I (SCARB1), also known as SR-BI, promotes DENV infections, necessitating further research in order to elucidate the functional importance of lipoproteins in dengue pathogenesis [87].
5. Targeting Host Proteins or Enzymes
Host proteins and enzymes play an integral role in Flaviviruses and are necessary for their entry into the host, as well as for replication and assembly. DENV dominates some processes to manipulate the host cell proteins and metabolic pathways. Post-translational modifications, especially the carbohydrate modification pathways (e.g., glycosylation), have been demonstrated as targets against Flaviviruses [94]. Toxicity and side-effects that were generated through the inhibition of proteins must therefore be carefully considered when targeting host proteins as antiviral therapeutics [99]. However, the potential of such compounds for screening purposes is tremendous. The host proteins are potential antiviral targets and have been shown in extant studies focusing on inhibiting such compounds so as to not be toxic for the host [100] (Table 1).
5.1. Targeting Host Protease
Host protease is an effective target for antiviral drug development, as it is essential for virus replication. Host proteases, such as furin and signalase, have been used to cleave the DENV RNA genome co- and post-translationally and were translated as a polyprotein [101]. Correct processing can be used to generate the polyproteins essential for the viral life cycle [100,102]. Since host protease required for virus polyprotein formation is a basis of DENV replication, it can be a strong target for antiviral production. Protease furin is enriched in the Golgi apparatus of the host cell, where it assists in cleaving the DENV prM proteins, resulting in the formation of mature active forms of the virion M protein that plays an important role during DENV infections [99,103]. The signal peptidase on endoplasmic reticulum (ER) membrane cleaves the C/E-prM junctions [104]. It processes many secretory proteins, but the inhibition is likely to have side-effects. Nonetheless, recent research indicates that peptidomimetic furin luteolin inhibits the viral maturation process in an uncompetitive manner [105]. Peptide compound 45 and 46, as well as cavinafangin, have also been used as protease inhibitors in DENV-infected cells (Table 1). This finding indicates that further study is needed for the development of host proteases as an antiviral target.
5.2. Targeting Host Kinases
Host kinases are involved in DENV assembly and secretion. Protein kinase inhibits the dengue replication cycle and, in the absence of a cytotoxicity cause, multilog decreases in the viral titer. Dasatinib and saracatinib (AZD0530) are inhibitors of the protein kinase c-Src [106]. Compound 16i is a kinase inhibitor that is ten times more potent than ribavirin. It can thus capture both the virus NS5-NS3 interaction and the host kinases c-Src/Fyn [107]. SFV785 and derivative compounds affect the neurotrophic receptor tyrosine kinase 1 (NTRK1) and MAP kinase-activated protein kinase 5 (MAPKAPK5) kinase activity and inhibit DENV propagation [108]. GNF-2 and imatinib inhibit DENV but are mediated by cellular Abl kinases [109]. Many compounds that inhibit kinase activity by regulating mitogen-activated protein kinase (MAPK or MAP kinase) have been developed. Examples of such compounds are CGP57380 that inhibits extracellular receptor kinase (ERK) and p38 pathways in DENV-2 infected cells; and PD98059, U0126, and FR180204 that inhibit the MAPK/ERK kinase (MEK); while AR-12 inhibits PI3K/JAKT pathway by expressing GRP-78 for all four DENV serotypes. Sunitinib and erlotinib, as well as isothiazolo[5,4-b] pyridines and Imidazo[1,2-b] pyridazine, are inhibitors of AAK1 and GAK pathways that inhibit DENV replication, whereas U0126 inhibits the ERK pathway to reduce the replication DENV-2 and -3 infected cells (Table 1). These findings provide pharmacological evidence that kinase has the potential to become a new class of antiviral target.
5.3. Glucosidase Inhibitors
Glucosidase is a type of host enzyme, liable for viral maturation and proper folding. It initiates the process of glycosylation in N-linked oligosaccharides of the viral prM and E glycoproteins [110]. DENV structural protein prM and E glycoproteins are translocated into the host endoplasmic reticulum lumen (ER). During this time, a high mannose-rich oligosaccharide -Glc-3-Man-9-GlcNAc-2- (a total 14 residue core unit) is added co-translationally [111,112]. The resulting N-linked glycans are generated through the help of enzyme glucosidases I and II, where glucosidase I removes the terminal α-(1, 2) linked glucose from Glc-3-Man-9-GlcNAc-2, and glucosidase II removes the second and possibly the third terminal α-(1, 3) linked glucose residues from the Glc-3-Man-9-GlcNAc-2 oligosaccharide precursor, whereby the process is denoted “glucose trimming”. After that, it leaves the protein monoglucosylated and binds to the endoplasmic reticulum chaperones (calnexin or calreticulin) for proper folding [113]. As DENV prME heterodimer formation is not influenced by the inhibition of glucose trimming, it helps in generating a less stable complex characterized by reduced folding efficiency. It has been shown that the folding, stability, secretion, and activity of DENV glycoproteins in the ER depends on the trimming of these N-linked carbohydrates at N-130 and N-207 [110,112,114], thus rendering the responsible cellular glucosidase a potential host target. Castanospermine, a type of naturally occurring iminosugar, and N-nonyl-deoxy-nojirimycin (NNDNJ) isolated from Bacillus, are effective glucosidase inhibitors that have been indicated by both in vitro and in vivo studies in mice. The α-glucosidase inhibitor celgosivir has been shown to inhibit DENV, and treatment with celgosivir have shown to causing an improved survival rate in DENV infected mouse. The efficacy analyses were performed in patients with dengue fever [115]. The compound celgosivir is generally safe and well-tolerated but does not seem to reduce the viral load or fever burden in patients with dengue. Celgosivir derivative of 6-O-butanoyl is an oral pro-drug of castanospermine that can cause strong inhibition of DENV-1–4 [115,116]. Iminosugar drug UV-4, derived from deoxynojirimycin, was reported to decrease mortality in an “antibody-dependent enhancement” model of secondary DENV infection [117]. It is also noteworthy that α-glucosidase substrate mimics, such as CM 9 to 78 and CM 1018 (Table 1), are currently under development [118].
6. Targeting Host Immunity and Inflammatory Pathways
After the virus infects the host, cell signals are generated to block the dissemination of DENV. DENV causes acute disease without persistent infection. All virus strains have developed strategies that bypass the innate and adaptive immune response. Therefore, DENV does not escape the host defense mechanism. Innate immunity is known as the first line of antiviral defense mechanism that uses cytokine interferon I (IFN-I) for host defense purposes. It also results in the rapid activation of adaptive immune responses, resulting in the complete elimination of the virus [119]. To achieve this beneficial response, the immune system induces various factors and a series of gene expressions, including interferon-stimulated genes (ISGs), inflammatory responses, plasma, and vascular endothelium leakage, along with the disease progress factors both in infected and uninfected cells [120]. Hence, a better understanding of the host immune response during DENV infection and the evasion mechanisms would have great importance for potential antiviral production.
6.1. Targeting Host Immune Factors Involved in DENV Sensing
Viral pathogenic factors can be recognized through the pattern recognition receptors (PRRs) in the host cells. The endosomal toll-like receptors (TLRs) that recognize double-strand RNA (dsRNA) in endosomes, and the cytoplasmic receptor family complex form DEAD box, DEAH and the SKI proteins (DExD/H box), RNA helicases, Retinoic-acid inducible gene I (RIG-I) and Melanoma differentiation-associated protein 5 (MDA5)) that recognize intracellular double-strand dsRNA or single-strand viral RNA (ssRNA) [121], are the most important sensors in human cells that are implicated in detecting viral nucleic acids. After viral recognition by these two molecules, the interferon regulatory factors and the NF-kB (Nuclear factor kappa light chain enhancer of activated B cells) transcriptional molecules are activated, and these signaling cascades aid in generating IFN-α/β and inflammatory cytokines to activate the DC for an antiviral response. In the case of DENV, infected cells do not express themselves as a viral ligand sensed by retinoic-acid inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5), which is the reason why viral antibody is not detected by RNA sensors and C-type lectin domain family 5 member A (CLEC5A) does not block DENV infection [122]. Other important molecules such as toll-like receptors (TLR3) also sense dsRNA and can restrict DENV replication in different cell lineages [123,124,125]. DENV produces excess IFN-I (essential for DENV-induced production) through toll-like receptors 7 (TLR7) in plasmacytoid-DC, which can also restrict DENV replication in different cell lineages, as TLR3 [126]. Additionally, increasing inflammatory and humoral responses that decrease DENV replication have been found, such as when TLR3 and TLR7/8 agonists administrated into rhesus macaques aided in enhanced antiviral mechanisms during primary DENV infection [127]. Gene expression analysis indicates that RIG-I and MDA5 receptors promote the sensing ability in DENV-infected cells. The infected cells interact with these two receptors and stimulate interferon regulatory factor 3 (IRF-3) and the nuclear factor NF-kB that produces interferon beta (IFN-β) promoters, resulting in impaired replication of DENV. Additionally, DENV with double or single RIG-I/MDA5-deficient fibroblasts triggers both responses [128]. Furthermore, early detection of antibodies via RNA sensors (for examples RIG-I) enhanced DENV infection by tissue-resident mast cells that produce type IFN-I, along with chemokine ligands (CCL4, 5) and C-X-C motif chemokine ligands 10 (CXCL10) [129]. Consequently, human brain microvascular endothelial cells are infected with DENV, and rapid production of type-I IFN and proinflammatory cytokines were revoked after inhibition of RIG-I. Later, it was found that IFN-β production is induced by RIG-I, MDA5, and TLR3 sensors (major PRRs recognize innate responses to DENV infection), contributing to impairing DENV replication in vitro [130]. Activation of these major pattern recognition receptors (PRRs) by DENV generates a strong type of IFN-I response during human natural infections. Elevated levels of IFN-α for long periods in pediatric patients after the decrement period has also been reported [131]. Extant evidence (experimented upon mice) further indicates that DENV primary infection utilizes the interferon α/β receptor (IFNAR)-dependent (including STAT1-dependent and STAT1-independent) control mechanisms—where the STAT1 (Signal transducer and activator of transcription 1)-dependent mechanism controls the primary steps of infection, while the STAT1-independent mechanism controls the latter antiviral process—aiding virus propagation and disease control. In DENV primary infection, cells typically utilize both mechanisms [132,133]. Anti-CLEC5A blocks DENV by releasing pro-inflammatory cytokines and does not affect IFN-I production in the infected cells [134]. As CLEC5A blocks DENV infections, future studies are necessary to enhance CLEC5A activation.
