Metagenomic Evaluation of Bacterial and Fungal Assemblages Enriched within Diffusion Chambers and Microbial Traps Containing Uraniferous Soils
Abstract
1. Introduction
2. Material and Methods
2.1. Soil Sample Collection
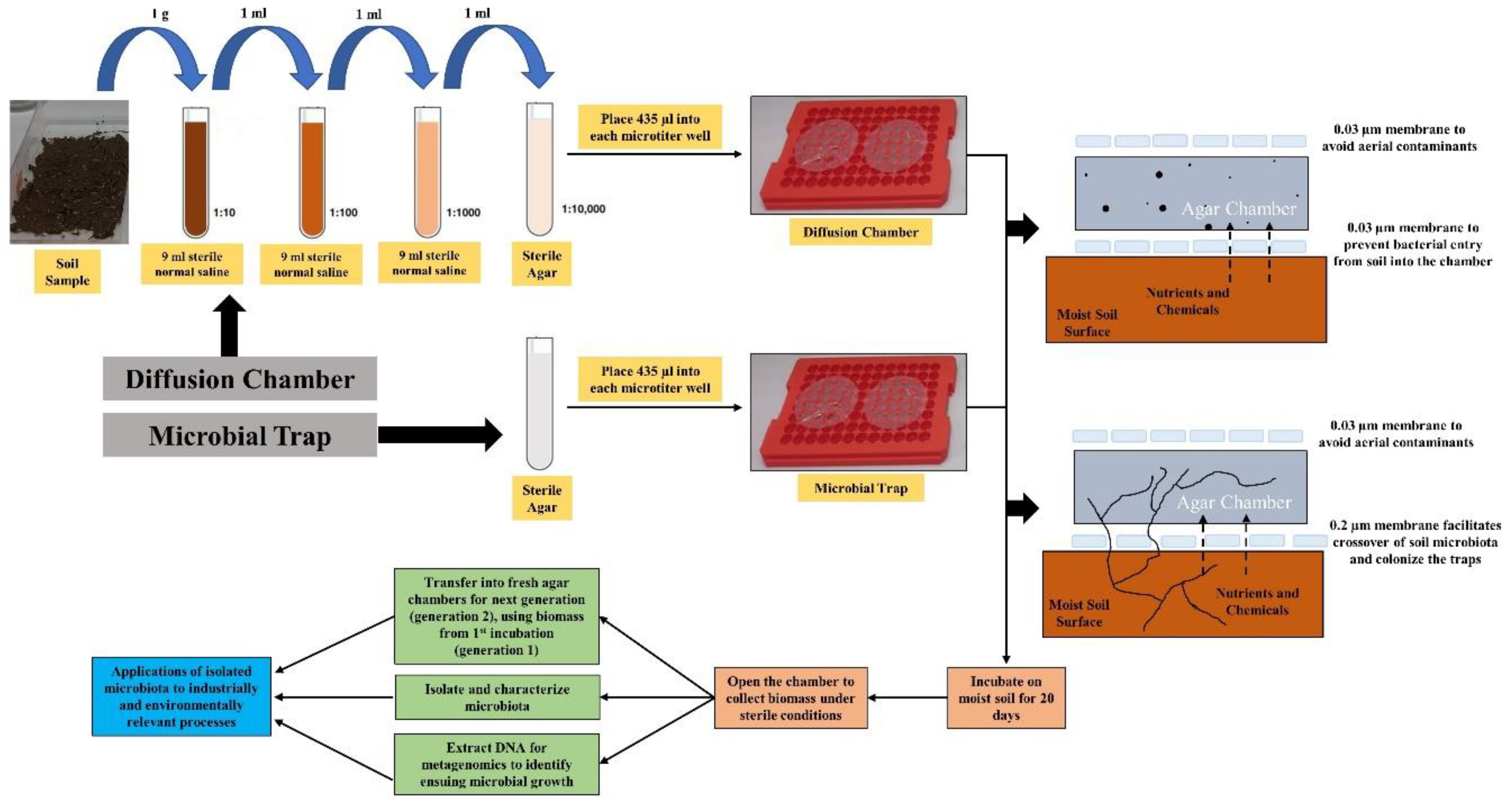
2.2. Establishment of Diffusion Chambers/Microbial Traps (DC/MT)
2.3. Microbiome Analysis
2.4. Metagenomic Sequence Accession Number
3. Results and Discussion
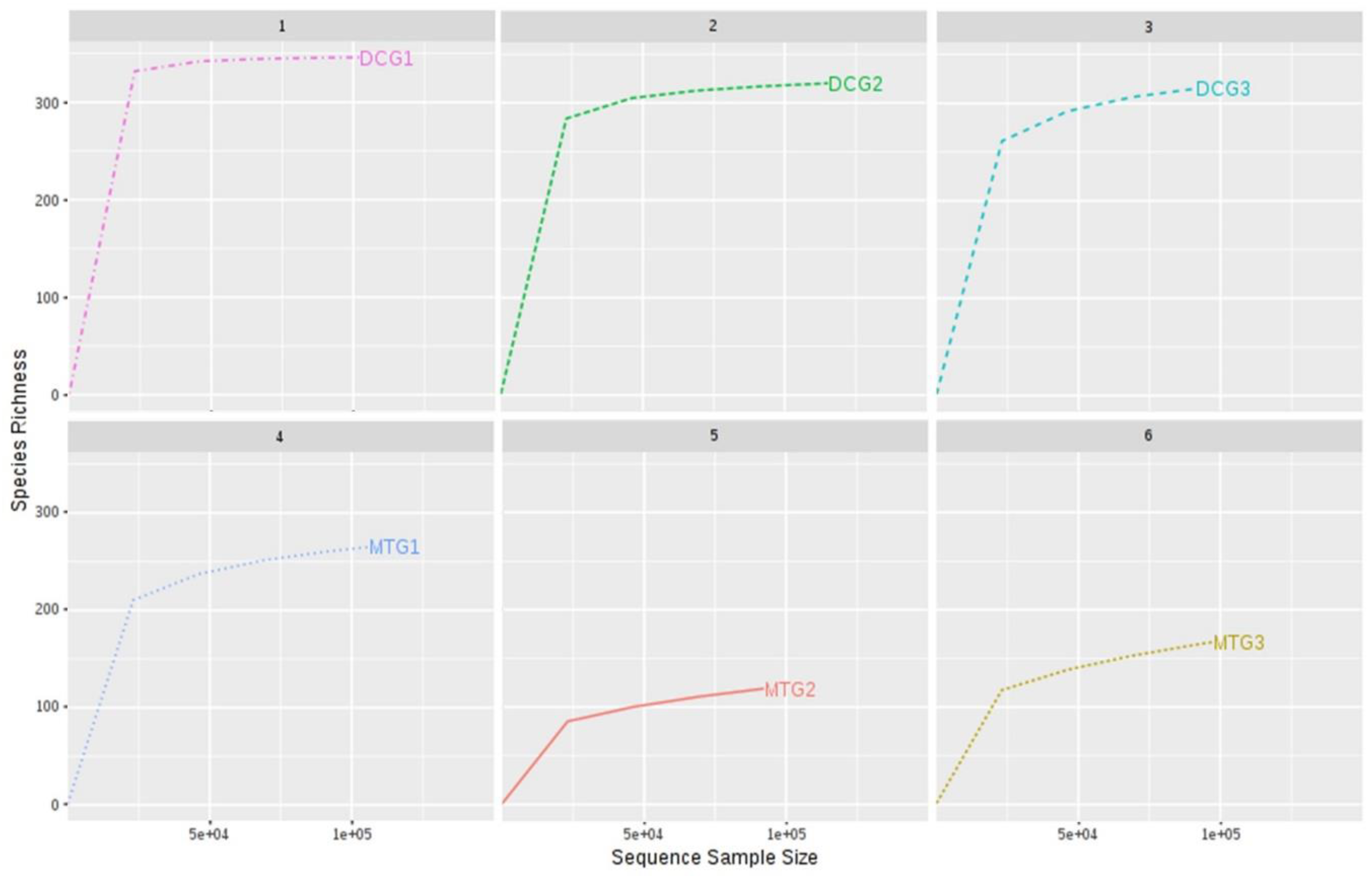
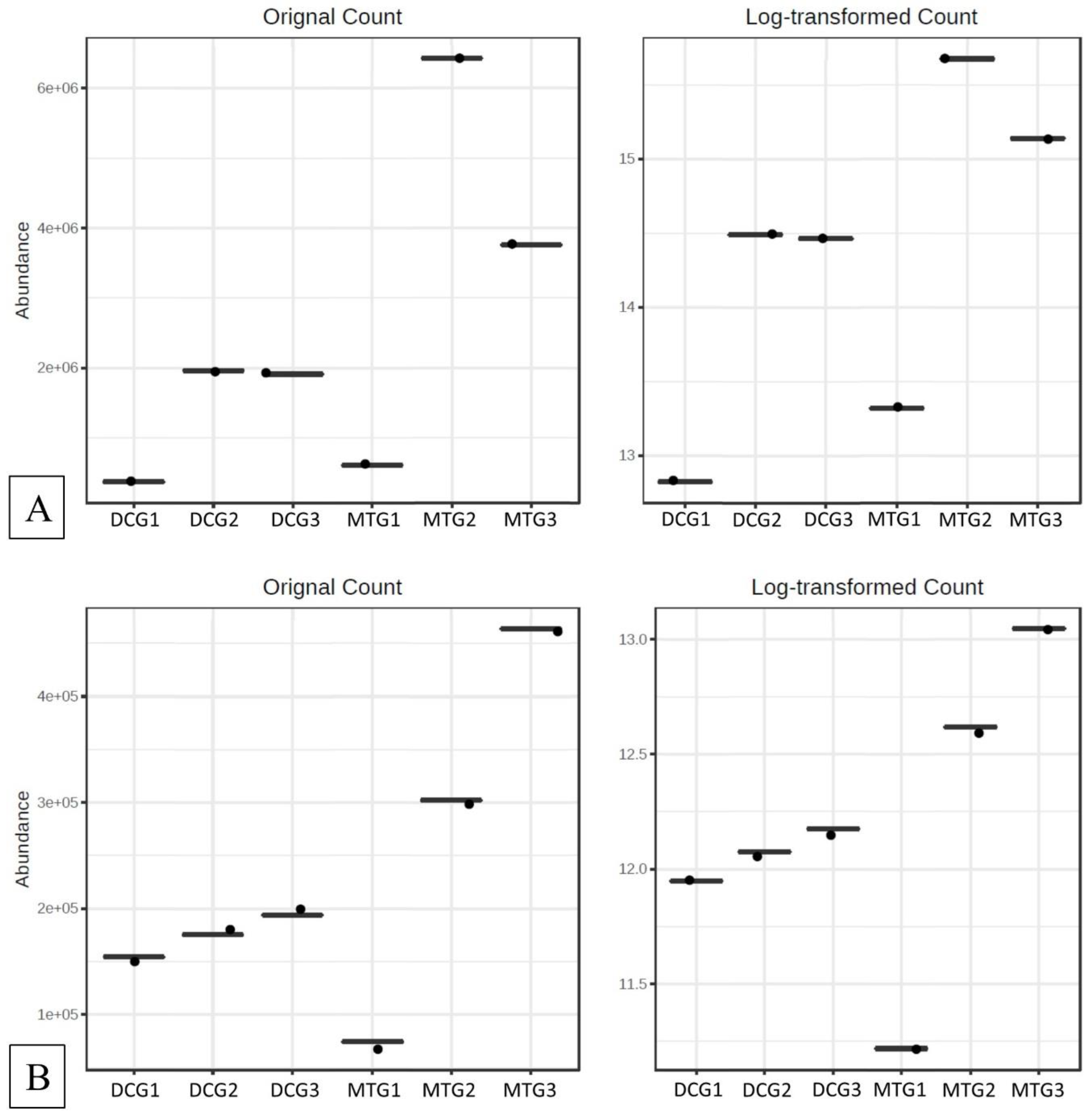
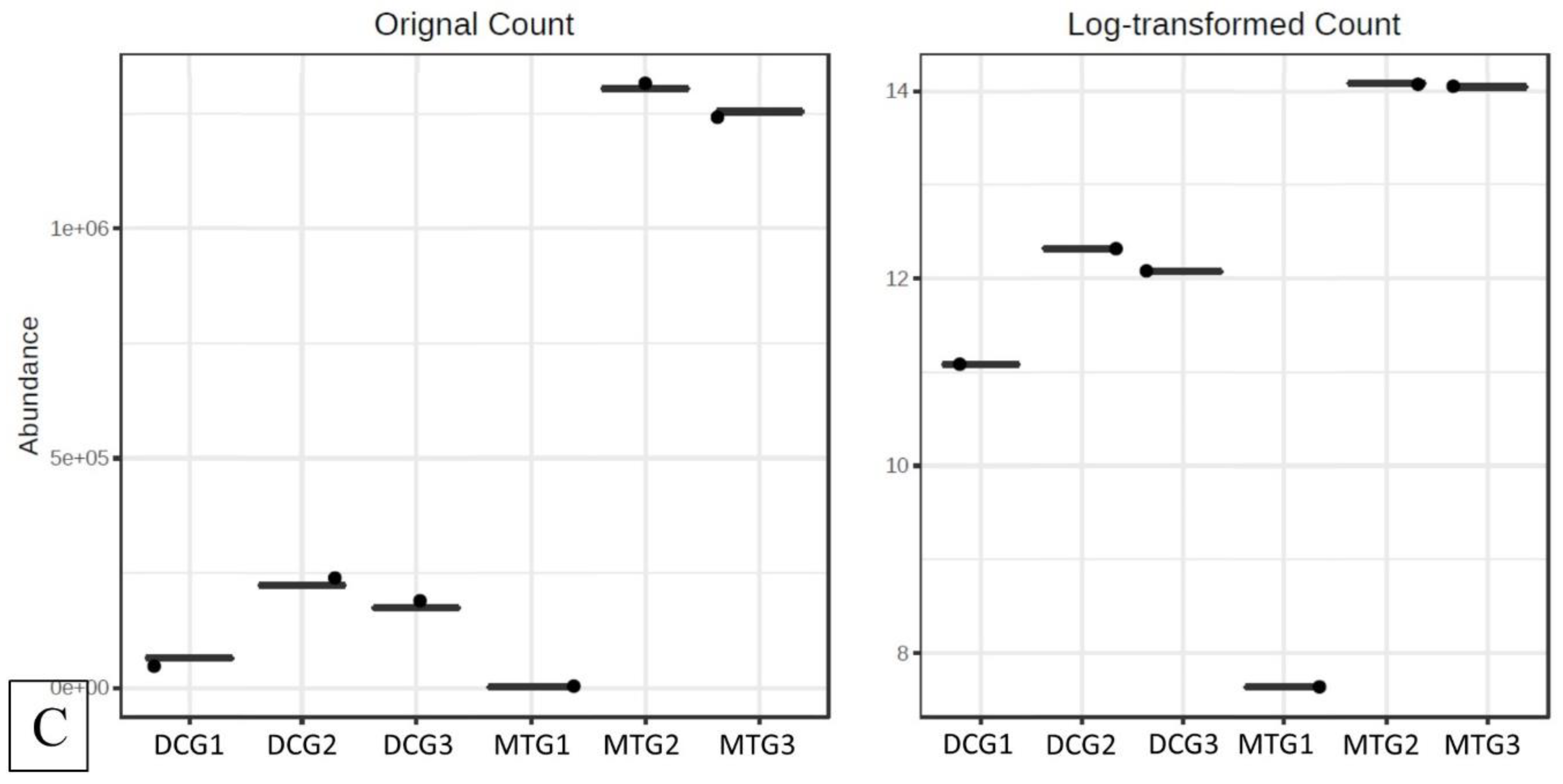
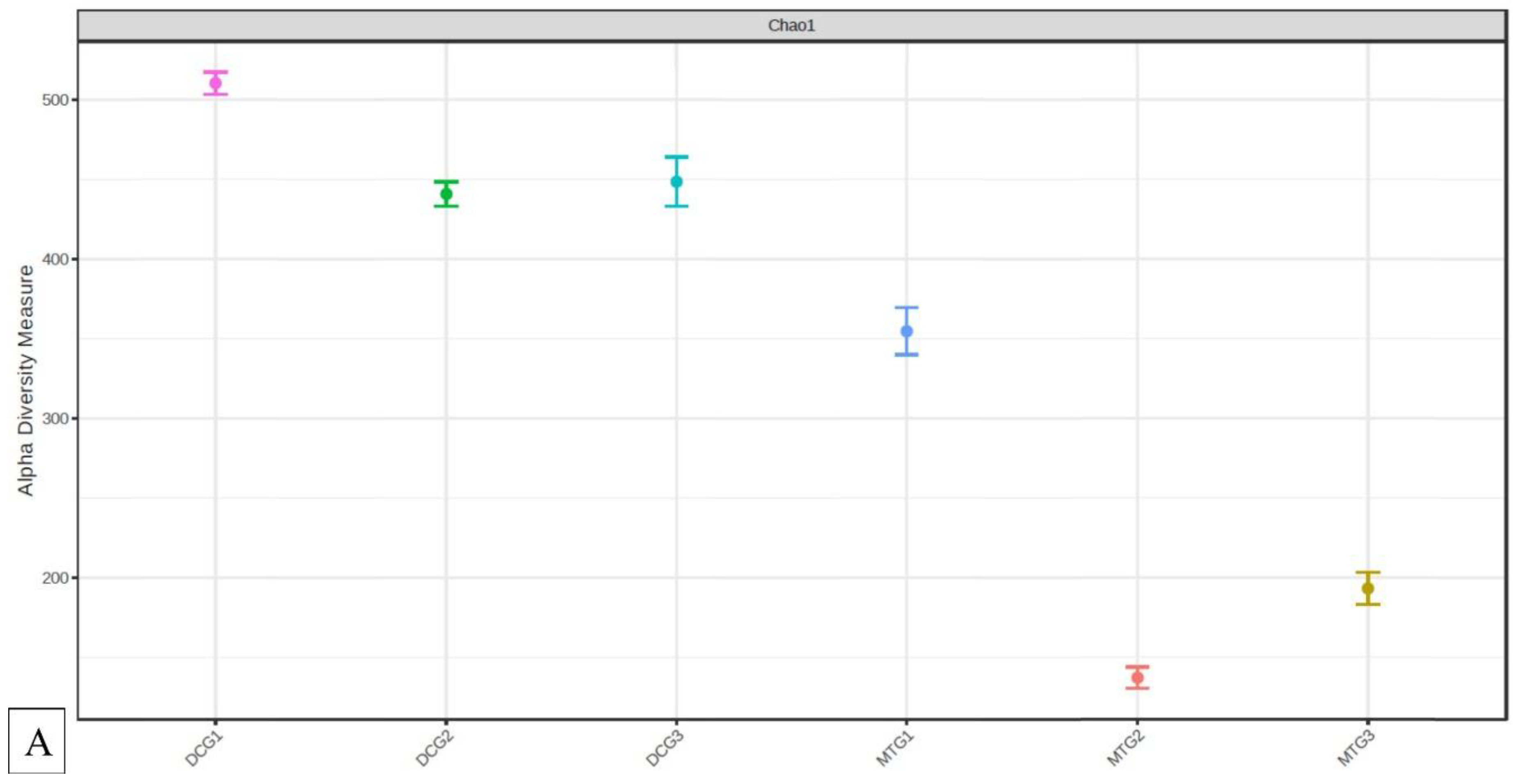
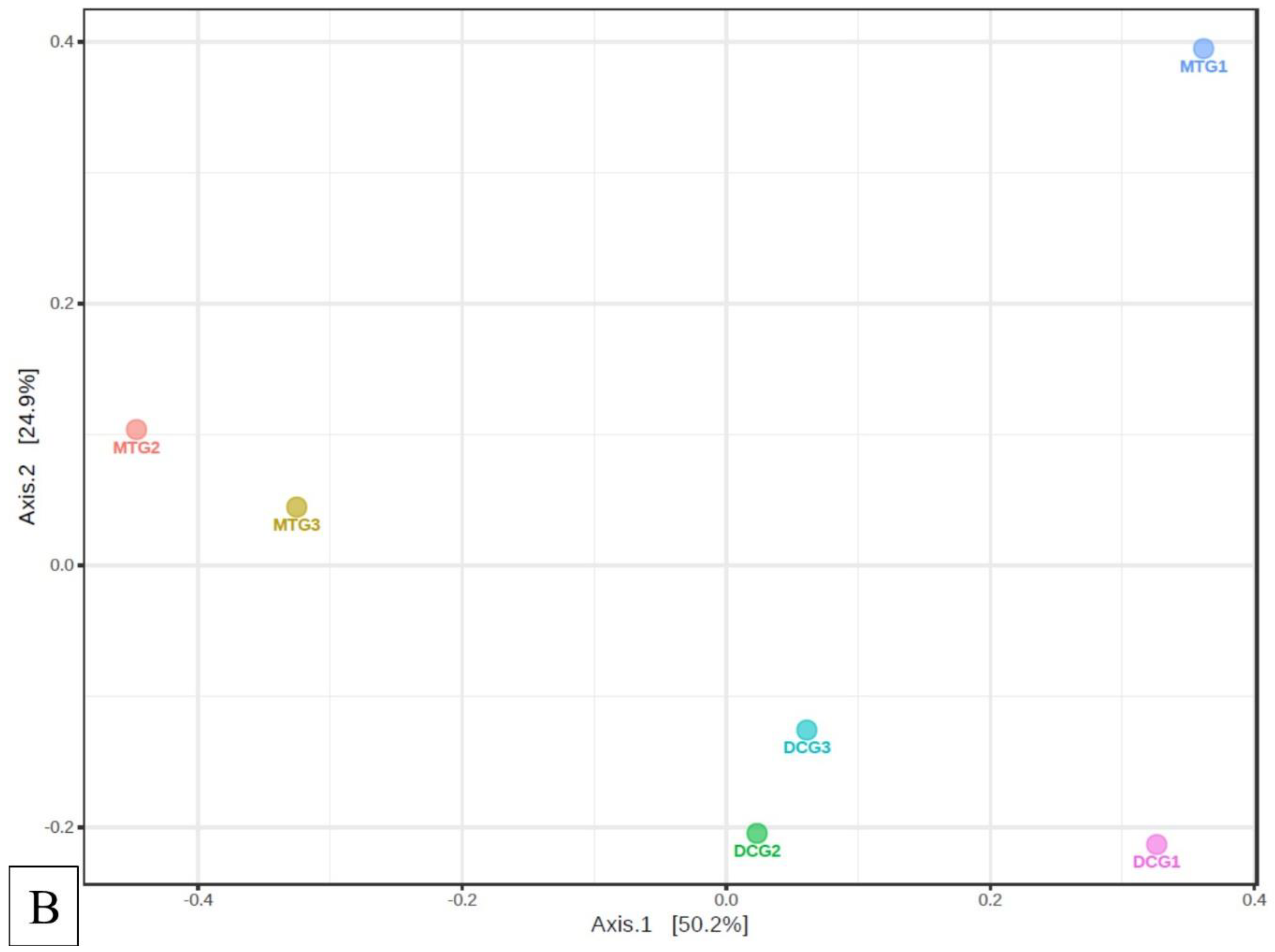
Changes in Bacterial and Fungal Communities Between the DC/MT Generations for Uranium Contaminated Soil
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Keller, M.; Zengler, K. Tapping into microbial diversity. Nat. Rev. Microbiol. 2004, 2, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.; Lewis, K.; Orjala, J.; Mo, S.; Ortenberg, R.; O’Connor, P.; Zhao, C.; Vouros, P.; Kaeberlein, T.; Epstein, S.S. Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl. Environ. Microbiol. 2008, 74, 4889–4897. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.J.; Hugenholtz, P.; Sangwan, P.; Osborne, C.A.; Janssen, P.H. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 2003, 69, 7210–7215. [Google Scholar] [CrossRef] [PubMed]
- Rappé, M.S.; Giovannoni, S.J. The uncultured microbial majority. Annu. Rev. Microbiol. 2003, 57, 369–394. [Google Scholar] [CrossRef] [PubMed]
- Zengler, K.; Toledo, G.; Rappe, M.; Elkins, J.; Mathur, E.J.; Short, J.M.; Keller, M. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 2002, 99, 15681–15686. [Google Scholar] [CrossRef] [PubMed]
