Survey of Biosynthetic Gene Clusters from Sequenced Myxobacteria Reveals Unexplored Biosynthetic Potential
Abstract
1. Introduction
2. Materials and Methods
3. Results
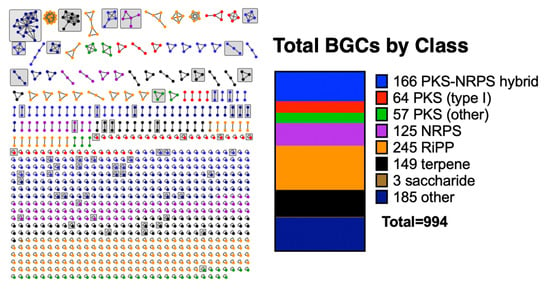
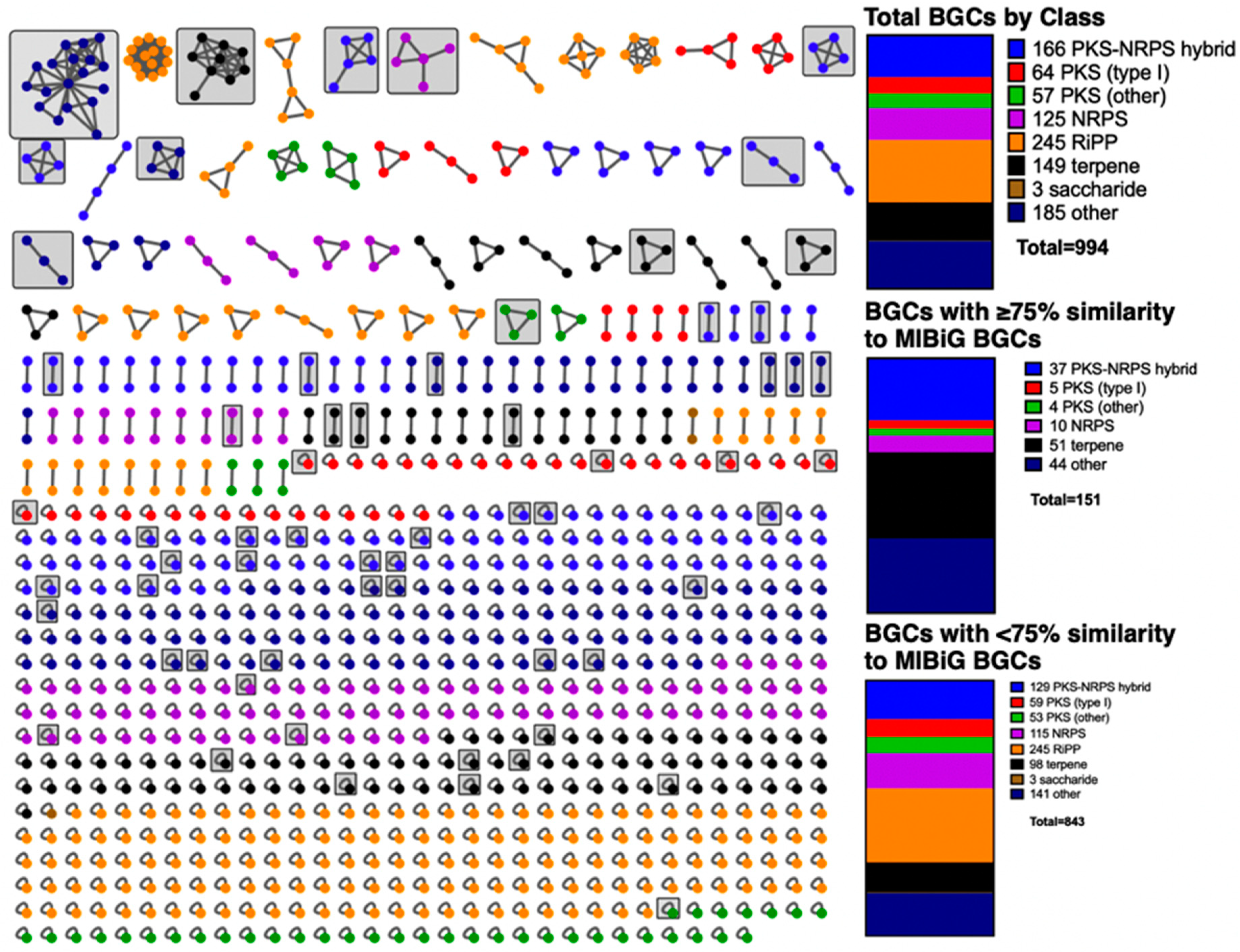
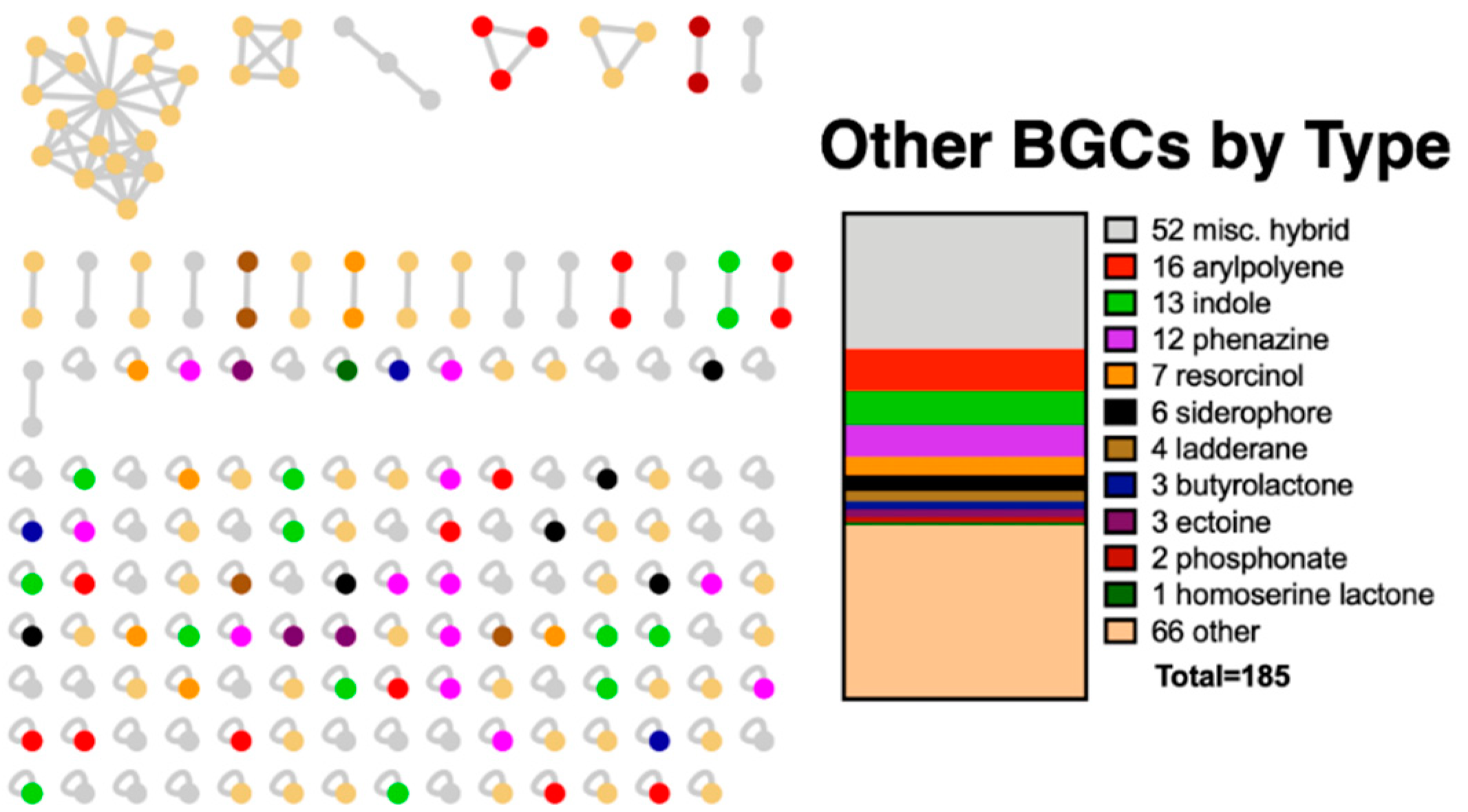
3.1. BiG-SCAPE Analysis of BGCs from Sequenced Myxobacteria
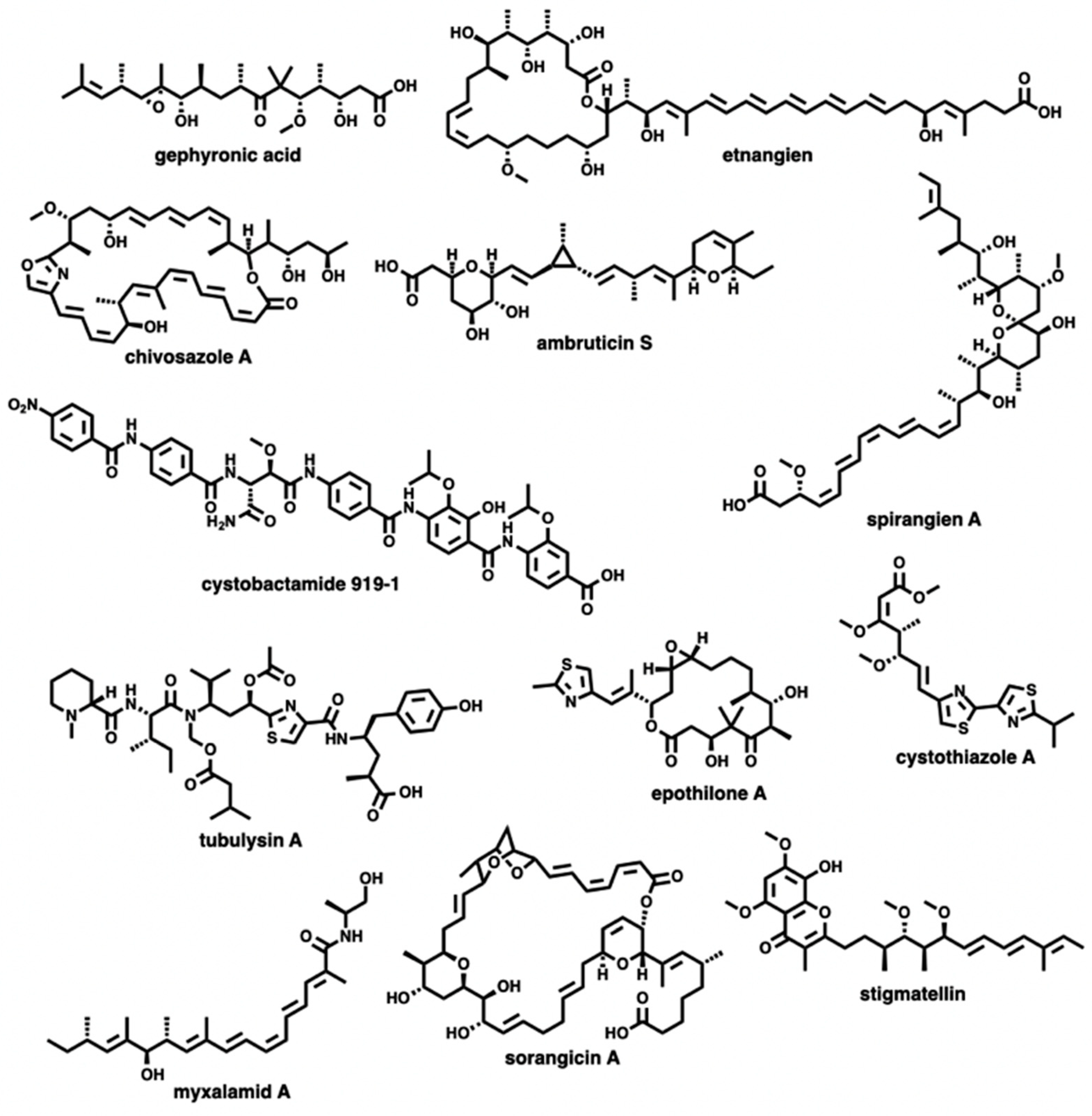
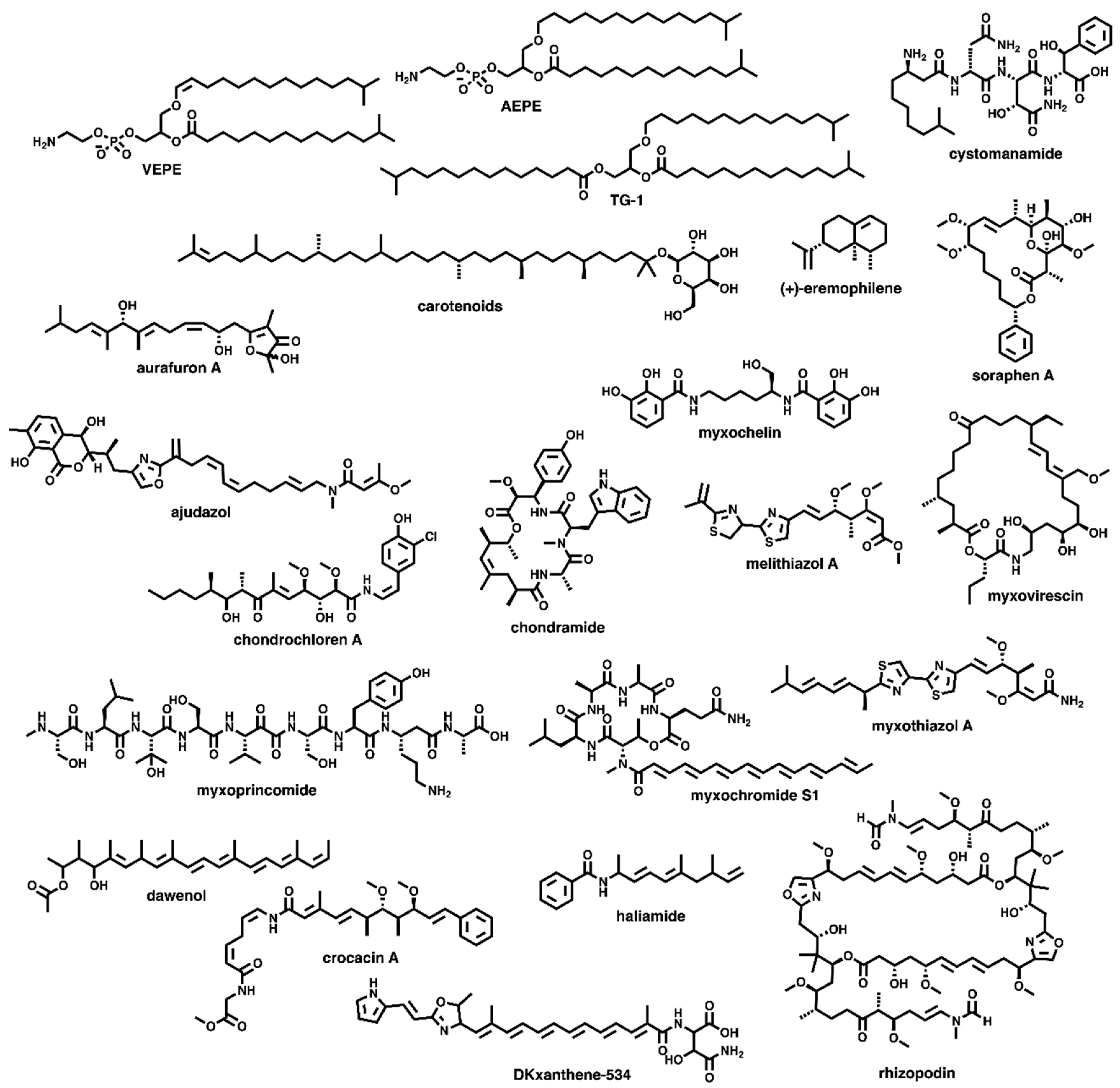
3.2. Discovered Metabolites from Myxobacteria and Associated BGCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brinkhoff, T.; Fischer, D.; Vollmers, J.; Voget, S.; Beardsley, C.; Thole, S.; Mussmann, M.; Kunze, B.; Wagner-Dobler, I.; Daniel, R.; et al. Biogeography and phylogenetic diversity of a cluster of exclusively marine myxobacteria. ISME J. 2012, 6, 1260–1272. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Dey, A.; Vassallo, C.N.; Wall, D. How Myxobacteria Cooperate. J. Mol. Biol. 2015, 427, 3709–3721. [Google Scholar] [CrossRef] [PubMed]
- Mohr, K.I. Diversity of Myxobacteria-We Only See the Tip of the Iceberg. Microorganisms 2018, 6, 84. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Dorado, J.; Marcos-Torres, F.J.; Garcia-Bravo, E.; Moraleda-Munoz, A.; Perez, J. Myxobacteria: Moving, Killing, Feeding, and Surviving Together. Front. Microbiol. 2016, 7, 781. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, J.; Fayad, A.A.; Müller, R. Natural products from myxobacteria: Novel metabolites and bioactivities. Nat. Prod. Rep. 2017, 34, 135–160. [Google Scholar] [CrossRef] [PubMed]
- Landwehr, W.; Wolf, C.; Wink, J. Actinobacteria and Myxobacteria-Two of the Most Important Bacterial Resources for Novel Antibiotics. Curr. Top. Microbiol. Immunol. 2016, 398, 273–302. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Natural product drug discovery in the genomic era: Realities, conjectures, misconceptions, and opportunities. J. Ind. Microbiol. Biotechnol. 2019, 46, 281–299. [Google Scholar] [CrossRef]
- Liu, R.; Deng, Z.; Liu, T. Streptomyces species: Ideal chassis for natural product discovery and overproduction. Metab. Eng. 2018, 50, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Pascal Andreu, V.; de Los Santos, E.L.C.; Del Carratore, F.; Lee, S.Y.; Medema, M.H.; Weber, T. The antiSMASH database version 2: A comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2019, 47, D625–D630. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef]