6.2. Targeting Anti-Inflammatory Cytokine Populations
It has been demonstrated that excessive inflammation contributes to the pathogenesis of the severe form of dengue disease. Tumor necrosis factor (TNF)-α; interleukin (IL)-6, 8, 10; chemokine ligand (CCL)-2, 3; CXCL-8, 10; and interferon (IFN)-γ, all of which are responsible for primary and secondary DENV infection in humans, are excessively elevated and have been reported in patients with severe dengue disease [132,135,136,137,138,139,140,141]. These factors are activated by IFN-I and PRRs and were discussed in the preceding section.
Cytokines are not responsible in causing any primary and secondary DENV infections in humans. Cytokine IFN-γ production in the host cell plays protective roles during primary DENV infection. In primary DENV infections, production of IL-12 and IL-18 proinflammatory cytokines precede IFN-γ release, and optimal IFN-γ production relies on the combined action of these two cytokines. For example, higher levels of IL-12 and 18 cytokines that are required for optimal IFN-γ production are usually recorded for DF patients, but in the case of DHF patients (Grade III and IV), the levels of this cytokine were non-detectable [142,143,144]. It has been demonstrated that IFN-γ controls nitric oxide synthase II-mediated nitric oxide production that assists the host in resistance against primary DENV infection [145], which was previously found to inhibit DENV replication [146]. Sustained IFN-γ production is necessary during the acute phase of illness to protect the host against fever and viremia [147]. With increased production of IFN-γ, the survival rates enhanced in DHF patients [138]. Hence, IFN-γ can be a potential target for the host to control DENV replication and resistance to infection. Enhanced proinflammatory cytokine TNF-α production is also associated with severity of dengue manifestation in humans. For example, T-cells isolated from patients are found to contain higher amounts of TNF-α after ex vivo stimulation with DENV antigens [148]. Hence, the blocking of enhanced proinflammatory cytokine TNF-α might reduce the pathology due to the primary [149], [150] and secondary [151] infections. The migration inhibitory factor (MIF) is indicative of a more severe disease form during primary DENV infections [138]. Experiments have shown that DENV primary infections were less severe in MIF−/− mice, and they exhibited a significant delay in lethality, indicating that reduced proinflammatory cytokine levels (such as TNF-α) are correlated with lower viral loads at the initial phases of infection. Therefore, elevated production of the proinflammatory cytokines TNF-α and MIF during the host response to DENV infection favors more severe disease [152].
The chemokine system that plays a protective and pathologic role during DENV infections produces CXCL10 and activates CXCR3 (C-X-C chemokine receptor type 3) to improve host resistance against DENV infection [7]. Clinical studies in endemic areas indicated the presence of a correlation between DENV outcome and the level of CCL2, 3, 4 concentrations that were related to hypotension, thrombocytopenia, and hemorrhagic shock. Another study also found a link between CCL5 and DENV-induced hepatic dysfunction [135,139,153]. Reduced lethality rates, liver damage, alleviated leukocyte activation, and lower production of IL-6 have been found in chemokine receptor type 2 and type 4 (CCR2−/−and CCR4−/−) knockout mice with primary DENV infection. However, no difference in viral load has been found in the case of CCR-deficient mice. Hence, we can conclude that CXCR3 expresses protective host responses, but CCR2 and CCR4 cause infection rather than providing protection against DENV infection [154].
Cellular populations are also important for DENV infection. For example, cluster of differentiation 8 (CD8+) T cells are crucial for the control of viral replication, whereas an invariant natural killer T (iNKT) cell is important in the pathogenesis of dengue disease [155].
6.3. Targeting Host Plasma and Vascular Endothelium Leakage
Plasma leakage is an important factor in dengue disease progression and can cause DHF/DSS. Endothelium (primary fluid barrier) is changed by DENV, inducing edema and hemorrhage because of cell barrier permeability [200]. Vascular leakage can be blocked by FX06 (28-AA cleavage product), which decreases primary dengue infection. The protective effect of FX06 has been found to be elevated when combined with src/Fyn kinase. After activation, the 28-AA (28-Amino Acid) cleavage product Fyn dissociates from vascular endothelial and is combined with p190-Rho-GAP, an antagonist of RhoA activation. Thus, blocking vascular leakage by stabilization of endothelial cell development is important for DENV infection prevention [201]. The MIF inhibitor ISO-1 reduces permeability in the human hepatoma cell line. ISO-1, or the phosphoinositide 3-kinase (PI3K/AKT)/Ras-Raf-MEK-ERK/JNK signaling pathway, can be partially inhibited through the tight junction protein zonula occludens-1 (ZO-1) [202]. Chemokines CCL2 [168], leukocyte metalloproteinases 9 and 2 [203], and Box1 (HMGB1) [204] proteins are also involved in increasing vascular permeability. It has also been shown that type I-IFNs, IFN-β, VEGFR2 (Vascular Endothelial Growth Factor Receptor 2), and INF-α inhibit plasma leakage, where this process occurs with the help of endothelial stabilization [205].
6.4. Targeting Immune Factor Progress Disease after DENV Infection
Platelet-activating factor receptors (PAFR) released from macrophages, which were obtained previously from patients that were primarily infected with DENV-1, were found to be involved in the pathogenesis of severe dengue. The inflammatory response has also been demonstrated in DENV-2 virus infection, whereby PAF/PAFR was reported to interact with leukocytes and other cells [206].
Asunaprevir [192] and salidroside [191], extracts from Uncaria tomentosa, Norantea brasiliensis Choisy, Uncaria guianensis [196,197], promyelocytic leukemia protein intrinsic, Ivermectin [198], extracts from Cissampelos pareira Linn [198], and BST2/tetherin [199] are some of natural products that have advantages over synthetic drug design [207] and also tested as DENV inhibitors. Interferon type I [129], human heme oxygenase I, purinergic receptor P2X7 [195], and helicase with zinc linger 2 [194] are other examples of DENV inhibitors that have been experimentally studied to ascertain their dependence on host immunity, inflammation, pathogenesis of disease, and other disease progress factors (Table 1).
7. Conclusions and Remarks
Several remarkable points arise in the way of host inhibition processes that interrupt DENV replication along with infections. Host metabolic pathways serve as a source of energy, and molecular building blocks are required for the multiplication of DENV. For example, the primary glycolysis is conservatively required by DENV and exogenous glucose, and glutamine deprivation decreases DENV production, which could lead to the development of novel broad-spectrum antiviral therapies. The fatty acid metabolic pathway induces the activation of autophagy in DENV-infected cells by increased β-oxidation of fatty acids, and helps to bind with C proteins during virion assembly. An extensive body of research has been dedicated to the role of lipid and fatty acid metabolism during DENV infection, and the extant findings indicate that modulating lipid metabolisms in the host can be a viable anti-dengue therapeutic approach. Cellular nucleoside biosynthesis pathways of the host supply necessary nucleosides required for DENV replication. Hence, targeting the host nucleoside biosynthesis pathways can assist in blocking the essential functions of DENV as another avenue for antiviral drug development.
In the case of viral infection, initial attachment to the target cell is necessary to continue the viral life cycle. This process can occur in DENV through the interaction between viral surface proteins and host attachment factors, or receptor molecules present at the host cell surface. These factors are responsible for the binding of a viral protein that leads to viral cell entry and subsequent genome release into the cytoplasm. If the early steps of DENV infection cycle can be blocked by targeting host attachment factors or receptor molecules, this would be significant progress in the development of antiviral drugs.
Without utilizing host proteins and enzymes, DENV would be unable to propagate rapidly in host cells. Numerous host proteins are found to be essential in DENV replication. For example, host proteases aid in the RNA genome cleavage and polyprotein formation, while host kinases help in DENV assembly, and glucosidase is used in DENV maturation and folding. Every single step mentioned utilizes host proteins and enzymes, which are the most important factors for DENV multiplications. Hence, targeting one of these can reduce DENV production and may lead to an effective antiviral drug.
The inflammatory response is activated in host cells during DENV infection to clear the pathogen from the host immune system. Whenever the host senses the presence of the DENV virus, activation of innate and inflammatory pathways occurs as a means of eliminating the disease. Alternation of host responses is a hallmark of dengue infection, whereby weak innate immunity and inflammatory response may lead to parasite growth and disease advancement. Again, excessive inflammation may be the reason behind the pathogenesis of severe dengue disease. In that case, a reduction of proinflammatory molecules can help to decrease dengue-induced vascular leakage. In summary, we require a better understanding of host innate and inflammatory pathways, as this information can help to identify appropriate targets. Targeting such inhibitors may result in antiviral drug development (focusing on blocking inflammation and endothelial barrier permeability) without interfering with the host immune mechanisms. Resolving the issues discussed in this work can yield more comprehensive knowledge about DENV and related host factors that can be utilized in novel therapeutic targets for the development of anti-DENV drugs.
Author Contributions
All authors contributed equally to the writing of the review. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
This work was supported by the Ministry of Education Malaysia (MOE) through FRGS/1/2016/STG04/UIAM/02/1.