- Bollmann, A.; Palumbo, A.V.; Lewis, K.; Epstein, S.S. Isolation and physiology of bacteria from contaminated subsurface sediments. Appl. Environ. Microbiol. 2010, 76, 7413–7419. [Google Scholar] [CrossRef] [PubMed]
- Bruns, A.; Cypionka, H.; Overmann, J. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 2002, 68, 3978–3987. [Google Scholar] [CrossRef] [PubMed]
- Tamaki, H.; Hanada, S.; Sekiguchi, Y.; Tanaka, Y.; Kamagata, Y. Effect of gelling agent on colony formation in solid cultivation of microbial community in lake sediment. Environ. Microbiol. 2009, 11, 1827–1834. [Google Scholar] [CrossRef]
- Davis, K.E.R.; Joseph, S.J.; Janssen, P.H. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 2005, 71, 826–834. [Google Scholar] [CrossRef]
- Kato, S.; Yamagishi, A.; Daimon, S.; Kawasaki, K.; Tamaki, H.; Kitagawa, W.; Abe, A.; Tanaka, M.; Sone, T.; Asano, K.; et al. Isolation of Previously Uncultured Slow-Growing Bacteria by Using a Simple Modification in the Preparation of Agar Media. Appl. Environ. Microbiol. 2018, 84, e00807-18. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Seo, C.; Ji, M.; Paik, M.-J.; Myung, S.-W.; Kim, J. Effective soil extraction method for cultivating previously uncultured soil bacteria. Appl. Environ. Microbiol. 2018, 84, e01145-18. [Google Scholar] [CrossRef] [PubMed]