- Baral, B.; Akhgari, A.; Metsa-Ketela, M. Activation of microbial secondary metabolic pathways: Avenues and challenges. Synth. Syst. Biotechnol. 2018, 3, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Okada, B.K.; Wu, Y.; Xu, F.; Seyedsayamdost, M.R. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr. Opin. Microbiol. 2018, 45, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.; Tryon, J.H.; Parkinson, E.I.; de los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A computational framework for systematic exploration of biosynthetic diversity from large-scale genomic data. Biorxiv 2018. [Google Scholar] [CrossRef]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Richardson, L.J.; Rawlings, N.D.; Salazar, G.A.; Almeida, A.; Haft, D.R.; Ducq, G.; Sutton, G.G.; Finn, R.D. Genome properties in 2019: A new companion database to InterPro for the inference of complete functional attributes. Nucleic Acids Res. 2019, 47, D564–D572. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de Los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Bhat, S.; Ahrendt, T.; Dauth, C.; Bode, H.B.; Shimkets, L.J. Two lipid signals guide fruiting body development of Myxococcus xanthus. MBio 2014, 5, e00939-13. [Google Scholar] [CrossRef]
- Botella, J.A.; Murillo, F.J.; Ruiz-Vazquez, R. A cluster of structural and regulatory genes for light-induced carotenogenesis in Myxococcus xanthus. Eur. J. Biochem 1995, 233, 238–248. [Google Scholar] [CrossRef]
- Buntin, K.; Weissman, K.J.; Müller, R. An unusual thioesterase promotes isochromanone ring formation in ajudazol biosynthesis. Chembiochem 2010, 11, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, M.; Murillo, F.J. Role for vitamin B(12) in light induction of gene expression in the bacterium Myxococcus xanthus. J. Bacteriol. 2002, 184, 2215–2224. [Google Scholar] [CrossRef] [PubMed]
- Cortina, N.S.; Krug, D.; Plaza, A.; Revermann, O.; Müller, R. Myxoprincomide: A natural product from Myxococcus xanthus discovered by comprehensive analysis of the secondary metabolome. Angew. Chem. Int. Ed. Engl. 2012, 51, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Etzbach, L.; Plaza, A.; Garcia, R.; Baumann, S.; Müller, R. Cystomanamides: Structure and biosynthetic pathway of a family of glycosylated lipopeptides from myxobacteria. Org. Lett. 2014, 16, 2414–2417. [Google Scholar] [CrossRef] [PubMed]