Acknowledgments
Our appreciation goes to the Department of Biotechnology Engineering, Faculty of Engineering, International Islamic University Malaysia, Virology Unit, Institute for Medical Research and Faculty of Veterinary Medicine, Malaysia, and University Malaysia Kelantan.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Buchmeier, M.J.; Peters, C.J.; de La, T.J.F.V. Arenaviridae: The Virus and Their Replication. Fields Virol. 2007, 1792–1827. [Google Scholar]
- Teles, F.R.R.; Prazeres, D.M.F.; Lima-Filho, J.L. Trends in Dengue Diagnosis. Rev. Med. Virol. 2005, 15, 287–302. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, E.R.A.; Mohana-Borges, R.; de Alencastro, R.B.; Horta, B.A.C. The Flavivirus Capsid Protein: Structure, Function and Perspectives towards Drug Design. Virus Res. 2017, 227, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Miller, N. Recent Progress in Dengue Vaccine Research and Development. Curr. Opin. Mol. 2010, 12, 31–38. [Google Scholar]
- Fontaine, K.A.; Sanchez, E.L.; Camarda, R.; Lagunoff, M. Dengue Virus Induces and Requires Glycolysis for Optimal Replication. J. Virol. 2015, 89, 2358–2366. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, S.S.; Blaney, J.E.; Durbin, A.P.; Murphy, B.R. Prospects for a Dengue Virus Vaccine. Nat. Rev. Microbiol. 2007, 5, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Halstead, S.B. Antibody, Macrophages, Dengue Virus Infection, Shock, and Hemorrhage: A Pathogenetic Cascade. Rev. Infect. Dis. 1989, 11, 830–839. [Google Scholar] [CrossRef]
- Christie, T. Remarks on “Kidinga Pepo”: A Peculiar Form of Exan-Thematous Disease. Br. Med. J. 1872, 1, 577–579. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J. Epidemic Dengue/Dengue Hemorrhagic Fever as a Public Health, Social and Economic Problem in the 21st Century. Trends Microbiol. 2002, 10, 100–103. [Google Scholar] [CrossRef]
- Carey, D.E. Chikungunya and Dengue: A Case of Mistaken Identity? J. Hist. Med. Allied Sci. 1971, 26, 243–262. [Google Scholar] [CrossRef] [PubMed]
- Mattingly, P.F. Ecological Aspects of the Evolution of Mosquito-Borne Virus Diseases. Trans. R. Soc. Trop. Med. Hyg. 1960, 54, 97–112. [Google Scholar] [CrossRef]
- Maas, O. On the Aetiology of Stuttering. J. Ment. Sci. 1946, 92, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Ashburn, P.M.; Craig, C.F.; Lieutenant, F.; SA, T. Experimental investigations regarding the etiology of dengue fever. Infect. Dis. 1907, 4, 440–475. [Google Scholar] [CrossRef]
- Cleland, J.B.; Bradley, B.; MacDonald, W. Further Experiments in the Etiology of Dengue Fever. J. Hyg. 1919, 18, 217–254. [Google Scholar] [CrossRef] [PubMed]
- Nishiura, H.; Halstead, S.B. Natural History of Dengue Virus (DENV) 1 and DENV 4 Infections. J. Infect. Dis. 2007, 195, 1007–1013. [Google Scholar] [CrossRef]
- Siler, J.F.; Hall, M.W.; Hitchens, A.P. Dengue: Its History, Epidemiology, Mechanism of Transmission, Etiology, Clinical Manifestations, Immunity, and Prevention. Philipp. J. Sci 1926, 29, 1–2. [Google Scholar]
- Davenport, F.M. Viral and Rickettsial Infections of Man. Am. J. Public Heal. Nations Heal. 2008, 49, 971. [Google Scholar] [CrossRef]
- Halstead, S.B. Mosquito-Borne Haemorrhagic Fevers of South and South-East Asia. Bull. World Health Organ. 1966, 35, 3–15. [Google Scholar]
- Konishi, E.; Kuno, G. In Memoriam: Susumu Hotta (1918–2011). Emerg. Infect. Dis. 2013, 19, 843–845. [Google Scholar] [CrossRef]
- Messina, J.P.; Brady, O.J.; Scott, T.W.; Zou, C.; Pigott, D.M.; Duda, K.A.; Simmons, C.P. Global Spread of Dengue Virus Types: Mapping the 70 Year History. Trends Microbiol. 2014, 22, 138–146. [Google Scholar] [CrossRef]
- Ebi, K.L.; Nealon, J. Dengue in a Changing Climate. Environ. Res. 2016, 151, 115–123. [Google Scholar] [CrossRef]
- Henchal, E.A.; Putnak, J.R. The dengue viruses. Clin. Microbiol. Rev. 1990, 3, 376–396. [Google Scholar] [CrossRef] [PubMed]
- Khetarpal, N.; Khanna, I. Dengue Fever: Causes, Complications, and Vaccine Strategies. J. Immunol. Res. 2016, 2016, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.H.; Mary, E. Non-Vector Transmission of Dengue and Other Mosquito-Borne Flaviviruses. Dengue Bull. 2005, 29, 18–31. [Google Scholar]
- Wazieres, D.; Elite, A.S.; Clinical, M.J. Nosocomial transmission of dengue from a needlestick injury. Lancet 1998, 351, 498. [Google Scholar] [CrossRef]
- Hirsch, J.F.; Deschamps, C.; Lhuillier, M. Transmission MŽtropolitaine d’une Dengue Par Inoculation Accidentelle Hospitalière. Ann. Med. Intern. 1990, 141, 629. [Google Scholar]
- Langgartner, J.; Audebert, F.; Schölmerich, J.; Glück, T. Dengue Virus Infection Transmitted by Needle Stick Injury. J. Infect. 2002, 44, 269–270. [Google Scholar] [CrossRef] [PubMed]
- Nemes, Z.; Kiss, G.; Madarassi, E.P.; Peterfi, Z.; Ferenczi, E.; Bakonyi, T.; Ternak, G. Nosocomial Transmission of Dengue. Emerg. Infect. Dis. 2004, 10, 1880. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.; De With, K.; Huzly, D.; Hufert, F.; Weidmann, M.; Breisinger, S.; Eppinger, S.; Kern, W.V.; Bauer, T.M. Nosocomial Acquisition of Dengue. Emerg. Infect. Dis. 2004, 10, 1872–1873. [Google Scholar] [CrossRef]
- Berberian, G.; Farina, D.; Rosanova, M.T.; Hidalgo, S.; Enria, D.; Mitchenko, A.; Moreno, J.; Sánchez, I.S. Dengue Perinatal; Perinatal Dengue Infection. Arch. Argent. Pediatr. 2011, 109, 232–236. [Google Scholar]
- Rigau-Pérez, J.G.; Vorndam, A.V.; Clark, G.G. The Dengue and Dengue Hemorrhagic Fever Epidemic in Puerto Rico, 1994–1995. Am. J. Trop. Med. Hyg. 2001, 64, 67–74. [Google Scholar] [CrossRef]
- Chen, L.H.; Wilson, M.E. Transmission of Dengue Virus without a Mosquito Vector: Nosocomial Mucocutaneous Transmission and Other Routes of Transmission. Clin. Infect. Dis. 2004, 39, e56–e60. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Vasilakis, N. Dengue-Quo Tu et Quo Vadis? Viruses 2011, 3, 1562–1608. [Google Scholar] [CrossRef]
- Holmes, E.C.; Twiddy, S.S. The Origin, Emergence and Evolutionary Genetics of Dengue Virus. Infect. Genet. Evol. 2003, 3, 19–28. [Google Scholar] [CrossRef]
- Diallo, M.; Ba, Y.; Sall, A.A.; Diop, O.M.; Ndione, J.A.; Mondo, M.; Girault, L.; Mathiot, C. Amplification of the Sylvatic Cycle of Dengue Virus Type 2, Senegal, 1999-2000: Entomologic Findings and Epidemiologic Considerations. Emerg. Infect. Dis. 2003, 9, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Hervy, J.P.; Legros, F.; Roche, J.C.; Monteny, N.; Diaco, B. Circulation Du Virus Dengue 2 Dans Plusieurs Milieux Boisés des Savanes Soudaniennes de La Région de Bobo-Dioulasso (Burkina Faso). Cahiers Orstom. Série Entomologie Médicale et Parasitologie 1984, 22, 135–143. [Google Scholar]
- Rodhain, F. The role of monkeys in the biology of dengue and yellow fever. Comp. Immunol. Microbiol. Infect. Dis. 1991, 14, 9–19. [Google Scholar] [CrossRef]
- Robin, Y.; Cornet, M.; Heme, G.; Le Gonidec, G. Isolation of Dengue Virus from Senegal/Isolement Du Virus de La Dengue Au Senegal. Ann. Virol. E 1980, 131, 149–154. [Google Scholar]
- Roche, J.C.; Cordellier, R.; Hervy, J.P.; Digoutte, J.P.; Monteny, N. Isolement de 96 Souches de Virus Dengue 2 à Partir de Moustiques Capturés En Cote-d’ivoire et Haute-Volta. Ann. L’institut Pasteur Virol. 1983, 134, 233–244. [Google Scholar] [CrossRef]
- Rudnick, A. Dengue Virus Ecology in Malaysia. Inst. Med. Res. Malays. Bull. 1986, 23, 3. [Google Scholar]
- Black IV, W.C.; Bennett, K.E.; Gorrochótegui-Escalante, N.; Barillas-Mury, C.V.; Fernández-Salas, I.; Muñoz, M.D.L.; Farfán-Alé, J.A.; Olson, K.E.; Beaty, B.J. Flavivirus Susceptibility in Aedes Aegypti. Arch. Med. Res. 2002, 33, 379–388. [Google Scholar] [CrossRef]
- Yuwono, J.; Suharyono, W.; Koiman, I.; Tsuchiya, Y.; Tagaya, I. Seroepidemiological Survey on Dengue and Japanese Encephalitis Virus Infections in Asian Monkeys. South. Asian J. Trop. Med. Public Health 1984, 15, 194–200. [Google Scholar]
- Rosen, L. Observations on the Epidemiology of Dengue in Panama. Am. J. Epidemiol. 1958, 68, 45–58. [Google Scholar] [CrossRef]
- Roberts, D.; Peyton, E.; Pinheiro, F.; Balderrama, F. Associations of Arbovirus Vectors with Gallery Forests and Domestic Environments in Southeastern Bolivia; Department of Entomology, Walter Reed Army Institute of Research: Washington, DC, USA, 1984. [Google Scholar]