- Mumtaz, S.; Streten-Joyce, C.; Parry, D.L.; McGuinness, K.A.; Lu, P.; Gibb, K.S. Fungi outcompete bacteria under increased uranium concentration in culture media. J. Environ. Radioact. 2013, 120, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Maddela, N.R.; Masabanda, M.; Leiva-Mora, M. Novel diesel-oil-degrading bacteria and fungi from the Ecuadorian Amazon rainforest. Water Sci. Technol. 2015, 71, 1554–1561. [Google Scholar] [CrossRef] [PubMed]
- Oladipo, O.G.; Awotoye, O.O.; Olayinka, A.; Bezuidenhout, C.C.; Maboeta, M.S. Heavy metal tolerance traits of filamentous fungi isolated from gold and gemstone mining sites. Braz. J. Microbiol. 2018, 49, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Rajapaksha, R.M.C.P.; Tobor-Kapłon, M.A.; Bååth, E. Metal Toxicity Affects Fungal and Bacterial Activities in Soil Differently. Appl. Environ. Microbiol. 2004, 70, 2966–2973. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Pathak, A.; Rathore, R.S.; Prakash, O.; Singh, R.; Jaswal, R.; Seaman, J.; Chauhan, A. Proteogenomic Analysis of Burkholderia Species Strains 25 and 46 Isolated from Uraniferous Soils Reveals Multiple Mechanisms to Cope with Uranium Stress. Cells 2018, 7, 269. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.; Pathak, A.; Jaswal, R.; Edwards III, B.; Chappell, D.; Ball, C.; Garcia-Sillas, R.; Stothard, P.; Seaman, J. Physiological and Comparative Genomic Analysis of Arthrobacter sp. SRS-W-1-2016 Provides Insights on Niche Adaptation for Survival in Uraniferous Soils. Genes 2018, 9, 31. [Google Scholar] [CrossRef]
- Jaswal, R.; Pathak, A.; Edwards III, B.; Lewis III, R.; Seaman, J.C.; Stothard, P.; Krivushin, K.; Blom, J.; Rupp, O.; Chauhan, A. Metagenomics-Guided Survey, Isolation, and Characterization of Uranium Resistant Microbiota from the Savannah River Site, USA. Genes 2019, 10, 325. [Google Scholar] [CrossRef]
