Does Soil Contribute to the Human Gut Microbiome?
Abstract
1. Introduction
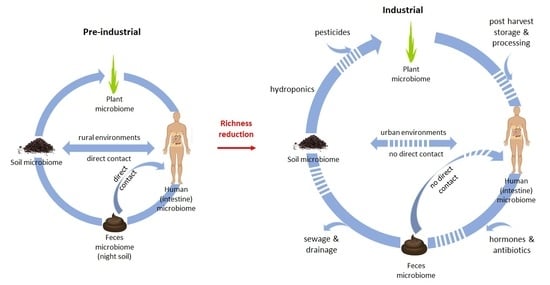
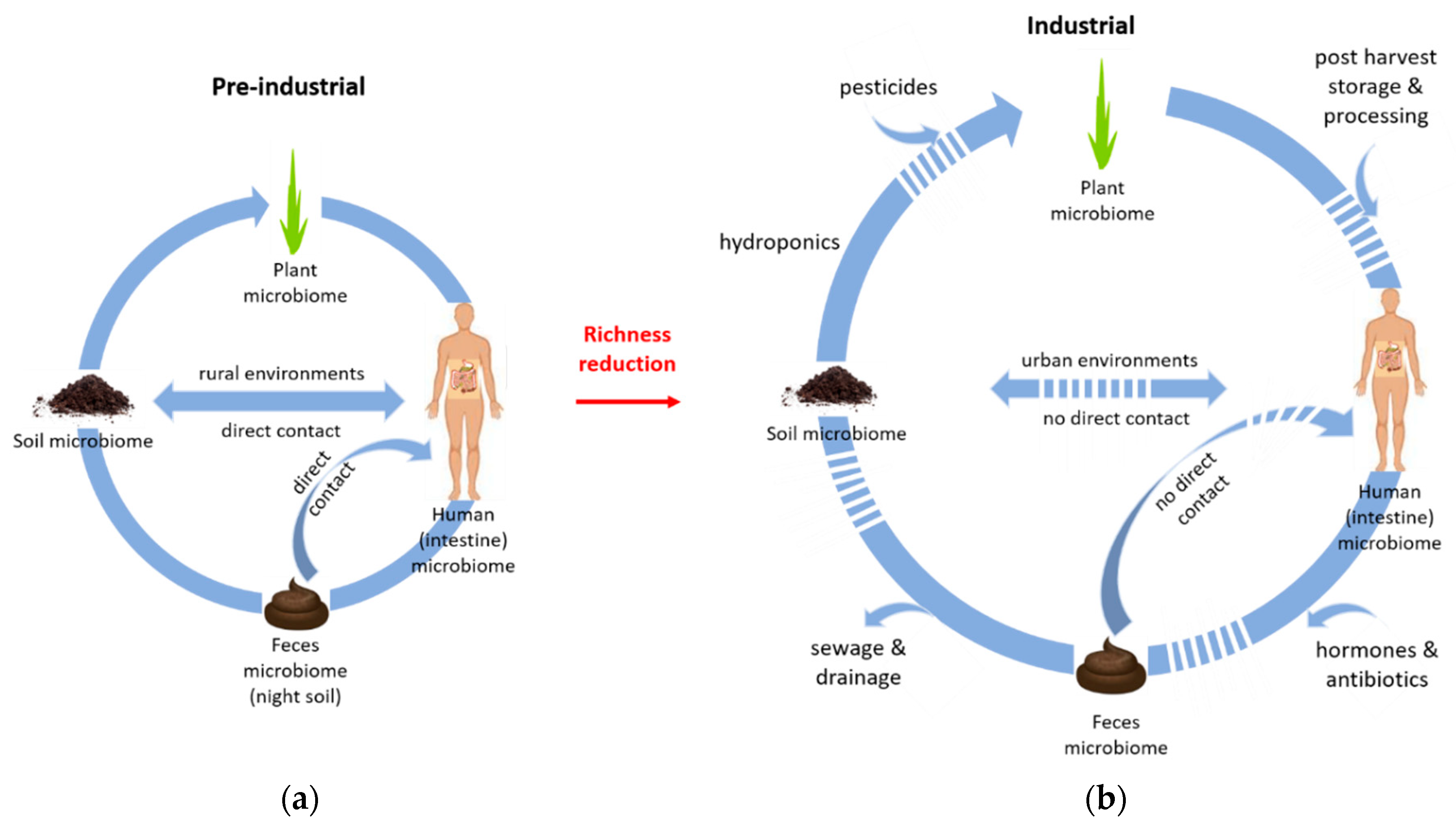
2. The Complex Relationship between the Soil Microbiome and the Human Intestinal Microbiome
3. The Human Intestinal Microbiome—Its Development and Evolution
4. Human Intestinal Microbiome and the Environment—Lifestyle
5. Human Intestinal Microbiome and Diet/Nutrition
6. Soil Microbiome as a Functional Ecosystem—Potential Links to the Gut Microbiome
7. Summary
8. Conclusions and Outlook
Funding
Acknowledgments
Conflicts of Interest
References
- Brevik, E.; Pereg, L. History of soils in relation to animal and human health. In The Nexus of Soils, Plants, Animals and Human Health; Catena Soil Sciences: Stuttgart, Germeny, 2017. [Google Scholar]
- Sing, D.; Sing, C.F. Impact of direct soil exposures from airborne dust and geophagy on human health. Int. J. Environ. Res. Public Health 2010, 7, 1205–1223. [Google Scholar] [CrossRef] [PubMed]
- Blum, H.E. The microbiome: A key player in human health and disease. J. Healthc. Commun. 2017, 2, 1–5. [Google Scholar] [CrossRef]
- Nishida, A.H.; Ochman, H. Rates of gut microbiome divergence in mammals. Mol. Ecol. 2018, 27, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Shamayim, T.; Luis, E.; Berenice, J.M.; Luis, M.B.; Mónica, R.; Julio, M.; Marco, A.R.; Ernesto, O.O.; Esperanza, M.R. Gut and root microbiota commonalities. Appl. Environ. Microbiol. 2013, 79, 2–9. [Google Scholar]
- Zhou, D.R.; Bai, Z.M.; Zhang, H.L.; Li, N.; Bai, Z.Y.; Cheng, F.D.; Jiang, H.T.; Mao, C.B.; Sun, X.; Lu, Z.H. Soil is a key factor influencing gut microbiota and its effect is comparable to that exerted by diet for mice. F1000Research 2018, 7, 1588. [Google Scholar] [CrossRef]
- Laura, E.G.; Marie, J.E.; Susan, C.A.; Ran, B.; Gideon, B.; Jenny, T.; Elizabeth, A.A. Genes, geology and germs: Gut microbiota across a primate hybrid zone are explained by site soil properties, not host species. Proc. R. Soc. B: Biol. Sci. 2019, 286, doi. [Google Scholar]
- Lederberg, J. Microbiology’s World Wide Web. [Project Syndicate The Worlds’s Opinion Page]. 2001. Available online: https://www.project-syndicate.org/commentary/microbiology-s-world-wide-web?barrier = accesspaylog (accessed on 18 December 2018).
- Marcel, G.A.; Van Der, H.; Martin, H. Networking in the Plant. Microbiome. PLoS Biol. 2016, 14, e1002378. [Google Scholar]
- Lynch, S.V.; Pedersen, O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Berg, R.D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996, 4, 430–435. [Google Scholar] [CrossRef]
- Daniel, R. The metagenomics of soil. Nat. Rev. Microbiol. 2005, 3, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Roesch, L.F.W.; Fulthorpe, R.R.; Riva, A.; Casella, G.; Hadwin, A.K.M.; Kent, A.D.; Daroub, S.H.; Camargo, F.A.O.; Farmerie, W.G.; Triplett, E.W. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007, 1, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Lennon, J.T. The generation and maintenance of diversity in microbial communities. Am. J. Bot. 2011, 98, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Lennon, J.T.; Jones, S.E. Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 2011, 9, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef]