- Frank, B.; Wenzel, S.C.; Bode, H.B.; Scharfe, M.; Blocker, H.; Müller, R. From genetic diversity to metabolic unity: Studies on the biosynthesis of aurafurones and aurafuron-like structures in myxobacteria and streptomycetes. J. Mol. Biol. 2007, 374, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Gaitatzis, N.; Kunze, B.; Müller, R. In vitro reconstitution of the myxochelin biosynthetic machinery of Stigmatella aurantiaca Sg a15: Biochemical characterization of a reductive release mechanism from nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. USA 2001, 98, 11136–11141. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Weissman, K.J.; Müller, R. Myxochelin biosynthesis: Direct evidence for two- and four-electron reduction of a carrier protein-bound thioester. J. Am. Chem. Soc. 2008, 130, 7554–7555. [Google Scholar] [CrossRef] [PubMed]
- Ligon, J.; Hill, S.; Beck, J.; Zirkle, R.; Molnar, I.; Zawodny, J.; Money, S.; Schupp, T. Characterization of the biosynthetic gene cluster for the antifungal polyketide soraphen A from Sorangium cellulosum So ce26. Gene 2002, 285, 257–267. [Google Scholar] [CrossRef]
- Lopez-Rubio, J.J.; Elias-Arnanz, M.; Padmanabhan, S.; Murillo, F.J. A repressor-antirepressor pair links two loci controlling light-induced carotenogenesis in Myxococcus xanthus. J. Biol. Chem. 2002, 277, 7262–7270. [Google Scholar] [CrossRef]
- Lorenzen, W.; Ahrendt, T.; Bozhuyuk, K.A.; Bode, H.B. A multifunctional enzyme is involved in bacterial ether lipid biosynthesis. Nat. Chem. Biol. 2014, 10, 425–427. [Google Scholar] [CrossRef]
- Meiser, P.; Weissman, K.J.; Bode, H.B.; Krug, D.; Dickschat, J.S.; Sandmann, A.; Müller, R. DKxanthene biosynthesis--understanding the basis for diversity-oriented synthesis in myxobacterial secondary metabolism. Chem. Biol. 2008, 15, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Rachid, S.; Hoffmann, T.; Surup, F.; Volz, C.; Zaburannyi, N.; Müller, R. Biosynthesis of crocacin involves an unusual hydrolytic release domain showing similarity to condensation domains. Chem. Biol. 2014, 21, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Osswald, C.; Zaburannyi, N.; Burgard, C.; Hoffmann, T.; Wenzel, S.C.; Müller, R. A highly unusual polyketide synthase directs dawenol polyene biosynthesis in Stigmatella aurantiaca. J. Biotechnol. 2014, 191, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Perez-Marin, M.C.; Padmanabhan, S.; Polanco, M.C.; Murillo, F.J.; Elias-Arnanz, M. Vitamin B12 partners the CarH repressor to downregulate a photoinducible promoter in Myxococcus xanthus. Mol. Microbiol. 2008, 67, 804–819. [Google Scholar] [CrossRef] [PubMed]