- Feng, G.; Xie, T.; Wang, X.; Bai, J.; Tang, L.; Zhao, H.; Wei, W.; Wang, M.; Zhao, Y. Metagenomic analysis of microbial community and function involved in cd-contaminated soil. BMC Microbiol 2018, 18, 11. [Google Scholar] [CrossRef]
- Hemme, C.L.; Tu, Q.; Shi, Z.; Qin, Y.; Gao, W.; Deng, Y.; Nostrand, J.D.V.; Wu, L.; He, Z.; Chain, P.S.G.; et al. Comparative metagenomics reveals impact of contaminants on groundwater microbiomes. Front. Microbiol. 2015, 6, 1205. [Google Scholar] [CrossRef]
- Bollmann, A.; Lewis, K.; Epstein, S.S. Incubation of Environmental Samples in a Diffusion Chamber Increases the Diversity of Recovered Isolates. Appl. Environ. Microbiol. 2007, 73, 6386–6390. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.G.; Bauer, L.R.; Haselow, J.S.; Hayes, D.W.; Martin, H.L.; McDowell, W.L.; Pickett, J.B. Uranium in the Savannah River Site Environment; Westinghouse Savannah River Co.: Aiken, SC, USA, December 1992. [Google Scholar]
- Gavrish, E.; Bollmann, A.; Epstein, S.; Lewis, K. A trap for in situ cultivation of filamentous actinobacteria. J. Microbiol. Methods 2008, 72, 257–262. [Google Scholar] [CrossRef] [PubMed]
- United States Environmental Protection Agency. National Primary Drinking Water Regulations. Available online: https://www.epa.gov/ground-water-and-drinking-water/national-primary-drinking-water-regulations (accessed on 25 October 2018).