- Flandroy, L.; Poutahidis, T.; Berg, G.; Clarke, G.; Dao, M.C.; Decaestecker, E.; Furman, E.; Haahtela, T.; Massart, S.; Plovier, H.; et al. The impact of human activities and lifestyles on the interlinked microbiota and health of humans and of ecosystems. Sci. Total Environ. 2018, 627, 1018–1038. [Google Scholar] [CrossRef] [PubMed]
- Raynaud, X.; Nunan, N. Spatial ecology of bacteria at the microscale in soil. PLoS ONE 2014, 9, e87217. [Google Scholar] [CrossRef] [PubMed]
- Torsvik, V.; Øvreås, L. Microbial diversity and function in soil: From genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Saleem, M.; Traw, M.; Pervaiz, Z. Microbiome Community Ecology: Fundamentals and Applications; Springer: New York, NY, USA, 2015. [Google Scholar]
- Ziglio, G.; Andreottola, G.; Barbesti, S.; Boschetti, G.; Bruni, L.; Foladori, P.; Villa, R. Assessment of activated sludge viability with flow cytometry. Water Res. 2002, 36, 460–468. [Google Scholar] [CrossRef]
- Vaahtovuo, J.; Korkeamäki, M.; Munukka, E.; Viljanen, M.K.; Toivanen, P. Quantification of bacteria in human feces using 16S rRNA-hybridization, DNA-staining and flow cytometry. J. Microbiol. Methods 2005, 63, 276–286. [Google Scholar] [CrossRef] [PubMed]
- van der Waaij, L.A.; Limburg, P.C.; Mesander, G.; van der Waaij, D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut 1996, 38, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Franks, A.H.; Harmsen, H.J.; Raangs, G.C.; Jansen, G.J.; Schut, F.; Welling, G.W. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 1998, 64, 3336–3345. [Google Scholar]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Ley, R.E.; Lozupone, C.A.; Hamady, M.; Knight, R.; Gordon, J.I. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008, 6, 776–788. [Google Scholar] [CrossRef]
- Haque, S.Z.; Haque, M. The ecological community of commensal, symbiotic, and pathogenic gastrointestinal microorganisms - an appraisal. Clin. Exp. Gastroenterol. 2017, 10, 91–103. [Google Scholar] [CrossRef]
- Walter, J.; Britton, R.A.; Roos, S. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc. Natl. Acad. Sci. USA 2011, 108, 4645–4652. [Google Scholar] [CrossRef]
- Moeller, A.H.; Li, Y.; Mpoudi Ngole, E.; Ahuka-Mundeke, S.; Lonsdorf, E.V.; Pusey, A.E.; Peeters, M.; Hahn, B.H.; Ochman, H. Rapid changes in the gut microbiome during human evolution. Proc. Natl. Acad. Sci. USA 2014, 111, 16431–16435. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Martínez-Romero, E. bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-Infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 2018, 24, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Ungar, P.S. Evolution of the Human Diet: The Known, the Unknown, and the Unknowable; Oxford University Press: New York, NY, USA, 2007. [Google Scholar]