- Pistorius, D.; Müller, R. Discovery of the rhizopodin biosynthetic gene cluster in Stigmatella aurantiaca Sg a15 by genome mining. Chembiochem 2012, 13, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Rachid, S.; Scharfe, M.; Blocker, H.; Weissman, K.J.; Müller, R. Unusual chemistry in the biosynthesis of the antibiotic chondrochlorens. Chem. Biol. 2009, 16, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Schifrin, A.; Ly, T.T.; Gunnewich, N.; Zapp, J.; Thiel, V.; Schulz, S.; Hannemann, F.; Khatri, Y.; Bernhardt, R. Characterization of the gene cluster CYP264B1-geoA from Sorangium cellulosum So ce56: Biosynthesis of (+)-eremophilene and its hydroxylation. Chembiochem 2015, 16, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Silakowski, B.; Schairer, H.U.; Ehret, H.; Kunze, B.; Weinig, S.; Nordsiek, G.; Brandt, P.; Blocker, H.; Hofle, G.; Beyer, S.; et al. New lessons for combinatorial biosynthesis from myxobacteria. The myxothiazol biosynthetic gene cluster of Stigmatella aurantiaca DW4/3-1. J. Biol. Chem. 1999, 274, 37391–37399. [Google Scholar] [CrossRef]
- Simunovic, V.; Zapp, J.; Rachid, S.; Krug, D.; Meiser, P.; Müller, R. Myxovirescin A biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl-CoA synthases, and trans-acting acyltransferases. Chembiochem 2006, 7, 1206–1220. [Google Scholar] [CrossRef]
- Sun, Y.; Tomura, T.; Sato, J.; Iizuka, T.; Fudou, R.; Ojika, M. Isolation and Biosynthetic Analysis of Haliamide, a New PKS-NRPS Hybrid Metabolite from the Marine Myxobacterium Haliangium ochraceum. Molecules 2016, 21, 59. [Google Scholar] [CrossRef]
- Weinig, S.; Hecht, H.J.; Mahmud, T.; Müller, R. Melithiazol biosynthesis: Further insights into myxobacterial PKS/NRPS systems and evidence for a new subclass of methyl transferases. Chem. Biol. 2003, 10, 939–952. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, S.C.; Kunze, B.; Hofle, G.; Silakowski, B.; Scharfe, M.; Blocker, H.; Müller, R. Structure and biosynthesis of myxochromides S1-3 in Stigmatella aurantiaca: Evidence for an iterative bacterial type I polyketide synthase and for module skipping in nonribosomal peptide biosynthesis. Chembiochem 2005, 6, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Rachid, S.; Krug, D.; Kunze, B.; Kochems, I.; Scharfe, M.; Zabriskie, T.M.; Blocker, H.; Müller, R. Molecular and biochemical studies of chondramide formation-highly cytotoxic natural products from Chondromyces crocatus Cm c5. Chem. Biol. 2006, 13, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Baumann, S.; Herrmann, J.; Raju, R.; Steinmetz, H.; Mohr, K.I.; Huttel, S.; Harmrolfs, K.; Stadler, M.; Müller, R. Cystobactamids: Myxobacterial topoisomerase inhibitors exhibiting potent antibacterial activity. Angew. Chem. Int. Ed. Engl. 2014, 53, 14605–14609. [Google Scholar] [CrossRef] [PubMed]
- Beyer, S.; Kunze, B.; Silakowski, B.; Müller, R. Metabolic diversity in myxobacteria: Identification of the myxalamid and the stigmatellin biosynthetic gene cluster of Stigmatella aurantiaca Sg a15 and a combined polyketide-(poly)peptide gene cluster from the epothilone producing strain Sorangium cellulosum So ce90. Biochim. Biophys. Acta 1999, 1445, 185–195. [Google Scholar] [PubMed]
- Feng, Z.; Qi, J.; Tsuge, T.; Oba, Y.; Kobayashi, T.; Suzuki, Y.; Sakagami, Y.; Ojika, M. Construction of a bacterial artificial chromosome library for a myxobacterium of the genus Cystobacter and characterization of an antibiotic biosynthetic gene cluster. Biosci. Biotechnol. Biochem. 2005, 69, 1372–1380. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Frank, B.; Knauber, J.; Steinmetz, H.; Scharfe, M.; Blocker, H.; Beyer, S.; Müller, R. Spiroketal polyketide formation in Sorangium: Identification and analysis of the biosynthetic gene cluster for the highly cytotoxic spirangienes. Chem. Biol. 2007, 14, 221–233. [Google Scholar] [CrossRef]
- Irschik, H.; Kopp, M.; Weissman, K.J.; Buntin, K.; Piel, J.; Müller, R. Analysis of the sorangicin gene cluster reinforces the utility of a combined phylogenetic/retrobiosynthetic analysis for deciphering natural product assembly by trans-AT PKS. Chembiochem 2010, 11, 1840–1849. [Google Scholar] [CrossRef]
- Julien, B.; Shah, S.; Ziermann, R.; Goldman, R.; Katz, L.; Khosla, C. Isolation and characterization of the epothilone biosynthetic gene cluster from Sorangium cellulosum. Gene 2000, 249, 153–160. [Google Scholar] [CrossRef]