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. In PCR-Protocols and Applications-A Laboratory Manual; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. ISBN 978-0-12-372180-8. [Google Scholar]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinform. 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinform. 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Tikhonov, M.; Leach, R.W.; Wingreen, N.S. Interpreting 16S metagenomic data without clustering to achieve sub-OTU resolution. ISME J. 2015, 9, 68–80. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Yan, X.; Luo, X.; Zhao, M. Metagenomic analysis of microbial community in uranium-contaminated soil. Appl. Microbiol. Biotechnol. 2016, 100, 299–310. [Google Scholar] [CrossRef]
- Akob, D.M.; Mills, H.J.; Kostka, J.E. Metabolically active microbial communities in uranium-contaminated subsurface sediments. FEMS Microbiol. Ecol. 2007, 59, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Green, S.J.; Prakash, O.; Jasrotia, P.; Overholt, W.A.; Cardenas, E.; Hubbard, D.; Tiedje, J.M.; Watson, D.B.; Schadt, C.W.; Brooks, S.C.; et al. Denitrifying bacteria from the genus Rhodanobacter dominate bacterial communities in the highly contaminated subsurface of a nuclear legacy waste site. Appl. Environ. Microbiol. 2012, 78, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, B.; Chariton, A.A.; Harford, A.J.; Hose, G.C.; Greenfield, P.; Elbourne, L.D.H.; Oytam, Y.; Stephenson, S.; Midgley, D.J.; Paulsen, I.T. Effects of uranium concentration on microbial community structure and functional potential. Environ. Microbiol. 2017, 19, 3323–3341. [Google Scholar] [CrossRef] [PubMed]