- Germain, M.; Mouchet, J.; Cordellier, R.; Chippaux, A.; Cornet, M.; Herve, J.P.; Sureau, P.; Fabre, J.; Robin, Y. Epidemiology of yellow fever in Africa. Med. Infect. Dis. 1978, 8, 69–77. [Google Scholar]
- Rudnick, A.L.B.E.R.T. Ecology of Dengue Virus. Asian J. Infect. Dis. 1978, 2, 156–160. [Google Scholar]
- Freier, J.E.; Rosen, L. Vertical Transmission of Dengue Viruses by Aedes Mediovittatus. Am. J. Trop. Med. Hyg. 1988, 39, 218–222. [Google Scholar] [CrossRef]
- WHO. Dengue and Dengue Haemorrhagic Fever. 1990–1994. Relev. Epidemiol. Hebd. 1995, 70, 334–335. [Google Scholar]
- Gubler, D.J. Dengue and Dengue Hemorrhagic Fever, 1996. Epidemiol. Bull. 1996, 17, 12–14. [Google Scholar]
- Tai, A.W.; Benita, Y.; Peng, L.F.; Kim, S.; Sakamoto, N.; Xavier, R.J.; Chung, R.T. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 2009, 5, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.M.; Aizaki, H.; Machida, K.; Ou, J.-H.J.; Lai, M.M.C. Hepatitis C Virus Translation Preferentially Depends on Active RNA Replication. PLoS ONE 2012, 7, e43600. [Google Scholar] [CrossRef]
- Brass, A.L.; Dykxhoorn, D.M.; Benita, Y.; Yan, N.; Engelman, A.; Xavier, R.J.; Lieberman, J.; Elledge, S.J. Identification of Host Proteins Required for HIV Infection through a Functional Genomic Screen. Science 2008, 319, 921–926. [Google Scholar] [CrossRef] [PubMed]
- König, R.; Zhou, Y.; Elleder, D.; Diamond, T.L.; Bonamy, G.M.C.; Irelan, J.T.; Chiang, C.-Y.; Tu, B.P.; De Jesus, P.D.; Lilley, C.E.; et al. Global Analysis of Host-Pathogen Interactions That Regulate Early-Stage HIV-1 Replication. Cell 2008, 135, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xu, M.; Huang, Q.; Gates, A.T.; Zhang, X.D.; Castle, J.C.; Stec, E.; Ferrer, M.; Strulovici, B.; Hazuda, D.J.; et al. Genome-Scale RNAi Screen for Host Factors Required for HIV Replication. Cell Host Microbe 2008, 4, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Stertz, S.; Zhou, Y.; Inoue, A.; Hoffmann, H.; Bhattacharyya, S.; Alamares, J.G.; Tscherne, D.M.; Ortigoza, M.B.; Liang, Y.; Gao, Q.; et al. Human Host Factors Required for Influenza Virus Replication. Nature 2010, 463, 813–817. [Google Scholar]
- Hao, L.; Sakurai, A.; Watanabe, T.; Sorensen, E.; Nidom, C.A.; Newton, M.A.; Ahlquist, P.; Kawaoka, Y. Drosophila RNAi Screen Identifies Host Genes Important for Influenza Virus Replication. Nature 2008, 454, 890–893. [Google Scholar] [CrossRef] [PubMed]
- Martín-Acebes, M.A.; Blázquez, A.B.; Jiménez de Oya, N.; Escribano-Romero, E.; Saiz, J.C. West Nile Virus Replication Requires Fatty Acid Synthesis but Is Independent on Phosphatidylinositol-4-Phosphate Lipids. PLoS ONE 2011, 6, e24970. [Google Scholar] [CrossRef]
- Krishnan, M.N.; Ng, A.; Sukumaran, B.; Gilfoy, F.D.; Uchil, P.D.; Sultana, H.; Brass, A.L.; Adametz, R.; Tsui, M.; Qian, F.; et al. RNA Interference Screen for Human Genes Associated with West Nile Virus Infection. Nature 2008, 455, 242–245. [Google Scholar] [CrossRef]
- Sessions, O.M.; Barrows, N.J.; Souza-Neto, J.A.; Robinson, T.J.; Hershey, C.L.; Rodgers, M.A.; Ramirez, J.L.; Dimopoulos, G.; Yang, P.L.; Pearson, J.L.; et al. Discovery of Insect and Human Dengue Virus Host Factors. Nature 2009, 458, 1047–1050. [Google Scholar] [CrossRef]
- Jordan, T.X.; Randall, G. Flavivirus Modulation of Cellular Metabolism. Curr. Opin. Virol. 2016, 19, 7–10. [Google Scholar] [CrossRef]
- Yu, Y.; Clippinger, A.J.; Alwine, J.C. Viral effects on metabolism: Changes in glucose and glutamine utilization during human cytomegalovirus infection. Trends Microbiol. 2011, 19, 360–367. [Google Scholar] [CrossRef]
- Datan, E.; Roy, S.G.; Germain, G.; Zali, N.; McLean, J.E.; Golshan, G.; Harbajan, S.; Lockshin, R.A.; Zakeri, Z. Dengue-Induced Autophagy, Virus Replication and Protection from Cell Death Require ER Stress (PERK) Pathway Activation. Cell Death Dis. 2016, 7, e2127. [Google Scholar] [CrossRef] [PubMed]
- Heaton, N.S.; Randall, G. Dengue Virus-Induced Autophagy Regulates Lipid Metabolism. Cell Host Microbe 2010, 8, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Khakpoor, A.; Panyasrivanit, M.; Wikan, N.; Smith, D.R. A Role for Autophagolysosomes in Dengue Virus 3 Production in HepG2 Cells. J. Gen. Virol. 2009, 90, 1093–1103. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.R.; Hu, H.Y.; Kuo, S.H.; Lei, H.Y.; Lin, Y.S.; Yeh, T.M.; Liu, C.C.; Liu, H.S. Dengue Virus Infection Induces Autophagy: An in Vivo Study. J. Biomed. Sci. 2013, 20, 65. [Google Scholar] [CrossRef] [PubMed]
- Mateo, R.; Nagamine, C.M.; Spagnolo, J.; Méndez, E.; Rahe, M.; Gale, M.; Yuan, J.; Kirkegaard, K. Inhibition of Cellular Autophagy Deranges Dengue Virion Maturation. J. Virol. 2013, 87, 1312–1321. [Google Scholar] [CrossRef] [PubMed]
- Jordan, T.X.; Randall, G. Flaviviruses and Autophagy. In Autophagy, Infection, and the Immune Response; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 81–100. [Google Scholar]
- Rothwell, C.; LeBreton, A.; Young Ng, C.; Lim, J.Y.H.; Liu, W.; Vasudevan, S.; Labow, M.; Gu, F.; Gaither, L.A. Cholesterol Biosynthesis Modulation Regulates Dengue Viral Replication. Virology 2009, 389, 8–19. [Google Scholar] [CrossRef]
- Wang, Q.Y.; Bushell, S.; Qing, M.; Xu, H.Y.; Bonavia, A.; Nunes, S.; Zhou, J.; Poh, M.K.; Florez de Sessions, P.; Niyomrattanakit, P.; et al. Inhibition of Dengue Virus through Suppression of Host Pyrimidine Biosynthesis. J. Virol. 2011, 85, 6548–6556. [Google Scholar] [CrossRef]
- Liu, S.; Neidhardt, E.A.; Grossman, T.H.; Ocain, T.; Clardy, J. Structures of Human Dihydroorotate Dehydrogenase in Complex with Antiproliferative Agents. Structure 2000, 8, 25–33. [Google Scholar] [CrossRef]
- McLean, J.E.; Neidhardt, E.A.; Grossman, T.H.; Hedstrom, L. Multiple Inhibitor Analysis of the Brequinar and Leflunomide Binding Sites on Human Dihydroorotate Dehydrogenase. Biochemistry 2001, 40, 2194–2200. [Google Scholar] [CrossRef]
- Navarro-Sanchez, E.; Altmeyer, R.; Amara, A.; Schwartz, O.; Fieschi, F.; Virelizier, J.L.; Arenzana-Seisdedos, F.; Desprès, P. Dendritic-Cell-Specific ICAM3-Grabbing Non-Integrin Is Essential for the Productive Infection of Human Dendritic Cells by Mosquito-Cell-Derived Dengue Viruses. EMBO Rep. 2003, 4, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Tassaneetrithep, B.; Burgess, T.H.; Granelli-Piperno, A.; Trumpfheller, C.; Finke, J.; Sun, W.; Eller, M.A.; Pattanapanyasat, K.; Sarasombath, S.; Birx, D.L.; et al. DC-SIGN (CD209) Mediates Dengue Virus Infection of Human Dendritic Cells. J. Exp. Med. 2003, 197, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Hung, J.J.; Hsieh, M.T.; Young, M.J.; Kao, C.L.; King, C.C.; Chang, W. An External Loop Region of Domain III of Dengue Virus Type 2 Envelope Protein Is Involved in Serotype-Specific Binding to Mosquito but Not Mammalian Cells. J. Virol. 2004, 78, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Jindadamrongwech, S.; Thepparit, C.; Smith, D.R. Identification of GRP 78 (BiP) as a Liver Cell Expressed Receptor Element for Dengue Virus Serotype 2. Arch. Virol. 2004, 149, 915–927. [Google Scholar] [CrossRef] [PubMed]
- Meertens, L.; Carnec, X.; Lecoin, M.P.; Ramdasi, R.; Guivel-Benhassine, F.; Lew, E.; Lemke, G.; Schwartz, O.; Amara, A. The TIM and TAM Families of Phosphatidylserine Receptors Mediate Dengue Virus Entry. Cell Host Microbe 2012, 12, 544–557. [Google Scholar] [CrossRef]
- Ichiyama, K.; Gopala Reddy, S.B.; Zhang, L.F.; Chin, W.X.; Muschin, T.; Heinig, L.; Suzuki, Y.; Nanjundappa, H.; Yoshinaka, Y.; Ryo, A.; et al. Sulfated Polysaccharide, Curdlan Sulfate, Efficiently Prevents Entry/Fusion and Restricts Antibody-Dependent Enhancement of Dengue Virus Infection In Vitro: A Possible Candidate for Clinical Application. PLoS Negl. Trop. Dis. 2013, 7, e2188. [Google Scholar] [CrossRef]
- Kato, D.; Era, S.; Watanabe, I.; Arihara, M.; Sugiura, N.; Kimata, K.; Suzuki, Y.; Morita, K.; Hidari, K.I.P.J.; Suzuki, T. Antiviral Activity of Chondroitin Sulphate E Targeting Dengue Virus Envelope Protein. Antivir. Res. 2010, 88, 236–243. [Google Scholar] [CrossRef]
- Chen, Y.; Maguire, T.; Hileman, R.; Fromm, J.; Esko, J.; Linhardt, R.; Marks, R. Dengue Virus Infectivity Depends Envelope Binding Heparan Sulfate. Nat. Med. 1997, 3, 866–871. [Google Scholar] [CrossRef]