- Julien, B.; Tian, Z.Q.; Reid, R.; Reeves, C.D. Analysis of the ambruticin and jerangolid gene clusters of Sorangium cellulosum reveals unusual mechanisms of polyketide biosynthesis. Chem. Biol. 2006, 13, 1277–1286. [Google Scholar] [CrossRef]
- Menche, D.; Arikan, F.; Perlova, O.; Horstmann, N.; Ahlbrecht, W.; Wenzel, S.C.; Jansen, R.; Irschik, H.; Müller, R. Stereochemical determination and complex biosynthetic assembly of etnangien, a highly potent RNA polymerase inhibitor from the myxobacterium Sorangium cellulosum. J. Am. Chem. Soc. 2008, 130, 14234–14243. [Google Scholar] [CrossRef] [PubMed]
- Molnar, I.; Schupp, T.; Ono, M.; Zirkle, R.; Milnamow, M.; Nowak-Thompson, B.; Engel, N.; Toupet, C.; Stratmann, A.; Cyr, D.D.; et al. The biosynthetic gene cluster for the microtubule-stabilizing agents epothilones A and B from Sorangium cellulosum So ce90. Chem. Biol. 2000, 7, 97–109. [Google Scholar] [CrossRef]
- Perlova, O.; Gerth, K.; Kaiser, O.; Hans, A.; Müller, R. Identification and analysis of the chivosazol biosynthetic gene cluster from the myxobacterial model strain Sorangium cellulosum So ce56. J. Biotechnol. 2006, 121, 174–191. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, A.; Sasse, F.; Müller, R. Identification and analysis of the core biosynthetic machinery of tubulysin, a potent cytotoxin with potential anticancer activity. Chem. Biol. 2004, 11, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Silakowski, B.; Nordsiek, G.; Kunze, B.; Blocker, H.; Müller, R. Novel features in a combined polyketide synthase/non-ribosomal peptide synthetase: The myxalamid biosynthetic gene cluster of the myxobacterium Stigmatella aurantiaca Sga15. Chem. Biol. 2001, 8, 59–69. [Google Scholar] [CrossRef]
- Tang, L.; Shah, S.; Chung, L.; Carney, J.; Katz, L.; Khosla, C.; Julien, B. Cloning and heterologous expression of the epothilone gene cluster. Science 2000, 287, 640–642. [Google Scholar] [CrossRef] [PubMed]
- Young, J.; Stevens, D.C.; Carmichael, R.; Tan, J.; Rachid, S.; Boddy, C.N.; Müller, R.; Taylor, R.E. Elucidation of gephyronic acid biosynthetic pathway revealed unexpected SAM-dependent methylations. J. Nat. Prod. 2013, 76, 2269–2276. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.P.; Li, Z.F.; Sun, X.; Li, S.G.; Li, Y.Z. Characteristics and activity analysis of epothilone operon promoters from Sorangium cellulosum strains in Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 6857–6866. [Google Scholar] [CrossRef]