- O’Connell, J.F.; Hawkes, K.; Jones, N.G.B. Grandmothering and the evolution of Homo erectus. J. Hum. Evol. 1999, 36, 461–485. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.D. The gut microbiome and its role in obesity. Nutr. Today 2016, 51, 167–174. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Clemente, J.C.; Pehrsson, E.C.; Blaser, M.J.; Sandhu, K.; Gao, Z.; Wang, B.; Magris, M.; Hidalgo, G.; Contreras, M.; Noya-Alarcón, Ó.; et al. The microbiome of uncontacted Amerindians. Sci. Adv. 2015, 1, e1500183. [Google Scholar] [CrossRef]
- Hanski, I.; von Hertzen, L.; Fyhrquist, N.; Koskinen, K.; Torppa, K.; Laatikainen, T.; Karisola, P.; Auvinen, P.; Paulin, L.; Mäkelä, M.J.; et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc. Natl. Acad. Sci. USA 2012, 109, 8334–8339. [Google Scholar] [CrossRef]
- Tasnim, N.; Abulizi, N.; Pither, J.; Hart, M.M.; Gibson, D.L. Linking the Gut Microbial Ecosystem with the Environment: Does Gut Health Depend. on Where We Live? Front. Microbiol. 2017, 8, 1935. [Google Scholar] [CrossRef]
- Stein, M.M.; Hrusch, C.L.; Gozdz, J.; Igartua, C.; Pivniouk, V.; Murray, S.E.; Ledford, J.G.; Marques dos Santos, M.; Anderson, R.L.; Metwali, N.; et al. Innate immunity and asthma risk in amish and hutterite farm. Child. New Engl. J. Med. 2016, 375, 411–421. [Google Scholar] [CrossRef]
- Von Hertzen, L.; Hanski, I.; Haahtela, T. Natural immunity. EMBO Rep. 2011, 12, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Rook, G.A.W. Review series on helminths, immune modulation and the hygiene hypothesis: The broader implications of the hygiene hypothesis. Immunology 2009, 126, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Wall, D.H.; Nielsen, U.N.; Six, J. Soil biodiversity and human health. Nature 2015, 528, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Zhang, H.; Bai, Z.; Zhang, A.; Bai, F.; Luo, X.; Hou, Y.; Ding, X.; Sun, B.; Sun, X.; et al. Exposure to soil, house dust and decaying plants increases gut microbial diversity and decreases serum immunoglobulin E levels in BALB/c mice. Environ. Microbiol. 2015, 18, 1326–1337. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.L.; Song, S.J.; Morton, J.T.; Weiss, S.; Seguin-Orlando, A.; Joly, F.; Feh, C.; Taberlet, P.; Coissac, E.; Amir, A.; et al. Evaluating the impact of domestication and captivity on the horse gut microbiome. Sci. Rep. 2017, 7, 15497. [Google Scholar] [CrossRef] [PubMed]
- Muegge, B.D.; Kuczynski, J.; Knights, D.; Clemente, J.C.; González, A.; Fontana, L.; Henrissat, B.; Knight, R.; Gordon, J.I. Diet. drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 2011, 332, 970–974. [Google Scholar] [CrossRef] [PubMed]
- Bender, S.F.; Wagg, C.; Van Der Heijden, M.G.A. An underground revolution: Biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol. Evol. 2016, 31, 440–452. [Google Scholar] [CrossRef] [PubMed]
- UN-DESA. World Population Prospects: The 2017 Revision, Key Findings and Advance Tables; D.o.E.a.S.A. United Nations, Ed.; Population Division: New York, NY, USA, 2017.
- Turner, W.R.; Nakamura, T.; Dinetti, M. Global urbanization and the separation of humans from nature. BioScience 2004, 54, 585–590. [Google Scholar] [CrossRef]