- Lee, E.; Pavy, M.; Young, N.; Freeman, C.; Lobigs, M. Antiviral Effect of the Heparan Sulfate Mimetic, PI-88, against Dengue and Encephalitic Flaviviruses. Antivir. Res. 2006, 69, 31–38. [Google Scholar] [CrossRef]
- Hung, S.L.; Lee, P.L.; Chen, H.W.; Chen, L.K.; Kao, C.L.; King, C.C. Analysis of the Steps Involved in Dengue Virus Entry into Host Cells. Virology 1999, 257, 156–167. [Google Scholar] [CrossRef]
- Hidari, K.I.P.J.; Takahashi, N.; Arihara, M.; Nagaoka, M.; Morita, K.; Suzuki, T. Structure and Anti-Dengue Virus Activity of Sulfated Polysaccharide from a Marine Alga. Biochem. Biophys. Res. Commun. 2008, 376, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Vervaeke, P.; Alen, M.; Noppen, S.; Schols, D.; Oreste, P.; Liekens, S. Sulfated Escherichia Coli K5 Polysaccharide Derivatives Inhibit Dengue Virus Infection of Human Microvascular Endothelial Cells by Interacting with the Viral Envelope Protein E Domain III. PLoS ONE 2013, 8, e74035. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Tang, W.; Tong, X.; Ding, K.; Zuo, J. Structure Elucidation and Sulfated Derivatives Preparation of Two α-d-Glucans from Gastrodia Elata Bl. and Their Anti-Dengue Virus Bioactivities. Carbohydr. Res. 2007, 342, 2230–2236. [Google Scholar] [CrossRef] [PubMed]
- Alen, M.M.F.; Kaptein, S.J.F.; De Burghgraeve, T.; Balzarini, J.; Neyts, J.; Schols, D. Antiviral Activity of Carbohydrate-Binding Agents and the Role of DC-SIGN in Dengue Virus Infection. Virology 2009, 387, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Talarico, L.B.; Damonte, E.B. Interference in Dengue Virus Adsorption and Uncoating by Carrageenans. Virology 2007, 363, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Reyes-del Valle, J.; Salas-Benito, J.; Soto-Acosta, R.; del Angel, R.M. Dengue Virus Cellular Receptors and Tropism. Curr. Trop. Med. Rep. 2014, 1, 36–43. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.H.; Engering, A.; Van Kooyk, Y. DC-SIGN, a C-Type Lectin on Dendritic Cells That Unveils Many Aspects of Dendritic Cell Biology. J. Leukoc. Biol. 2002, 271, 921–931. [Google Scholar]
- Liu, P.; Ridilla, M.; Patel, P.; Betts, L.; Gallichotte, E.; Shahidi, L.; Thompson, N.L.; Jacobson, K. Beyond Attachment: Roles of DC-SIGN in Dengue Virus Infection. Traffic 2017, 18, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Flipse, J.; Wilschut, J.; Traffic, J.S. Molecular mechanisms involved in antibody-dependent enhancement of dengue virus infection in humans. Traffic 2013, 14, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Lozach, P.Y.; Burleigh, L.; Staropoli, I.; Navarro-Sanchez, E.; Harriague, J.; Virelizier, J.L.; Rey, F.A.; Desprès, P.; Arenzana-Seisdedos, F.; Amara, A. Dendritic Cell-Specific Intercellular Adhesion Molecule 3-Grabbing Non-Integrin (DC-SIGN)-Mediated Enhancement of Dengue Virus Infection Is Independent of DC-SIGN Internalization Signals. J. Biol. Chem. 2005, 280, 23698–23708. [Google Scholar] [CrossRef] [PubMed]
- Rodenhuis-Zybert, I.A.; Wilschut, J.; Smit, J.M. Dengue Virus Life Cycle: Viral and Host Factors Modulating Infectivity. Cell. Mol. Life Sci. 2010, 67, 2773–2786. [Google Scholar] [CrossRef] [PubMed]
- Pugach, P.; Ketas, T.J.; Michael, E.; Moore, J.P. Neutralizing Antibody and Anti-Retroviral Drug Sensitivities of HIV-1 Isolates Resistant to Small Molecule CCR5 Inhibitors. Virology 2008, 377, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Barragan, J.D.J.; del Angel, R.M. Identification of a Putative Coreceptor on Vero Cells That Participates in Dengue 4 Virus Infection. J. Virol. 2002, 75, 7818–7827. [Google Scholar] [CrossRef] [PubMed]
- Boonnak, K.; Slike, B.M.; Burgess, T.H.; Mason, R.M.; Wu, S.-J.; Sun, P.; Porter, K.; Rudiman, I.F.; Yuwono, D.; Puthavathana, P.; et al. Role of Dendritic Cells in Antibody-Dependent Enhancement of Dengue Virus Infection. J. Virol. 2008, 82, 3939–3951. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Zagórska, A.; Lew, E.D.; Shrestha, B.; Rothlin, C.V.; Naughton, J.; Diamond, M.S.; Lemke, G.; Young, J.A.T. Enveloped Viruses Disable Innate Immune Responses in Dendritic Cells by Direct Activation of TAM Receptors. Cell Host Microbe 2013, 14, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Perera-Lecoin, M.; Meertens, L.; Carnec, X.; Viruses, A.A. Flavivirus entry receptors: An update. Viruses 2014, 6, 69–88. [Google Scholar] [CrossRef] [PubMed]
- Reyes-del Valle, J.; Chavez-Salinas, S.; Medina, F.; del Angel, R.M. Heat Shock Protein 90 and Heat Shock Protein 70 Are Components of Dengue Virus Receptor Complex in Human Cells. J. Virol. 2005, 79, 4557–4567. [Google Scholar] [CrossRef] [PubMed]
- Elshuber, S.; Allison, S.L.; Heinz, F.X.; Mandl, C.W. Cleavage of Protein PrM Is Necessary for Infection of BHK-21 Cells by Tick-Borne Encephalitis Virus. J. Gen. Virol. 2003, 84, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Arakaki, T.L.; Fang, N.X.; Fairlie, D.P.; Young, P.R.; Martin, J.L. Catalytically Active Dengue Virus NS3 Protease Forms Aggregates That Are Separable by Size Exclusion Chromatography. Protein Expr. Purif. 2002, 25, 241–247. [Google Scholar] [CrossRef]
- Melino, S.; Paci, M. Progress for dengue virus diseases. FEBS J. 2007, 274, 2986–3002. [Google Scholar] [CrossRef]
- Gouvea, I.E.; Izidoro, M.A.; Judice, W.A.S.; Cezari, M.H.S.; Caliendo, G.; Santagada, V.; dos Santos, C.N.D.; Queiroz, M.H.; Juliano, M.A.; Young, P.R.; et al. Substrate Specificity of Recombinant Dengue 2 Virus NS2B-NS3 Protease: Influence of Natural and Unnatural Basic Amino Acids on Hydrolysis of Synthetic Fluorescent Substrates. Arch. Biochem. Biophys. 2007, 457, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Stadler, K.; Allison, S.L.; Schalich, J.; Heinz, F.X. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 1997, 71, 8475–8481. [Google Scholar] [PubMed]
- Chambers, T.; Nestorowicz, A.; Amberg, S.; Rice, C. Mutagenesis of the Yellow Fever Virus Nonstructural Polyportein: A Catalitically Active NS3 Proteinase Domain and NS2B Are Required for Cleavages at Dibasic Sites. J. Virol. 1993, 65, 6797–6807. [Google Scholar]
- Peng, M.; Watanabe, S.; Chan, K.W.K.; He, Q.; Zhao, Y.; Zhang, Z.; Lai, X.; Luo, D.; Vasudevan, S.G.; Li, G. Luteolin restricts dengue virus replication through inhibition of the proprotein convertase furin. Antivir. Res. 2017, 143, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.J.H.; Yang, P.L. C-Src Protein Kinase Inhibitors Block Assembly and Maturation of Dengue Virus. Proc. Natl. Acad. Sci. USA 2007, 104, 3520–3525. [Google Scholar] [CrossRef] [PubMed]
- de Wispelaere, M.; LaCroix, A.J.; Yang, P.L. The Small Molecules AZD0530 and Dasatinib Inhibit Dengue Virus RNA Replication via Fyn Kinase. J. Virol. 2013, 87, 7367–7381. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Hosoya, T.; Leong, K.M.; Onogi, H.; Okuno, Y.; Hiramatsu, T.; Koyama, H.; Suzuki, M.; Hagiwara, M.; Garcia-Blanco, M.A. The Kinase Inhibitor Sfv785 Dislocates Dengue Virus Envelope Protein from the Replication Complex and Blocks Virus Assembly. PLoS ONE 2011, 6, e23246. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.J.; Miduturu, C.; Schmidt, A.G.; Zhu, X.; Pitts, J.D.; Wang, J.; Potisopon, S.; Zhang, J.; Wojciechowski, A.; Hann Chu, J.J.; et al. GNF-2 Inhibits Dengue Virus by Targeting Abl Kinases and the Viral e Protein. Cell Chem. Biol. 2016, 23, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Courageot, M.P.; Frenkiel, M.P.; Dos Santos, C.D.; Deubel, V.; Desprès, P. Alpha-Glucosidase Inhibitors Reduce Dengue Virus Production by Affecting the Initial Steps of Virion Morphogenesis in the Endoplasmic Reticulum. J. Virol. 2000, 74, 564–572. [Google Scholar] [CrossRef]
- Chambers, T. Flavivirus Genome Organization, Expression, And Replication. Annu. Rev. Microbiol. 1990, 44, 649–688. [Google Scholar] [CrossRef]
- Whitby, K.; Pierson, T.C.; Geiss, B.; Lane, K.; Engle, M.; Zhou, Y.; Doms, R.W.; Diamond, M.S. Castanospermine, a Potent Inhibitor of Dengue Virus Infection In Vitro and In Vivo. J. Virol. 2005, 79, 8698–8706. [Google Scholar] [CrossRef] [PubMed]