- Hetrick, K.J.; van der Donk, W.A. Ribosomally synthesized and post-translationally modified peptide natural product discovery in the genomic era. Curr. Opin. Chem. Biol. 2017, 38, 36–44. [Google Scholar] [CrossRef]
- Hudson, G.A.; Mitchell, D.A. RiPP antibiotics: Biosynthesis and engineering potential. Curr. Opin. Microbiol. 2018, 45, 61–69. [Google Scholar] [CrossRef]
- Ortega, M.A.; van der Donk, W.A. New Insights into the Biosynthetic Logic of Ribosomally Synthesized and Post-translationally Modified Peptide Natural Products. Cell Chem. Biol. 2016, 23, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Viehrig, K.; Surup, F.; Volz, C.; Herrmann, J.; Abou Fayad, A.; Adam, S.; Kohnke, J.; Trauner, D.; Müller, R. Structure and Biosynthesis of Crocagins: Polycyclic Posttranslationally Modified Ribosomal Peptides from Chondromyces crocatus. Angew. Chem. Int. Ed. Engl. 2017, 56, 7407–7410. [Google Scholar] [CrossRef] [PubMed]
- Flatt, P.M.; Mahmud, T. Biosynthesis of aminocyclitol-aminoglycoside antibiotics and related compounds. Nat. Prod. Rep. 2007, 24, 358–392. [Google Scholar] [CrossRef] [PubMed]
- Kudo, F.; Eguchi, T. Aminoglycoside Antibiotics: New Insights into the Biosynthetic Machinery of Old Drugs. Chem. Rec. 2016, 16, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Q.; Deng, Z. Parallel pathways in the biosynthesis of aminoglycoside antibiotics. F1000Res 2017, 6. [Google Scholar] [CrossRef]
- Biarnes-Carrera, M.; Breitling, R.; Takano, E. Butyrolactone signalling circuits for synthetic biology. Curr. Opin. Chem. Biol. 2015, 28, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Camilli, A.; Bassler, B.L. Bacterial small-molecule signaling pathways. Science 2006, 311, 1113–1116. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Polkade, A.V.; Mantri, S.S.; Patwekar, U.J.; Jangid, K. Quorum Sensing: An Under-Explored Phenomenon in the Phylum Actinobacteria. Front. Microbiol. 2016, 7, 131. [Google Scholar] [CrossRef][Green Version]
- Brotherton, C.A.; Medema, M.H.; Greenberg, E.P. luxR Homolog-Linked Biosynthetic Gene Clusters in Proteobacteria. mSystems 2018, 3. [Google Scholar] [CrossRef]
- Lloyd, D.G.; Whitworth, D.E. The Myxobacterium Myxococcus xanthus Can Sense and Respond to the Quorum Signals Secreted by Potential Prey Organisms. Front. Microbiol. 2017, 8, 439. [Google Scholar] [CrossRef] [PubMed]