- Martínez, I.; Stegen, J.C.; Maldonado-Gómez, M.X.; Eren, A.M.; Siba, P.M.; Greenhill, A.R.; Walter, J. The gut microbiota of rural papua new guineans: composition, diversity patterns, and ecological processes. Cell Rep. 2015, 11, 527–538. [Google Scholar]
- Schnorr, S.L.; Candela, M.; Rampelli, S.; Centanni, M.; Consolandi, C.; Basaglia, G.; Turroni, S.; Biagi, E.; Peano, C.; Severgnini, M.; et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014, 5, 1–12. [Google Scholar] [CrossRef]
- Graf, D.; Di Cagno, R.; Fåk, F.; Flint, H.J.; Nyman, M.; Saarela, M.; Watzl, B. Contribution of diet to the composition of the human gut microbiota. Microb. Ecol. Health Dis. 2015, 26, 26164. [Google Scholar] [CrossRef]
- Reischer, G.H.; Haider, J.M.; Sommer, R.; Stadler, H.; Keiblinger, K.M.; Hornek, R.; Zerobin, W.; Mach, R.L.; Farnleitner, A.H. Quantitative microbial faecal source tracking with sampling guided by hydrological catchment dynamics. Environ. Microbiol. 2008, 10, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Burtscher, M.M.; Köllner, K.E.; Sommer, R.; Keiblinger, K.; Farnleitner, A.H.; Mach, R.L. Development of a novel amplified fragment length polymorphism (AFLP) typing method for enterococci isolates from cattle faeces and evaluation of the single versus pooled faecal sampling approach. J. Microbiol. Methods 2006, 67, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Vierheilig, J.; Farnleitner, A.H.; Kollanur, D.; Blöschl, G.; Reischer, G.H. High abundance of genetic Bacteroidetes markers for total fecal pollution in pristine alpine soils suggests lack in specificity for feces. J. Microbiol. Methods 2012, 88, 433–435. [Google Scholar] [CrossRef] [PubMed]
- Blum, W.E.H. Functions of Soil for Society and the Environment. Rev. Environ. Sci. Bio/Technol. 2005, 4, 75–79. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Penakalapati, G.; Swarthout, J.; Delahoy, M.J.; McAliley, L.; Wodnik, B.; Levy, K.; Freeman, M.C. Exposure to animal feces and human health: A systematic review and proposed research priorities. Environ. Sci. Technol. 2017, 51, 11537–11552. [Google Scholar] [CrossRef] [PubMed]
- Brindley, P.; Jorgensen, A.; Maheswaran, R. Domestic gardens and self-reported health: A national population study. Int. J. of Health Geogr. 2018, 17, 1–11. [Google Scholar] [CrossRef]
- Mills, J.G.; Brookes, J.D.; Gellie, N.J.C.; Liddicoat, C.; Lowe, A.J.; Sydnor, H.R.; Thomas, T.; Weinstein, P.; Weyrich, L.S.; Breed, M.F. Relating urban. biodiversity to human health with the ‘holobiont’ concept. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Alcock, J.; Maley, C.C.; Aktipis, C.A. Is eating behavior manipulated by the gastrointestinal microbiota? Evolutionary pressures and potential mechanisms. BioEssays 2014, 36, 940–949. [Google Scholar] [CrossRef] [PubMed]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.-N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Giovannetti, M.; Sbrana, C.; Turrini, A. The impact of genetically modified crops on soil microbial communities. Biotech. Forum. 2004, 98, 393–417. [Google Scholar]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2013, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [PubMed]
