Transcriptomic Responses to Thermal Stress and Varied Phosphorus Conditions in Fugacium kawagutii
Abstract
:1. Introduction
2. Materials and Methods
2.1. F. kawagutii Cultures, Sampling, and RNA Sequencing
2.2. Data Preprocessing
2.3. Reads Counting
2.4. Identification of Expressed Core Genes
2.5. Differential Gene Expression Analysis
2.6. Gene Ontology and KEGG Functional Enrichment
3. Results
3.1. Overall Differential Gene Expression Profile
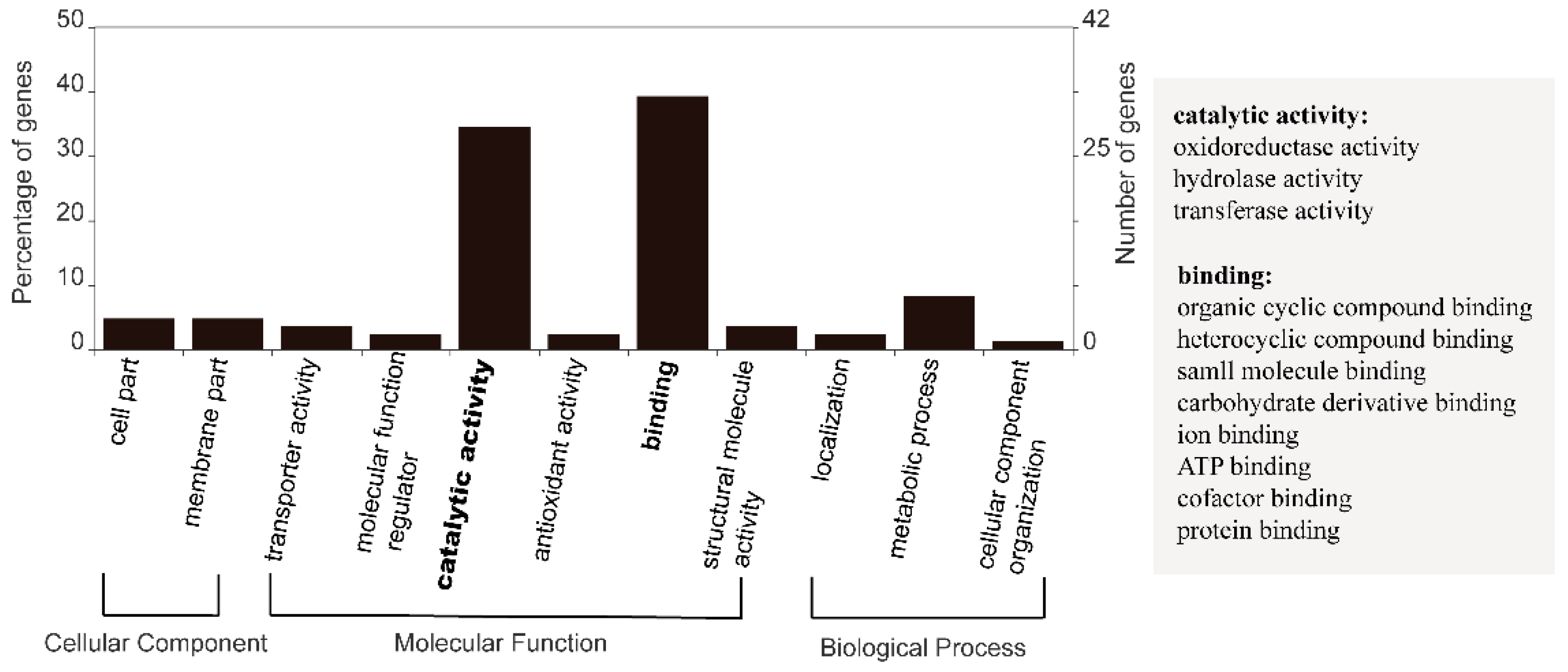