- Hebert, D.N.; Foellmer, B.; Helenius, A. Calnexin and Calreticulin Promote Folding, Delay Oligomerization and Suppress Degradation of Influenza Hemagglutinin in Microsomes. EMBO J. 1996, 15, 2961–2968. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.F.; Lee, C.J.; Liao, C.L.; Dwek, R.A.; Zitzmann, N.; Lin, Y.L. Antiviral Effects of an Iminosugar Derivative on Flavivirus Infections. J. Virol. 2002, 76, 3596–3604. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Rathore, A.P.S.; Sung, C.; Lu, F.; Khoo, Y.M.; Connolly, J.; Low, J.; Ooi, E.E.; Lee, H.S.; Vasudevan, S.G. Dose- and Schedule-Dependent Protective Efficacy of Celgosivir in a Lethal Mouse Model for Dengue Virus Infection Informs Dosing Regimen for a Proof of Concept Clinical Trial. Antivir. Res. 2012, 96, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.P.; Paradkar, P.N.; Watanabe, S.; Tan, K.H.; Sung, C.; Connolly, J.E.; Low, J.; Ooi, E.E.; Vasudevan, S.G. Celgosivir Treatment Misfolds Dengue Virus NS1 Protein, Induces Cellular pro-Survival Genes and Protects against Lethal Challenge Mouse Model. Antivir. Res. 2011, 92, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Perry, S.T.; Buck, M.D.; Plummer, E.M.; Penmasta, R.A.; Batra, H.; Stavale, E.J.; Warfield, K.L.; Dwek, R.A.; Butters, T.D.; Alonzi, D.S.; et al. An Iminosugar with Potent Inhibition of Dengue Virus Infection in Vivo. Antivir. Res. 2013, 98, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Schul, W.; Butters, T.D.; Yip, A.; Liu, B.; Goh, A.; Lakshminarayana, S.B.; Alonzi, D.; Reinkensmeier, G.; Pan, X.; et al. Combination of α-Glucosidase Inhibitor and Ribavirin for the Treatment of Dengue Virus Infection in Vitro and in Vivo. Antivir. Res. 2011, 89, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.F.; Saini, N.K.; Porcelli, S.A. Evasion of Innate and Adaptive Immunity by Mycobacterium Tuberculosis. Microbiol. Spectr. 2014, 2, 747–772. [Google Scholar] [CrossRef] [PubMed]
- Der, S.D.; Zhou, A.; Williams, B.R.G.; Silverman, R.H. Identification of Genes Differentially Regulated by Interferon α, β, or γ Using Oligonucleotide Arrays. Proc. Natl. Acad. Sci. USA 1998, 95, 15623–15628. [Google Scholar] [CrossRef] [PubMed]
- Meylan, E.; Tschopp, J.; Karin, M. Intracellular Pattern Recognition Receptors in the Host Response. Nature 2006, 442, 39–44. [Google Scholar] [CrossRef]
- Severa, M.; Fitzgerald, K.A. TLR-Mediated Activation of Type I IFN during Antiviral Immune Responses: Fighting the Battle to Win the War. Curr. Top. Microbiol. Immunol. 2007, 316, 167–192. [Google Scholar] [PubMed]
- Tsai, Y.T.; Chang, S.Y.; Lee, C.N.; Kao, C.L. Human TLR3 Recognizes Dengue Virus and Modulates Viral Replication in Vitro. Cell. Microbiol. 2009, 11, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Sen, G.C. dsRNA-activation of TLR3 and RLR signaling: gene induction-dependent and independent effects. J. Interferon Cytokine 2014, 34, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Wu, S.; Li, Y.; He, L.; Wu, M.; Jiang, L.; Feng, L.; Zhang, P.; Huang, X. Activation of Toll-Like Receptor 3 Impairs the Dengue Virus Serotype 2 Replication through Induction of IFN-β in Cultured Hepatoma Cells. PLoS ONE 2011, 6, e23346. [Google Scholar] [CrossRef] [PubMed]
- Sariol, C.A.; Martínez, M.I.; Rivera, F.; Rodríguez, I.V.; Pantoja, P.; Abel, K.; Arana, T.; Giavedoni, L.; Hodara, V.; White, L.J.; et al. Decreased Dengue Replication and an Increased Anti-Viral Humoral Response with the Use of Combined Toll-Like Receptor 3 and 7/8 Agonists in Macaques. PLoS ONE 2011, 6, e19323. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Ortiz, Z.G.; Warke, R.V.; Pacheco, L.; Xhaja, K.; Sarkar, D.; Fisher, P.B.; Shaw, S.K.; Martin, K.J.; Bosch, I. Discovering Innate Immunity Genes Using Differential Display: A Story of RNA Helicases. J. Cell. Physiol. 2006, 209, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.M.; Fornek, J.; Crochet, N.; Bajwa, G.; Perwitasari, O.; Martinez-Sobrido, L.; Akira, S.; Gill, M.A.; García-Sastre, A.; Katze, M.G.; et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008, 82, 335–345. [Google Scholar] [CrossRef]
- Brown, M.G.; McAlpine, S.M.; Huang, Y.Y.; Haidl, I.D.; Al-Afif, A.; Marshall, J.S.; Anderson, R. RNA Sensors Enable Human Mast Cell Anti-Viral Chemokine Production and IFN-Mediated Protection in Response to Antibody-Enhanced Dengue Virus Infection. PLoS ONE 2012, 7, e34055. [Google Scholar] [CrossRef]
- Nasirudeen, A.M.A.; Wong, H.H.; Thien, P.; Xu, S.; Lam, K.P.; Liu, D.X. RIG-I, MDA5 and TLR3 Synergistically Play an Important Role in Restriction of Dengue Virus Infection. PLoS Negl. Trop. Dis. 2011, 5, e926. [Google Scholar] [CrossRef]
- Kurane, I.; Innis, B.L.; Nimmannitya, S.; Nisalak, A.; Meager, A.; Ennis, F.A. High Levels of Interferon Alpha in the Sera of Children with Dengue Virus Infection. Am. J. Trop. Med. Hyg. 1993, 48, 222–229. [Google Scholar] [CrossRef]
- Perry, S.T.; Buck, M.D.; Lada, S.M.; Schindler, C.; Shresta, S. STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 2011, 7, e1001297. [Google Scholar] [CrossRef] [PubMed]
- Shresta, S.; Sharar, K. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J. Immunol. 2005, 175, 3946–3954. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.L.V.; Wee, L.J.K.; Khan, A.M.; Gil, L.H.V.G.; Marques, E.T.A.; Calzavara-Silva, C.E.; Tan, T.W. Classification of Dengue Fever Patients Based on Gene Expression Data Using Support Vector Machines. PLoS ONE 2010, 5, e11267. [Google Scholar] [CrossRef] [PubMed]
- Rothman, A.L. Immunity to Dengue Virus: A Tale of Original Antigenic Sin and Tropical Cytokine Storms. Nat. Rev. Immunol. 2011, 11, 532–543. [Google Scholar] [CrossRef] [PubMed]
- Medin, C.L.; Fitzgerald, K.A.; Rothman, A.L. Dengue Virus Nonstructural Protein NS5 Induces Interleukin-8 Transcription and Secretion. J. Virol. 2005, 79, 11053–11061. [Google Scholar] [CrossRef]
- Chaturvedi, U.C.; Agarwal, R.; Elbishbishi, E.A.; Mustafa, A.S. Cytokine cascade in dengue hemorrhagic fever: Implications for pathogenesis. FEMS Immunol. Med. Microbiol. 2000, 28, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.P.; Lu, H.L.; Lai, S.L.; Campanella, G.S.; Sung, J.M.; Lu, M.Y.; Wu-Hsieh, B.A.; Lin, Y.L.; Lane, T.E.; Luster, A.D.; et al. Dengue Virus Induces Expression of CXC Chemokine Ligand 10/IFN-Gamma-Inducible Protein 10, Which Competitively Inhibits Viral Binding to Cell Surface Heparan Sulfate. J. Immunol. 2006, 177, 3185–3192. [Google Scholar] [CrossRef]
- Bozza, F.A.; Cruz, O.G.; Zagne, S.M.; Azeredo, E.L.; Nogueira, R.M.; Assis, E.F.; Bozza, P.T.; Kubelka, C.F. Multiplex Cytokine Profile from Dengue Patients: MIP-1beta and IFN-Gamma as Predictive Factors for Severity. BMC Infect. Dis. 2008, 8, 86. [Google Scholar] [CrossRef]
- Brasier, A.R.; Scott, T.W.; Morrison, A.C.; Kochel, T.J.; Spratt, H.M.; Bazan, I.; Forshey, B.M.; Garcia, J.; Victor, S.S.; Rocha, C.; et al. A Three-Component Biomarker Panel for Prediction of Dengue Hemorrhagic Fever. Am. J. Trop. Med. Hyg. 2012, 86, 341–348. [Google Scholar] [CrossRef]
- Tolfvenstam, T.; Lindblom, A.; Schreiber, M.J.; Ling, L.; Chow, A.; Ooi, E.E.; Hibberd, M.L. Characterization of Early Host Responses in Adults with Dengue Disease. BMC Infect. Dis. 2011, 11, 209. [Google Scholar] [CrossRef]
- Fagundes, C.T.; Costa, V.V.; Cisalpino, D.; Amaral, F.A.; Souza, P.R.S.; Souza, R.S.; Ryffel, B.; Vieira, L.Q.; Silva, T.A.; Atrasheuskaya, A.; et al. IFN-γ Production Depends on IL-12 and IL-18 Combined Action and Mediates Host Resistance to Dengue Virus Infection in a Nitric Oxide-Dependent Manner. PLoS Negl. Trop. Dis. 2011, 5, e1449. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.V.; Fagundes, C.T.; Valadão, D.F.; Cisalpino, D.; Dias, A.C.F.; Silveira, K.D.; Kangussu, L.M.; Ávila, T.V.; Bonfim, M.R.Q.; Bonaventura, D.; et al. A Model of DENV-3 Infection That Recapitulates Severe Disease and Highlights the Importance of IFN-γ in Host Resistance to Infection. PLoS Negl. Trop. Dis. 2012, 6, e1663. [Google Scholar] [CrossRef] [PubMed]
- Pacsa, A.S.; Agarwal, R.; Elbishbishi, E.A.; Chaturvedi, U.C.; Nagar, R.; Mustafa, A.S. Role of Interleukin-12 in Patients with Dengue Hemorrhagic Fever. FEMS Immunol. Med. Microbiol. 2000, 28, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Neves-Souza, P.C.; Azeredo, E.L.; Zagne, S.M.; Valls-de-Souza, R.; Reis, S.R.; Cerqueira, D.I.; Nogueira, R.M.; Kubelka, C.F. Inducible nitric oxide synthase (iNOS) expression in monocytes during acute Dengue Fever in patients and during in vitro infection. BMC Infect. Dis. 2005, 5, 64. [Google Scholar] [CrossRef] [PubMed]