- Rillig, M.C.; Lehmann, A.; Lehmann, J.; Camenzind, T.; Rauh, C. Soil biodiversity effects from field to fork. Trends Plant Sci. 2018, 23, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Pedone-Bonfim, M.; Silva, F.; Maia, L. Production of secondary metabolites by mycorrhizal plants with medicinal or nutritional potential. Acta Physiol. Plantarum 2015, 37, 1. [Google Scholar] [CrossRef]
- Lori, M.; Symnaczik, S.; Mäder, P.; De Deyn, G.; Gattinger, A. Organic farming enhances soil microbial abundance and activity—A meta-analysis and meta-regression. PLoS ONE 2017, 12, e0180442. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626. [Google Scholar] [CrossRef]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef]
- Martiny, J.B.H.; Bohannan, B.J.M.; Brown, J.H.; Colwell, R.K.; Fuhrman, J.A.; Green, J.L.; Horner-Devine, M.C.; Kane, M.; Krumins, J.A.; Kuske, C.R.; et al. Microbial biogeography: Putting microorganisms on the map. Nat. Rev. Microbiol. 2006, 4, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Hobbie, J.E.; Hobbie, E.A. Microbes in nature are limited by carbon and energy: The starving-survival lifestyle in soil and consequences for estimating microbial rates. Front. Microbiol. 2013, 4, 324. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Hartman, K.; van der Heijden, M.G.; Roussely-Provent, V.; Walser, J.-C.; Schlaeppi, K. Deciphering composition and function of the root microbiome of a legume plant. Microbiome 2017, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Stone, M. Root and gut microbiomes are strikingly similar. Microbe 2016, 11, 107–110. [Google Scholar] [CrossRef]
- Mendes, R.; Raaijmakers, J.M. Cross-kingdom similarities in microbiome functions. ISME J. 2015, 9, 1905–1907. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Krause, R.; Mendes, R. Cross-Kingdom Similarities in Microbiome Ecology and Biocontrol of Pathogens. Front. Microbiol. 2015, 6, 1311. [Google Scholar] [CrossRef]
- Mallon, C.; Van, E.; Salles, J. Microbial Invasions: The process., patterns, and mechanisms. Trends Microbiol. 2015, 23, 719–729. [Google Scholar] [CrossRef]
- Adam, E.; Groenenboom, A.E.; Kurm, V.; Rajewska, M.; Schmidt, R.; Tyc, O.; Weidner, S.; Berg, G.; de Boer, W.; Falcão Salles, J. Controlling the microbiome: Microhabitat adjustments for successful biocontrol strategies in soil and human gut. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Frey, B.; Mayer, J.; Mäder, P.; Widmer, F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2015, 9, 1177–1194. [Google Scholar] [CrossRef] [PubMed]
- Blum, W.E.H. Soil and land resources for agricultural production: General trends and future scenarios-a worldwide perspective. Int. Soil Water Conserv. Res. 2013, 1, 1–14. [Google Scholar] [CrossRef]
- Macfarlane, S.; Bahrami, B.; Macfarlane, G.T. Chapter 4—Mucosal Biofilm Communities in the Human Intestinal Tract. In Advances in Applied Microbiology; Laskin, A.I., Sariaslani, S., Gadd, G.M., Eds.; Academic Press: Cambridge, MA, USA, 2011; pp. 111–143. [Google Scholar]