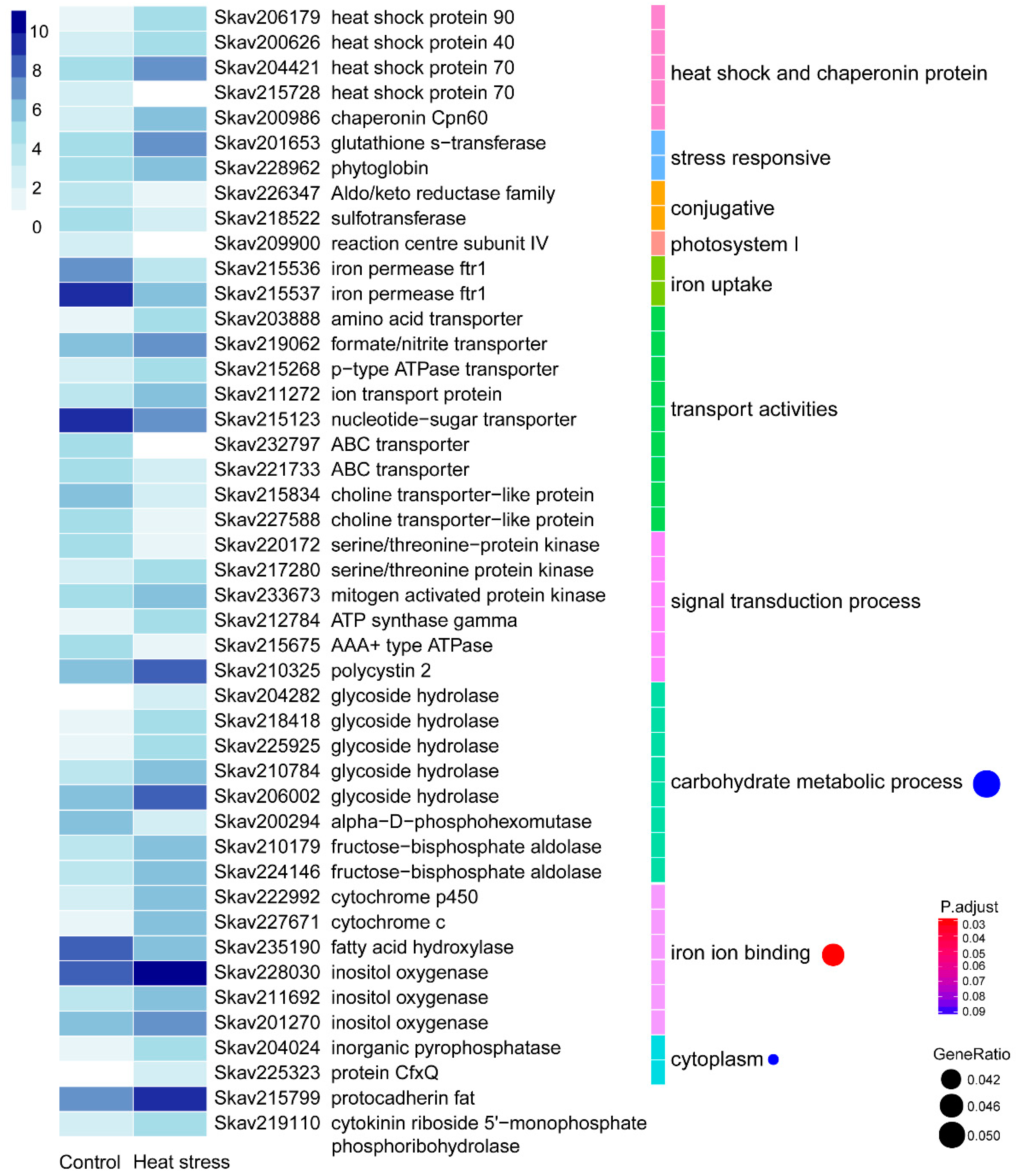
3.2. Functional Distribution of DEGs Responding to Heat Stress
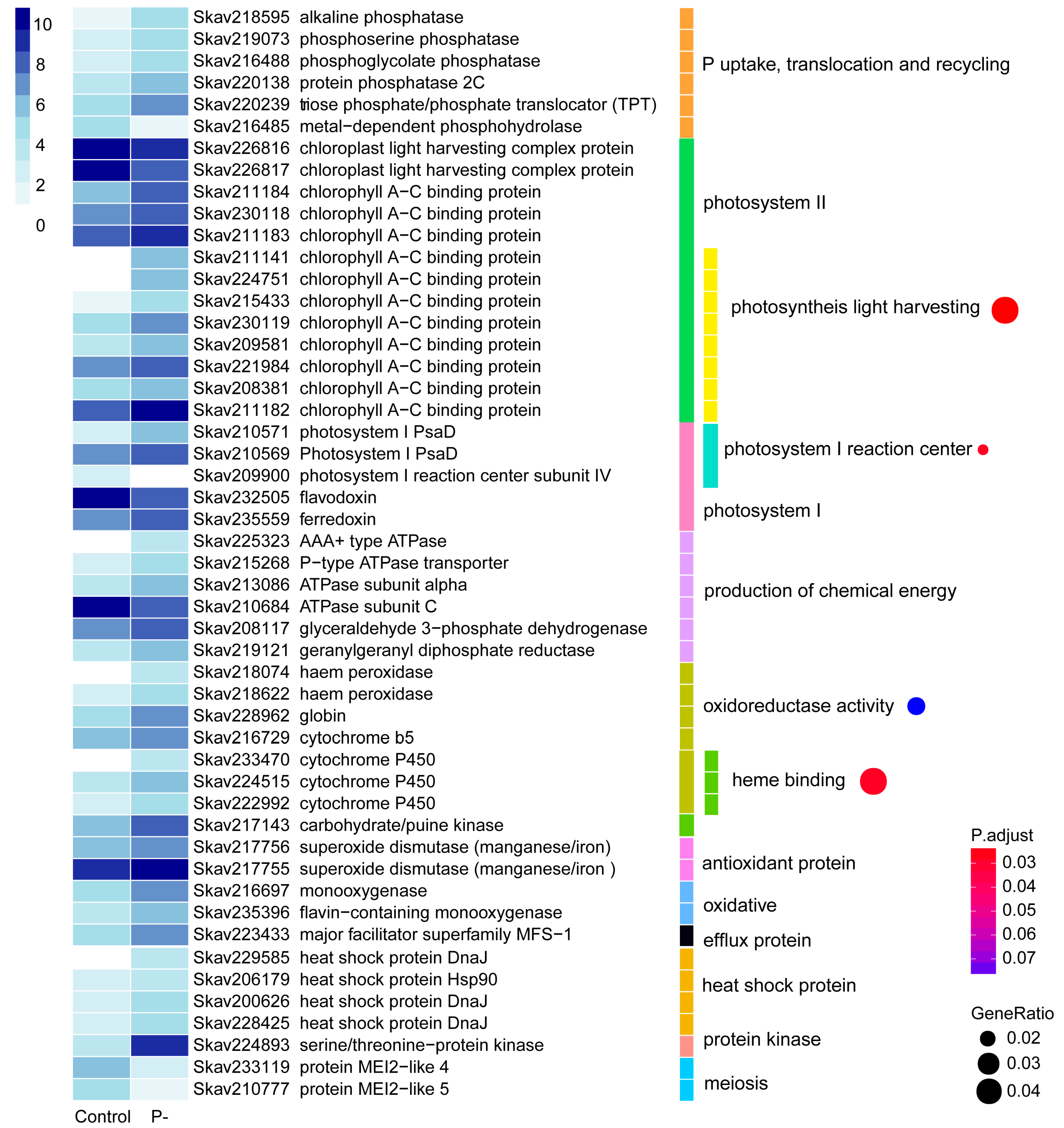
3.3. Functional Distribution of DEGs Responding to P Stress
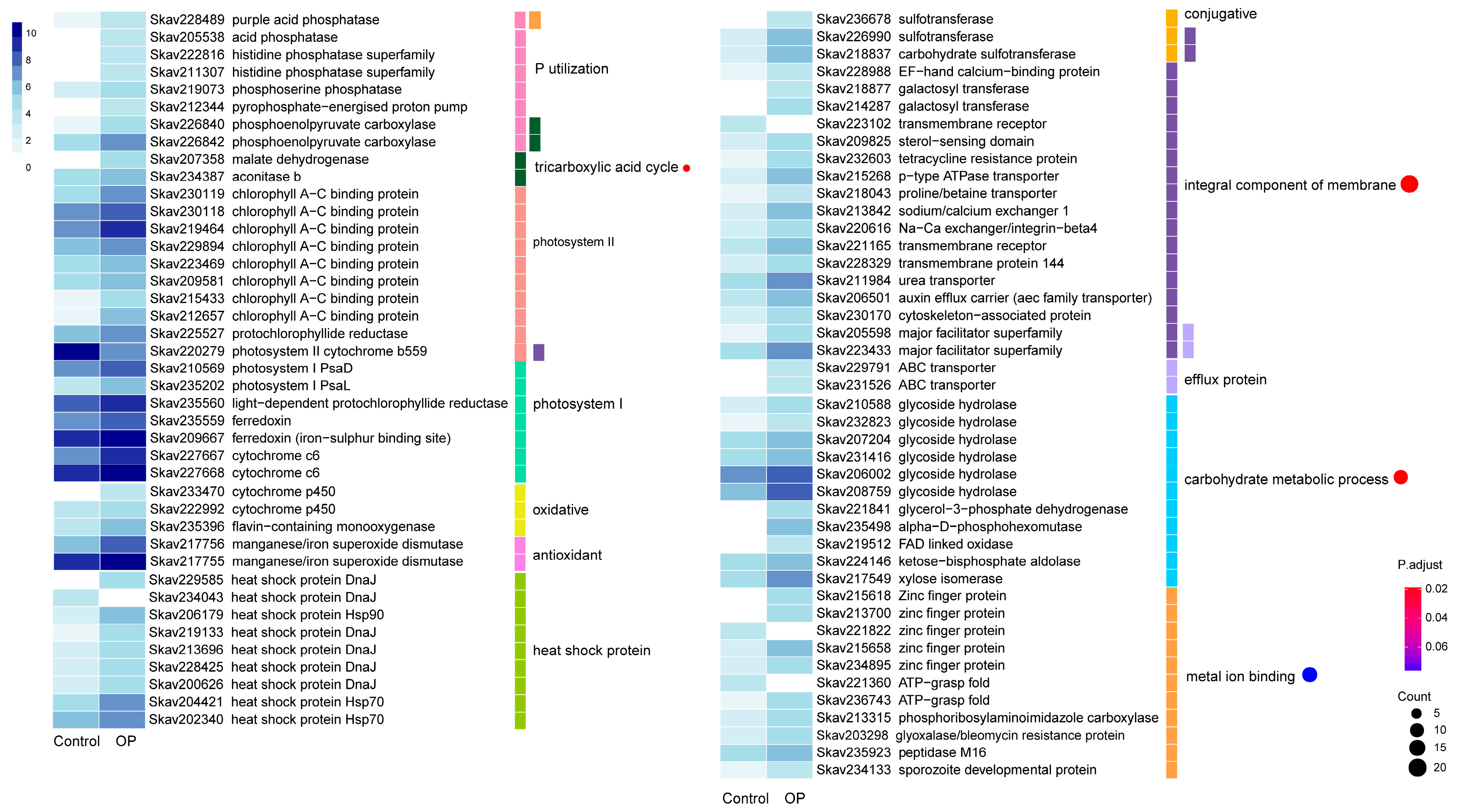
3.4. Functional Distribution of DEGs Responding to DOP Replacement
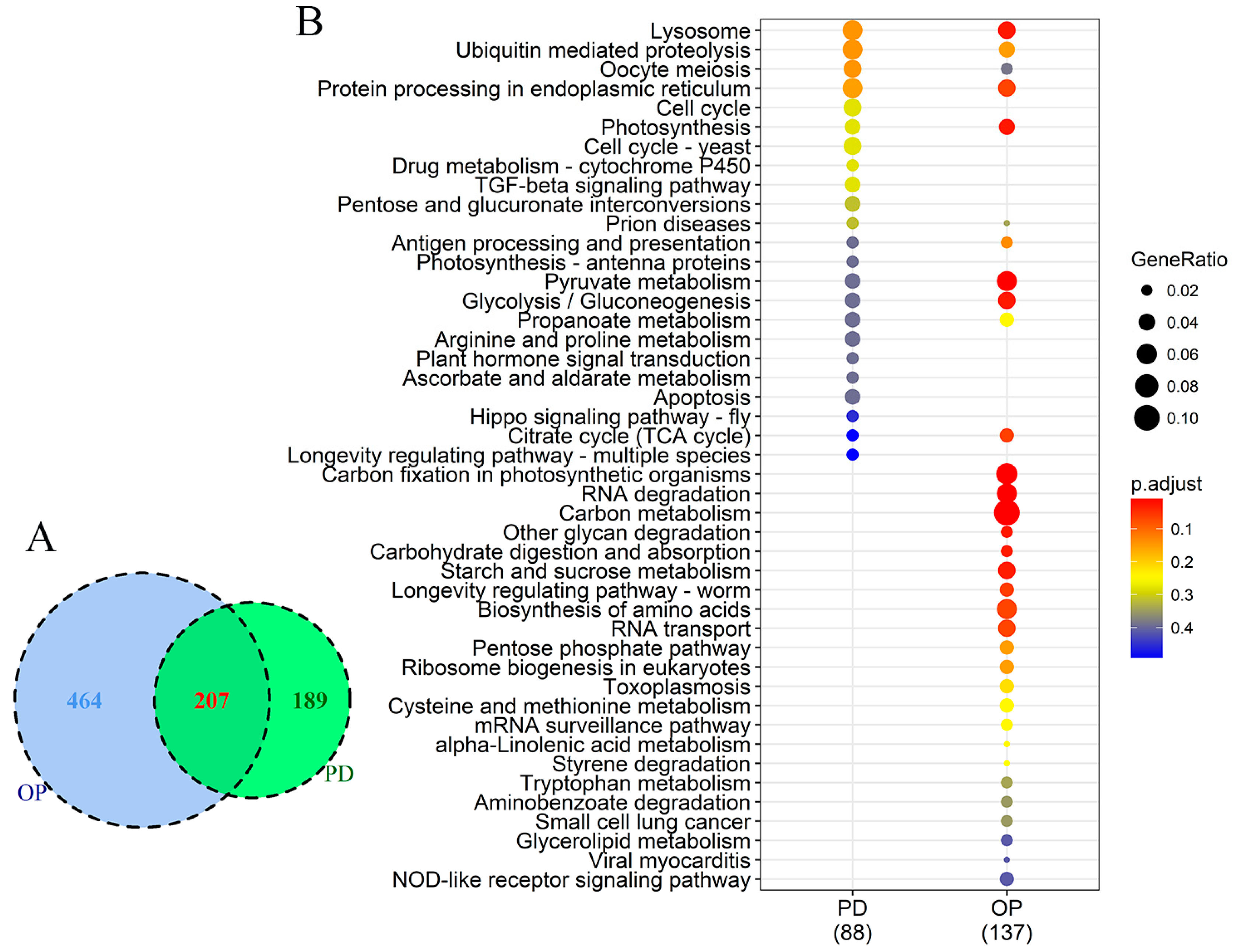
3.5. Comparison of DEGs between P Stress and DOP Replacement
4. Discussion
4.1. “Core” Genes and Responsive Gene Groups in F. kawagutii