- Pathak, A.; Chauhan, A.; Stothard, P.; Green, S.; Maienschein-Cline, M.; Jaswal, R.; Seaman, J. Genome-centric evaluation of Burkholderia sp. strain SRS-W-2-2016 resistant to high concentrations of uranium and nickel isolated from the Savannah River Site (SRS), USA. Genom. Data 2017, 12, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.S.; Handley, K.M.; Wrighton, K.C.; Frischkorn, K.R.; Thomas, B.C.; Banfield, J.F. Short-Read Assembly of Full-Length 16S Amplicons Reveals Bacterial Diversity in Subsurface Sediments. PLoS ONE 2013, 8, e56018. [Google Scholar] [CrossRef] [PubMed]
- Moreels, D.; Crosson, G.; Garafola, C.; Monteleone, D.; Taghavi, S.; Fitts, J.P.; van der Lelie, D. Microbial community dynamics in uranium contaminated subsurface sediments under biostimulated conditions with high nitrate and nickel pressure. Environ. Sci. Pollut. Res. Int. 2008, 15, 481–491. [Google Scholar] [CrossRef] [PubMed]
- North, N.N.; Dollhopf, S.L.; Petrie, L.; Istok, J.D.; Balkwill, D.L.; Kostka, J.E. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 2004, 70, 4911–4920. [Google Scholar] [CrossRef] [PubMed]
- Vishnivetskaya, T.A.; Brandt, C.C.; Madden, A.S.; Drake, M.M.; Kostka, J.E.; Akob, D.M.; Küsel, K.; Palumbo, A.V. Microbial community changes in response to ethanol or methanol amendments for U(VI) reduction. Appl. Environ. Microbiol. 2010, 76, 5728–5735. [Google Scholar] [CrossRef]
- Ormeño-Orrillo, E.; Rogel, M.A.; Chueire, L.M.O.; Tiedje, J.M.; Martínez-Romero, E.; Hungria, M. Genome sequences of Burkholderia sp. strains CCGE1002 and H160, isolated from legume nodules in Mexico and Brazil. J. Bacteriol. 2012, 194, 6927. [Google Scholar] [CrossRef]