- Srivastava, A.; Gupta, J.; Kumar, S.; Kumar, A. Gut biofilm forming bacteria in inflammatory bowel disease. Microb. Pathog. 2017, 112, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Ramey, B.E.; Koutsoudis, M.; Bodman, S.B.V.; Fuqua, C. Biofilm formation in plant–microbe associations. Curr. Opin. Microbiol. 2004, 7, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Rudrappa, T.; Biedrzycki, M.L.; Bais, H.P. Causes and consequences of plant-associated biofilms. FEMS Microbiol. Ecol. 2008, 64, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Shenkin, A. Micronutrients in health and disease. Postgrad. Med. J. 2006, 82, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, S.; Gardi, C. Soil biodiversity under threat—A review. Acta Soc. Zool. Bohem. 2010, 74, 7–12. [Google Scholar]
- Liu, D.; Keiblinger, K.M.; Schindlbacher, A.; Wegner, U.; Sun, H.; Fuchs, S.; Lassek, C.; Riedel, K.; Zechmeister-Boltenstern, S. Microbial functionality as affected by experimental warming of a temperate mountain forest soil—A metaproteomics survey. Appl. Soil Ecol. 2017, 117–118, 196–202. [Google Scholar] [CrossRef]
- Kuffner, M.; Hai, B.; Rattei, T.; Melodelima, C.; Schloter, M.; Zechmeister-Boltenstern, S.; Jandl, R.; Schindlbacher, A.; Sessitsch, A. Effects of season and experimental warming on the bacterial community in a temperate mountain forest soil assessed by 16S rRNA gene pyrosequencing. FEMS Microbiol. Ecol. 2012, 82, 551–562. [Google Scholar] [CrossRef]
- Pold, G.; Billings, A.F.; Blanchard, J.L.; Burkhardt, D.B.; Frey, S.D.; Melillo, J.M.; Schnabel, J.; van Diepen, L.T.; DeAngelis, K.M. Long-term warming alters carbohydrate degradation potential in temperate forest soils. Appl. Environ. Microbiol. 2016, 82, 6518–6530. [Google Scholar] [CrossRef]
| Habitat | Number of Cells per g | Number of Cells per mL | Species Diversity |
|---|---|---|---|
| Soil | 107–109 [22] | 1010 [23] | 4 × 103–5 × 104 species per g soil [22] |
| Sewage | 109 [24,25] | 25 per mL [24] | |
| Marine water | 105–106 [18] | ||
| Air | 1 (=106 cells/m³) [17] | ||
| Human gut | 1012 [26] | 4 × 102 species per g feces [27] | |
| 1011 [28] | 5 × 103 species per g feces [18] | ||
| Colon (large intestine) | 1011 [29] 1011–1012 [30] | ||
| Ileum (lower small intestine) | 108 [29] | ||
| Duodenum and jejunum (upper small intestine) | 103–104 [29] | ||
| Human mouth (saliva) | 108 [18] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blum, W.E.H.; Zechmeister-Boltenstern, S.; Keiblinger, K.M. Does Soil Contribute to the Human Gut Microbiome? Microorganisms 2019, 7, 287. https://doi.org/10.3390/microorganisms7090287
Blum WEH, Zechmeister-Boltenstern S, Keiblinger KM. Does Soil Contribute to the Human Gut Microbiome? Microorganisms. 2019; 7(9):287. https://doi.org/10.3390/microorganisms7090287
Chicago/Turabian StyleBlum, Winfried E.H., Sophie Zechmeister-Boltenstern, and Katharina M. Keiblinger. 2019. "Does Soil Contribute to the Human Gut Microbiome?" Microorganisms 7, no. 9: 287. https://doi.org/10.3390/microorganisms7090287
APA StyleBlum, W. E. H., Zechmeister-Boltenstern, S., & Keiblinger, K. M. (2019). Does Soil Contribute to the Human Gut Microbiome? Microorganisms, 7(9), 287. https://doi.org/10.3390/microorganisms7090287