- Charnsilpa, W.; Takhampunya, R.; Endy, T.P.; Mammen, M.P.; Libraty, D.H.; Ubol, S. Nitric Oxide Radical Suppresses Replication of Wild-Type Dengue 2 Viruses in Vitro. J. Med. Virol. 2005, 77, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Gunther, V.J.; Putnak, R.; Eckels, K.H.; Mammen, M.P.; Scherer, J.M.; Lyons, A.; Sun, W. A human challenge model for dengue infection reveals a possible protective role for sustained interferon gamma levels during the acute phase of illness. Vaccine 2011, 29, 3895–3904. [Google Scholar] [CrossRef] [PubMed]
- Mangada, M.M.; Endy, T.P.; Nisalak, A.; Chunsuttiwat, S.; Vaughn, D.W.; Libraty, D.H.; Green, S.; Ennis, F.A.; Rothman, A.L. Dengue-Specific T Cell Responses in Peripheral Blood Mononuclear Cells Obtained Prior to Secondary Dengue Virus Infections in Thai Schoolchildren. J. Infect. Dis. 2002, 185, 1697–1703. [Google Scholar] [CrossRef]
- Atrasheuskaya, A.; Petzelbauer, P.; Fredeking, T.M.; Ignatyev, G. Anti-TNF Antibody Treatment Reduces Mortality in Experimental Dengue Virus Infection. FEMS Immunol. Med. Microbiol. 2003, 35, 33–42. [Google Scholar] [CrossRef]
- Shresta, S.; Sharar, K.L.; Prigozhin, D.M.; Beatty, P.R.; Harris, E. Murine Model for Dengue Virus-Induced Lethal Disease with Increased Vascular Permeability. J. Virol. 2006, 80, 10208–10217. [Google Scholar] [CrossRef]
- Zellweger, R.M.; Prestwood, T.R.; Shresta, S. Supplemental Information Antibodies Enhance Infection of Liver Sinusoidal Endothelial Cells in a Mouse Model of Severe Dengue Disease. Cell Host Microbe 2010, 7, 128–139. [Google Scholar] [CrossRef]
- Assunção-Miranda, I.; Amaral, F.A.; Bozza, F.A.; Fagundes, C.T.; Sousa, L.P.; Souza, D.G.; Pacheco, P.; Barbosa-Lima, G.; Gomes, R.N.; Bozza, P.T.; et al. Contribution of Macrophage Migration Inhibitory Factor to the Pathogenesis of Dengue Virus Infection. FASEB J. 2010, 24, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Kyle, J.L.; Harris, E. Global Spread and Persistence of Dengue. Annu. Rev. Microbiol. 2008, 62, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Guabiraba, R.; Marques, R.E.; Besnard, A.G.; Fagundes, C.T.; Souza, D.G.; Ryffel, B.; Teixeira, M.M. Role of the Chemokine Receptors CCR1, CCR2 and CCR4 in the Pathogenesis of Experimental Dengue Infection in Mice. PLoS ONE. 2010, 5, e15680. [Google Scholar] [CrossRef] [PubMed]
- Renneson, J.; Guabiraba, R.; Maillet, I. A detrimental role for invariant natural killer T cells in the pathogenesis of experimental dengue virus infection. Am. J. Pathol. 2011, 179, 1872–1883. [Google Scholar] [CrossRef] [PubMed]
- Heaton, N.S.; Perera, R.; Berger, K.L.; Khadka, S.; LaCount, D.J.; Kuhn, R.J.; Randall, G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 17345–17350. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.K.; Miller, I.; Aja, S.; Landree, L.E.; Pinn, M.; McFadden, J.; Kuhajda, F.P.; Moran, T.H.; Ronnett, G.V. C75, a fatty acid synthase inhibitor, reduces food intake via hypothalamic AMP-activated protein kinase. J. Biol. Chem. 2004, 279, 19970–19976. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.M.; Khromykh, A.A.; Parton, R.G. Cholesterol Manipulation by West Nile Virus Perturbs the Cellular Immune Response. Cell Host Microbe 2007, 2, 229–239. [Google Scholar] [CrossRef]
- Martínez-Gutierrez, M.; Castellanos, J.E.; Gallego-Gómez, J.C. Statins Reduce Dengue Virus Production via Decreased Virion Assembly. Intervirology 2011, 54, 202–216. [Google Scholar] [CrossRef]
- Poh, M.K.; Shui, G.; Xie, X.; Shi, P.Y.; Wenk, M.R.; Gu, F. U18666A, an Intra-Cellular Cholesterol Transport Inhibitor, Inhibits Dengue Virus Entry and Replication. Antivir. Res. 2012, 93, 191–198. [Google Scholar] [CrossRef]
- Soto-Acosta, R.; Bautista-Carbajal, P.; Syed, G.H.; Siddiqui, A.; Del Angel, R.M. Nordihydroguaiaretic Acid (NDGA) Inhibits Replication and Viral Morphogenesis of Dengue Virus. Antivir. Res. 2014, 109, 132–140. [Google Scholar] [CrossRef]
- Hyrina, A.; Meng, F.; McArthur, S.J.; Eivemark, S.; Nabi, I.R.; Jean, F. Human Subtilisin Kexin Isozyme-1 (SKI-1)/Site-1 Protease (S1P) Regulates Cytoplasmic Lipid Droplet Abundance: A Potential Target for Indirect-Acting Anti-Dengue Virus Agents. PLoS ONE 2017, 12, e0174483. [Google Scholar] [CrossRef] [PubMed]
- Mazzucco, M.B.; Talarico, L.B.; Vatansever, S.; Carro, A.C.; Fascio, M.L.; D’Accorso, N.B.; García, C.C.; Damonte, E.B. Antiviral Activity of an N-Allyl Acridone against Dengue Virus. J. Biomed. Sci. 2015, 22, 29. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, H.-H.; Kunz, A.; Simon, V.A.; Palese, P.; Shaw, M.L. Broad-Spectrum Antiviral That Interferes with de Novo Pyrimidine Biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, 5777–5782. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.S.; Zachariah, M.; Harris, E. Mycophenolic Acid Inhibits Dengue Virus Infection by Preventing Replication of Viral RNA. Virology 2002, 304, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Furuta, Y.; Takahashi, K.; Shiraki, K.; Sakamoto, K.; Smee, D.F.; Barnard, D.L.; Gowen, B.B.; Julander, J.G.; Morrey, J.D. T-705 (Favipiravir) and Related Compounds: Novel Broad-Spectrum Inhibitors of RNA Viral Infections. Antivir. Res. 2009, 82, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Ono, L.; Wollinger, W.; Rocco, I.M.; Coimbra, T.L.M.; Gorin, P.A.J.; Sierakowski, M.R. In Vitro and in Vivo Antiviral Properties of Sulfated Galactomannans against Yellow Fever Virus (BeH111 Strain) and Dengue 1 Virus (Hawaii Strain). Antivir. Res. 2003, 60, 201–208. [Google Scholar] [CrossRef]
- Talarico, L.B.; Pujol, C.A.; Zibetti, R.G.M.; Faría, P.C.S.; Noseda, M.D.; Duarte, M.E.R.; Damonte, E.B. The Antiviral Activity of Sulfated Polysaccharides against Dengue Virus Is Dependent on Virus Serotype and Host Cell. Antivir. Res. 2005, 66, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Alen, M.M.F.; de Burghgraeve, T.; Kaptein, S.J.F.; Balzarini, J.; Neyts, J.; Schols, D. Broad Antiviral Activity of Carbohydrate-Binding Agents against the Four Serotypes of Dengue Virus in Monocyte-Derived Dendritic Cells. PLoS ONE 2011, 6, e21658. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; De Wispelaere, M.; Carocci, M.; Liu, Q.; Wang, J.; Yang, P.L.; Gray, N.S. Structure-Activity Relationship Study of QL47: A Broad-Spectrum Antiviral Agent. ACS Med. Chem. Lett. 2017, 8, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Varga, N.; Sutkeviciute, I.; Ribeiro-Viana, R.; Berzi, A.; Ramdasi, R.; Daghetti, A.; Vettoretti, G.; Amara, A.; Clerici, M.; Rojo, J.; et al. A Multivalent Inhibitor of the DC-SIGN Dependent Uptake of HIV-1 and Dengue Virus. Biomaterials 2014, 35, 4175–4184. [Google Scholar] [CrossRef] [PubMed]
- Kouretova, J.; Hammamy, M.Z.; Epp, A.; Hardes, K.; Kallis, S.; Zhang, L.; Hilgenfeld, R.; Bartenschlager, R.; Steinmetzer, T. Effects of NS2B-NS3 Protease and Furin Inhibition on West Nile and Dengue Virus Replication. J. Enzym. Inhib. Med. Chem. 2017, 32, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Hotokezaka, H.; Sakai, E.; Kanaoka, K.; Saito, K.; Matsuo, K.I.; Kitaura, H.; Nakayama, K. U0126 and PD98059, specific inhibitors of MEK, accelerate differentiation of RAW264. 7 cells into osteoclast-like cells. J. Biol. Chem. 2002, 277, 47366–47372. [Google Scholar] [CrossRef] [PubMed]
- Sreekanth, G.P.; Chuncharunee, A.; Sirimontaporn, A.; Panaampon, J.; Noisakran, S.; Yenchitsomanus, P.T.; Limjindaporn, T. SB203580 Modulates P38 MAPK Signaling and Dengue Virus-Induced Liver Injury by Reducing MAPKAPK2, HSP27, and ATF2 Phosphorylation. PLoS ONE 2016, 11, e0149486. [Google Scholar] [CrossRef] [PubMed]
- Roth, H.; Magg, V.; Uch, F.; Mutz, P.; Klein, P.; Haneke, K.; Lohmann, V.; Bartenschlager, R.; Fackler, O.T.; Locker, N.; et al. Flavivirus Infection Uncouples Translation Suppression from Cellular Stress Responses. MBio 2017, 8, e02150-16. [Google Scholar] [CrossRef] [PubMed]