- Oyetibo, G.O.; Ilori, M.O.; Obayori, O.S.; Amund, O.O. Biodegradation of petroleum hydrocarbons in the presence of nickel and cobalt. J. Basic Microbiol. 2013, 53, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pantoja, D.; Nikel, P.I.; Chavarría, M.; de Lorenzo, V. Endogenous stress caused by faulty oxidation reactions fosters evolution of 2,4-dinitrotoluene-degrading bacteria. PLoS Genet. 2013, 9, e1003764. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Moreno, Z.R.; Caballero-Mellado, J.; Coutinho, B.G.; Mendonça-Previato, L.; James, E.K.; Venturi, V. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 2012, 63, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, Z.; Chai, L.; Wang, Y.; Liu, Y.; Xiao, R. Bioleaching remediation of heavy metal-contaminated soils using Burkholderia sp. Z-90. J. Hazard Mater. 2016, 301, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Koribanics, N.M.; Tuorto, S.J.; Lopez-Chiaffarelli, N.; McGuinness, L.R.; Häggblom, M.M.; Williams, K.H.; Long, P.E.; Kerkhof, L.J. Spatial Distribution of an Uranium-Respiring Betaproteobacterium at the Rifle, CO Field Research Site. PLoS ONE 2015, 10, e0123378. [Google Scholar] [CrossRef] [PubMed]
- Gadd, G.M.; Fomina, M. Uranium and Fungi. Geomicrobiol. J. 2011, 28, 471–482. [Google Scholar] [CrossRef]

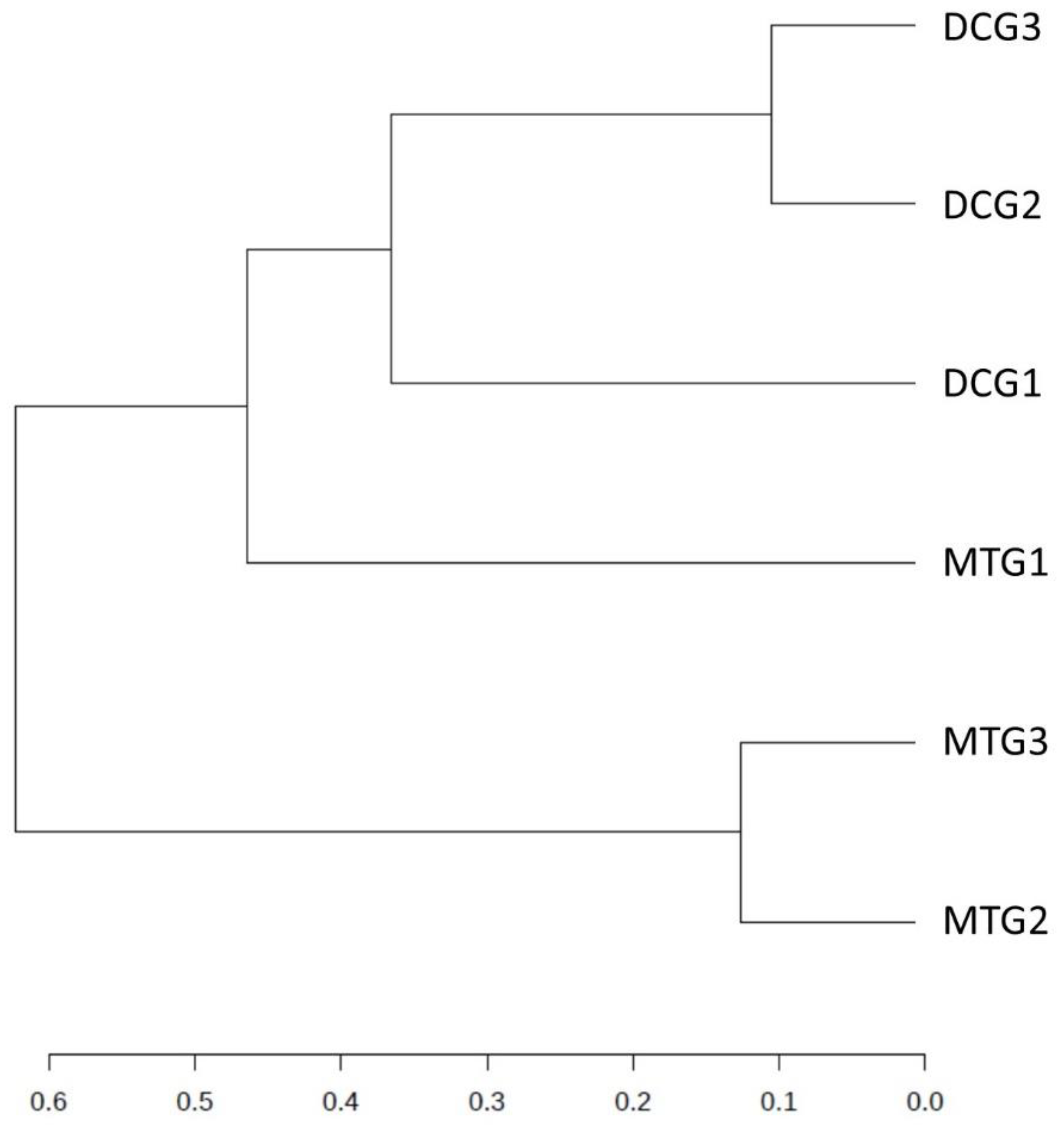
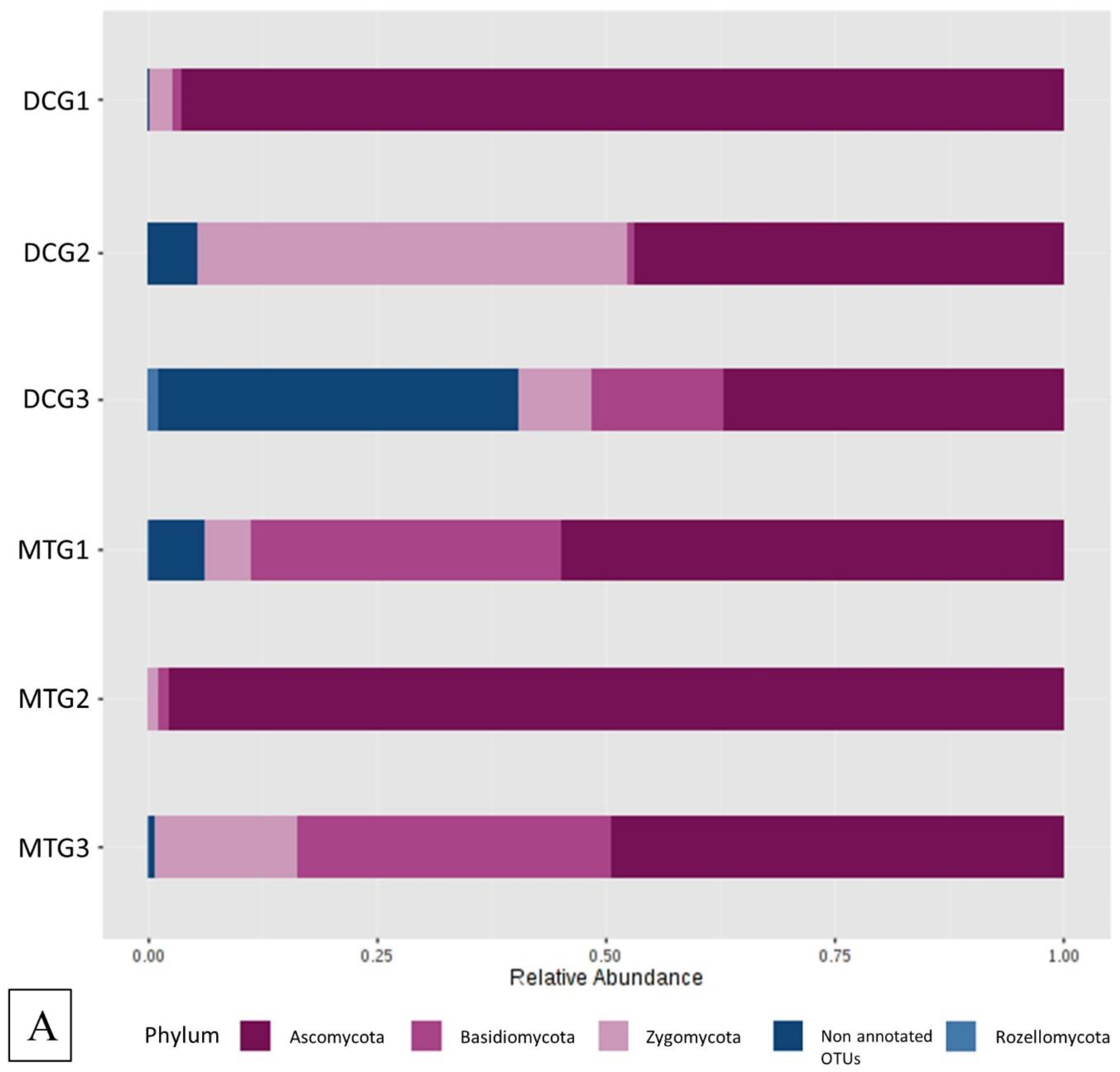
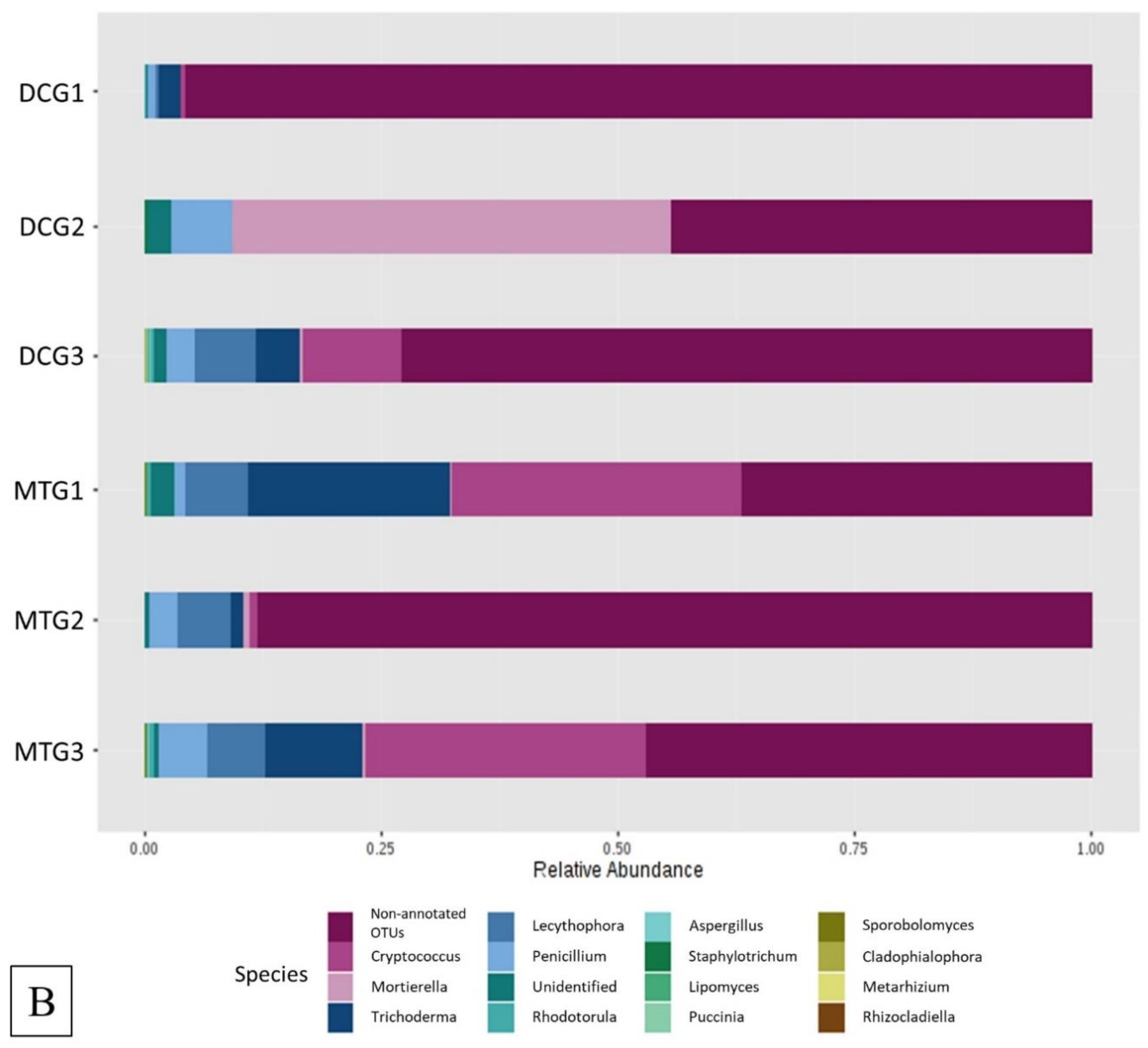
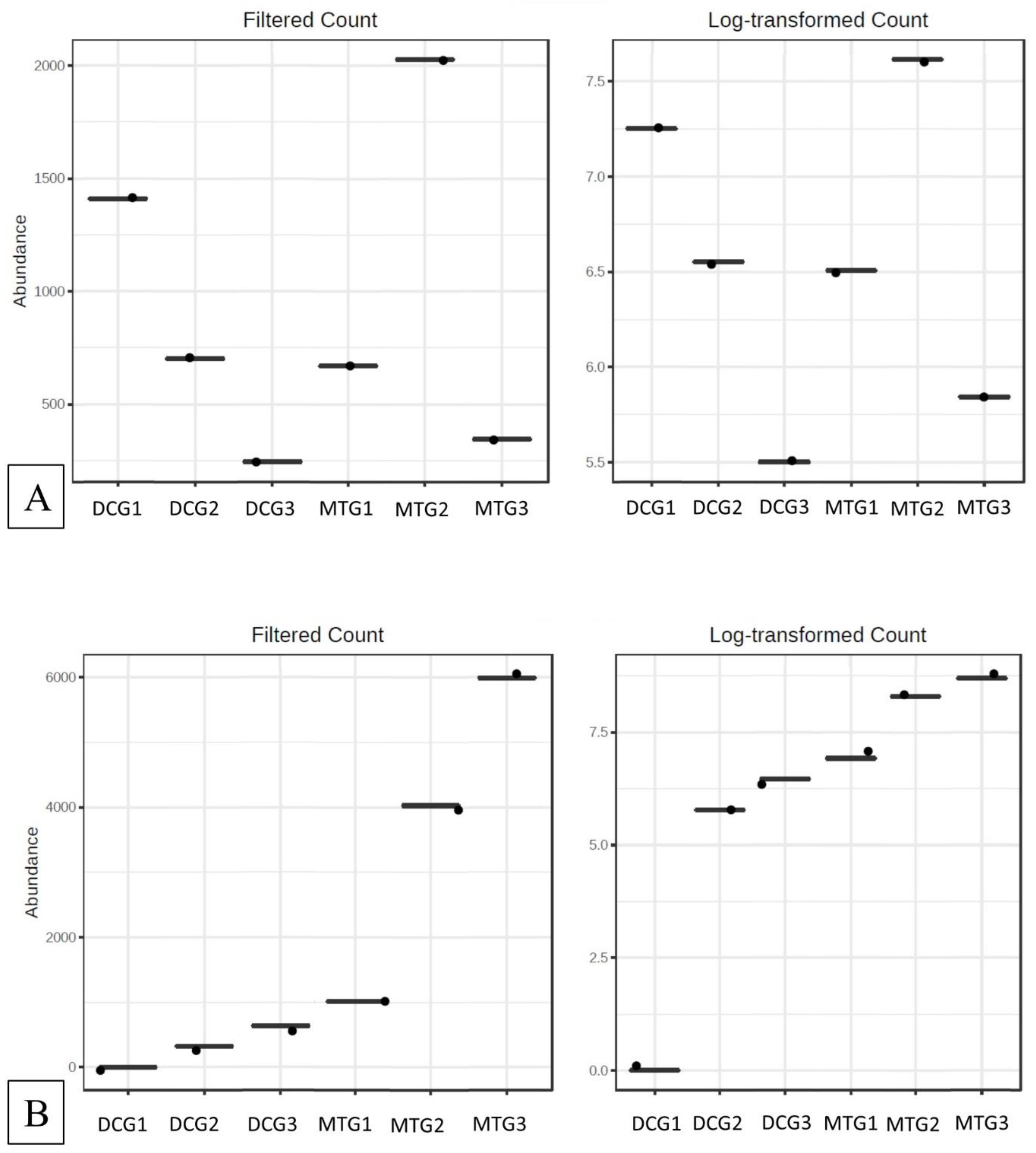
| Sample Name | Total Read Counts | Phylum Level Annotation | Genus Level Annotation |
|---|---|---|---|
| Domain Bacteria: | |||
| DC-generation 1 | 106,514 | 105,499 | 50,468 |
| DC-generation 2 | 120,145 | 119,179 | 67,456 |