- Amiri Moghaddam, J.; Crusemann, M.; Alanjary, M.; Harms, H.; Davila-Cespedes, A.; Blom, J.; Poehlein, A.; Ziemert, N.; Konig, G.M.; Schaberle, T.F. Analysis of the Genome and Metabolome of Marine Myxobacteria Reveals High Potential for Biosynthesis of Novel Specialized Metabolites. Sci. Rep. 2018, 8, 16600. [Google Scholar] [CrossRef] [PubMed]
- Bouhired, S.; Rupp, O.; Blom, J.; Schaberle, T.F.; Schiefer, A.; Kehraus, S.; Pfarr, K.; Goesmann, A.; Hoerauf, A.; Konig, G. Complete Genome Sequence of the Corallopyronin A-Producing Myxobacterium Corallococcus coralloides B035. Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.; Müller, R. Simulacricoccus ruber gen. nov., sp. nov., a microaerotolerant, non-fruiting, myxospore-forming soil myxobacterium and emended description of the family Myxococcaceae. Int. J. Syst. Evol. Microbiol. 2018, 68, 3101–3110. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, P.G.; Morphew, R.M.; Whitworth, D.E. Genome Sequencing and Pan-Genome Analysis of 23 Corallococcus spp. Strains Reveal Unexpected Diversity, With Particular Plasticity of Predatory Gene Sets. Front. Microbiol. 2018, 9, 3187. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gregory, K.; Salvador, L.A.; Akbar, S.; Adaikpoh, B.I.; Stevens, D.C. Survey of Biosynthetic Gene Clusters from Sequenced Myxobacteria Reveals Unexplored Biosynthetic Potential. Microorganisms 2019, 7, 181. https://doi.org/10.3390/microorganisms7060181
Gregory K, Salvador LA, Akbar S, Adaikpoh BI, Stevens DC. Survey of Biosynthetic Gene Clusters from Sequenced Myxobacteria Reveals Unexplored Biosynthetic Potential. Microorganisms. 2019; 7(6):181. https://doi.org/10.3390/microorganisms7060181
Chicago/Turabian StyleGregory, Katherine, Laura A. Salvador, Shukria Akbar, Barbara I. Adaikpoh, and D. Cole Stevens. 2019. "Survey of Biosynthetic Gene Clusters from Sequenced Myxobacteria Reveals Unexplored Biosynthetic Potential" Microorganisms 7, no. 6: 181. https://doi.org/10.3390/microorganisms7060181
APA StyleGregory, K., Salvador, L. A., Akbar, S., Adaikpoh, B. I., & Stevens, D. C. (2019). Survey of Biosynthetic Gene Clusters from Sequenced Myxobacteria Reveals Unexplored Biosynthetic Potential. Microorganisms, 7(6), 181. https://doi.org/10.3390/microorganisms7060181