- Kovackova, S.; Chang, L.; Bekerman, E.; Neveu, G.; Barouch-Bentov, R.; Chaikuad, A.; Heroven, C.; Šála, M.; De Jonghe, S.; Knapp, S.; et al. Selective Inhibitors of Cyclin G Associated Kinase (GAK) as Anti-Hepatitis C Agents. J. Med. Chem. 2015, 58, 3393–3410. [Google Scholar] [CrossRef] [PubMed]
- Bekerman, E.; Neveu, G.; Shulla, A.; Brannan, J.; Pu, S.Y.; Wang, S.; Xiao, F.; Barouch-Bentov, R.; Bakken, R.R.; Mateo, R.; et al. Anticancer Kinase Inhibitors Impair Intracellular Viral Trafficking and Exert Broad-Spectrum Antiviral Effects. J. Clin. Investig. 2017, 127, 1338–1352. [Google Scholar] [CrossRef] [PubMed]
- Pu, S.Y.; Xiao, F.; Schor, S.; Bekerman, E.; Zanini, F.; Barouch-Bentov, R.; Nagamine, C.M.; Einav, S. Feasibility and Biological Rationale of Repurposing Sunitinib and Erlotinib for Dengue Treatment. Antivir. Res. 2018, 155, 67–75. [Google Scholar] [CrossRef]
- Chen, H.H.; Chen, C.C.; Lin, Y.S.; Chang, P.C.; Lu, Z.Y.; Lin, C.F.; Chen, C.L.; Chang, C.P. AR-12 Suppresses Dengue Virus Replication by down-Regulation of PI3K/AKT and GRP78. Antivir. Res. 2017, 142, 158–168. [Google Scholar]
- Albarnaz, J.D.; De Oliveira, L.C.; Torres, A.A.; Palhares, R.M.; Casteluber, M.C.; Rodrigues, C.M.; Cardozo, P.L.; De Souza, A.M.R.; Pacca, C.C.; Ferreira, P.C.P.; et al. MEK/ERK Activation Plays a Decisive Role in Yellow Fever Virus Replication: Implication as an Antiviral Therapeutic Target. Antivir. Res. 2014, 111, 82–92. [Google Scholar] [CrossRef]
- Chang, J.; Schul, W.; Yip, A.; Xu, X.; Guo, J.T.; Block, T.M. Competitive Inhibitor of Cellular α-Glucosidases Protects Mice from Lethal Dengue Virus Infection. Antivir. Res. 2011, 92, 369–371. [Google Scholar] [CrossRef]
- Gu, B.; Mason, P.; Wang, L.; Norton, P.; Bourne, N.; Moriarty, R.; Mehta, A.; Despande, M.; Shah, R.; Block, T. Antiviral Profiles of Novel Iminocyclitol Compounds against Bovine Viral Diarrhea Virus, West Nile Virus, Dengue Virus and Hepatitis B Virus. Antivir. Chem. Chemother. 2007, 18, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.H.; Cheng, W.C.; Lee, Y.L.; Yu, H.P.; Wu, Y.T.; Lin, Y.L.; Wong, C.H. Novel Five-Membered Iminocyclitol Derivatives as Selective and Potent Glycosidase Inhibitors: New Structures for Antivirals and Osteoarthritis. ChemBioChem 2006, 7, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Wang, L.; Ma, D.; Qu, X.; Guo, H.; Xu, X.; Mason, P.M.; Bourne, N.; Moriarty, R.; Gu, B.; et al. Novel Imino Sugar Derivatives Demonstrate Potent Antiviral Activity against Flaviviruses. Antimicrob. Agents Chemother. 2009, 53, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Gill, T.; Wang, L.; Du, Y.; Ye, H.; Qu, X.; Guo, J.-T.; Cuconati, A.; Zhao, K.; Block, T.M.; et al. Design, Synthesis, and Biological Evaluation of N -Alkylated Deoxynojirimycin (DNJ) Derivatives for the Treatment of Dengue Virus Infection. J. Med. Chem. 2012, 55, 6061–6075. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Plummer, E.; Buck, M.D.; Sanchez, M.; Greenbaum, J.A.; Turner, J.; Grewal, R.; Klose, B.; Sampath, A.; Warfield, K.L.; Peters, B.; et al. Dengue Virus Evolution under a Host-Targeted Antiviral. J. Virol. 2015, 89, 5592–5601. [Google Scholar] [CrossRef]
- Tseng, C.K.; Lin, C.K.; Wu, Y.H.; Chen, Y.H.; Chen, W.C.; Young, K.C.; Lee, J.C. Human Heme Oxygenase 1 Is a Potential Host Cell Factor against Dengue Virus Replication. Sci. Rep. 2016, 6, 32176. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.S.; Wu, Y.H.; Tseng, C.K.; Lin, C.K.; Hsu, Y.C.; Chen, Y.H.; Lee, J.C. Schisandrin A Inhibits Dengue Viral Replication via Upregulating Antiviral Interferon Responses through STAT Signaling Pathway. Sci. Rep. 2017, 7, 45171. [Google Scholar] [CrossRef]
- Yu, J.S.; Tseng, C.K.; Lin, C.K.; Hsu, Y.C.; Wu, Y.H.; Hsieh, C.L.; Lee, J.C. Celastrol Inhibits Dengue Virus Replication via Up-Regulating Type I Interferon and Downstream Interferon-Stimulated Responses. Antivir. Res. 2017, 137, 49–57. [Google Scholar] [CrossRef]
- Pryke, K.; Abraham, J.; Sali, T.; Gall, B.; Archer, I. A novel agonist of the TRIF pathway induces a cellular state refractory to replication of Zika, chikungunya, and dengue viruses. MBio 2027, 8, e00452-17. [Google Scholar] [CrossRef]
- Sharma, N.; Mishra, K. Salidroside Exhibits Anti-Dengue Virus Activity by Upregulating Host Innate Immune Factors. Arch. Virol. 2016, 161, 3331–3344. [Google Scholar] [CrossRef]
- Tsai, W.L.; Cheng, J.S.; Shu, C.W.; Lai, K.H.; Chan, H.H.; Wu, C.C.; Wu, J.M.; Hsu, P.I.; Chung, R.T.; Chang, T.H. Asunaprevir Evokes Hepatocytes Innate Immunity to Restrict the Replication of Hepatitis C and Dengue Virus. Front. Microbiol. 2017, 8, 668. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.; Beljanski, V.; Yin, K. Sequence-specific modifications enhance the broad-spectrum antiviral response activated by RIG-I agonists. J. Virol. 2015, 89, 8011–8025. [Google Scholar] [CrossRef] [PubMed]
- Shafee, N.; AbuBakar, S. Zinc Accelerates Dengue Virus Type 2-Induced Apoptosis in Vero Cells. FEBS Lett. 2002, 524, 20–24. [Google Scholar] [CrossRef]
- Corrêa, G.; Lindenberg, C.D.A.; Fernandes-Santos, C.; Gandini, M.; Paiva, F.P.; Coutinho-Silva, R.; Kubelka, C.F. The Purinergic Receptor P2X7 Role in Control of Dengue Virus-2 Infection and Cytokine/Chemokine Production in Infected Human Monocytes. Immunobiology 2016, 221, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Mello, C.D.S.; Valente, L.M.M.; Wolff, T.; Lima-Junior, R.S.; Fialho, L.G.; Marinho, C.F.; Azeredo, E.L.; Oliveira-Pinto, L.M.; Pereira, R.D.C.A.; Siani, A.C.; et al. Decrease in Dengue Virus-2 Infection and Reduction of Cytokine/Chemokine Production by Uncaria Guianensis in Human Hepatocyte Cell Line Huh-7. Memórias do Instituto Oswaldo Cruz 2017, 112, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Reis, S.R.I.N.; Valente, L.M.M.; Sampaio, A.L.; Siani, A.C.; Gandini, M.; Azeredo, E.L.; D’Avila, L.A.; Mazzei, J.L.; Maria das Graças, M.H.; Kubelka, C.F. Immunomodulating and Antiviral Activities of Uncaria Tomentosa on Human Monocytes Infected with Dengue Virus-2. Int. Immunopharmacol. 2008, 8, 468–476. [Google Scholar] [CrossRef]
- Wagstaff, K.M.; Harrich, D.; Heaton, S.M.; Sivakumaran, H.; Jans, D.A. Ivermectin Is a Specific Inhibitor of Importin α/β-Mediated Nuclear Import Able to Inhibit Replication of HIV-1 and Dengue Virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.B.; Han, J.C.; Cong, X.; Wei, L. BST2/Tetherin Inhibits Dengue Virus Release from Human Hepatoma Cells. PLoS ONE 2012, 7, e51033. [Google Scholar] [CrossRef]
- Trung, D.T.; Wills, B. Systemic Vascular Leakage Associated with Dengue Infections—The Clinical Perspective; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Gröger, M.; Pasteiner, W.; Ignatyev, G.; Matt, U.; Knapp, S.; Atrasheuskaya, A.; Bukin, E.; Friedl, P.; Zinkl, D.; Hofer-Warbinek, R.; et al. Peptide Bβ15-42 Preserves Endothelial Barrier Function in Shock. PLoS ONE 2009, 4, e5391. [Google Scholar] [CrossRef]
- Chuang, Y.C.; Lei, H.Y.; Liu, H.S.; Lin, Y.S.; Fu, T.F.; Yeh, T.M. Macrophage Migration Inhibitory Factor Induced by Dengue Virus Infection Increases Vascular Permeability. Cytokine 2011, 54, 222–231. [Google Scholar] [CrossRef]
- Kubelka, C.; Azeredo, E.; Gandini, M.; Pinto, L. Metalloproteinases Are Produced during Dengue Fever and MMP9 Is Associated with Severity; The British Infection Association: Preston, UK, 2010. [Google Scholar]
- Ong, S.P.; Lee, L.M.; Leong, Y.F.I.; Ng, M.L.; Chu, J.J.H. Dengue Virus Infection Mediates HMGB1 Release from Monocytes Involving PCAF Acetylase Complex and Induces Vascular Leakage in Endothelial Cells. PLoS ONE 2012, 7, e41932. [Google Scholar] [CrossRef]
- Costa, V.V.; Fagundes, C.T.; Souza, D.G.; Teixeira, M.M. Inflammatory and Innate Immune Responses in Dengue Infection. Am. J. Pathol. 2013, 182, 1950–1961. [Google Scholar] [CrossRef]
- Souza, D.; Fagundes, C. Essential Role of Platelet-Activating Factor Receptor in the Pathogenesis of Dengue Virus Infection. Proc. Natl. Acad. Sci. USA 2009, 106, 14138–14143. [Google Scholar] [CrossRef]
- Rahman, S.M.; Atikullah, M.; Islam, M.N.; Mohaimenul, M.; Ahammad, F.; Islam, M.S.; Saha, B.; Rahman, M.H. Anti-inflammatory, antinociceptive and antidiarrhoeal activities of methanol and ethyl acetate extract of Hemigraphis alternata leaves in mice. Clin. Phytosci. 2019, 5, 16. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).