| DC-generation 3 | 92,696 | 92,121 | 58,014 |
| MT- generation 1 | 109,144 | 108,476 | 66,457 |
| MT- generation 2 | 99,139 | 98,791 | 84,591 |
| MT- generation 3 | 100,637 | 98,342 | 75,028 |
| Domain Fungi: | |||
| DC-generation 1 | 23,564 | 14,870 | 6186 |
| DC-generation 2 | 40,409 | 40,167 | 21,498 |
| DC-generation 3 | 30,051 | 28,305 | 17,923 |
| MT- generation 1 | 22,288 | 21,071 | 12,346 |
| MT- generation 2 | 23,073 | 23,058 | 2791 |
| MT- generation 3 | 27,535 | 27,488 | 1225 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaswal, R.; Pathak, A.; Chauhan, A. Metagenomic Evaluation of Bacterial and Fungal Assemblages Enriched within Diffusion Chambers and Microbial Traps Containing Uraniferous Soils. Microorganisms 2019, 7, 324. https://doi.org/10.3390/microorganisms7090324
Jaswal R, Pathak A, Chauhan A. Metagenomic Evaluation of Bacterial and Fungal Assemblages Enriched within Diffusion Chambers and Microbial Traps Containing Uraniferous Soils. Microorganisms. 2019; 7(9):324. https://doi.org/10.3390/microorganisms7090324
Chicago/Turabian StyleJaswal, Rajneesh, Ashish Pathak, and Ashvini Chauhan. 2019. "Metagenomic Evaluation of Bacterial and Fungal Assemblages Enriched within Diffusion Chambers and Microbial Traps Containing Uraniferous Soils" Microorganisms 7, no. 9: 324. https://doi.org/10.3390/microorganisms7090324
APA StyleJaswal, R., Pathak, A., & Chauhan, A. (2019). Metagenomic Evaluation of Bacterial and Fungal Assemblages Enriched within Diffusion Chambers and Microbial Traps Containing Uraniferous Soils. Microorganisms, 7(9), 324. https://doi.org/10.3390/microorganisms7090324