4.2. Genes and Encoded Functions Responsive to Heat Stress in F. kawagutii
4.3. Genes and Encoded Functions Responsive to P Deprivation in F. kawagutii
4.4. Genes and Encoded Functions Are Responsive to DOP Replacement in F. kawagutii
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baker, A.C. Flexibility and specificity in coral-algal symbiosis: Diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 661–689. [Google Scholar] [CrossRef]
- Pochon, X.; Gates, R.D. A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawaii. Mol. Phylogenet. Evol. 2010, 56, 492–497. [Google Scholar] [CrossRef]
- Aranda, M.; Li, Y.; Liew, Y.J.; Baumgarten, S.; Simakov, O.; Wilson, M.C.; Piel, J.; Ashoor, H.; Bougouffa, S.; Bajic, V.B.; et al. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 2016, 6, 39734. [Google Scholar] [CrossRef] [PubMed]
- Fournier, A. The story of symbiosis with zooxanthellae, or how they enable their host to thrive in a nutrient poor environment. Biosci. Master. Rev. 2013, pp, 1–8. [Google Scholar]
- Little, A.F.; van Oppen, M.J.H.; Willis, B.L. Flexibility in Algal endosymbioses shapes growth in reef corals. Science 2004, 304, 1492–1494. [Google Scholar] [CrossRef]
- Stat, M.; Morris, E.; Gates, R.D. Functional diversity in coral-dinoflagellate symbiosis. Proc. Natl. Acad. Sci. USA 2008, 105, 9256–9261. [Google Scholar] [CrossRef]
- Stat, M.; Gates, R.D. Clade D Symbiodinium in Scleractinian Corals: A “Nugget” of Hope, a Selfish Opportunist, an Ominous Sign, or All of the Above? J. Mar. Biol. 2011, 1, 9. [Google Scholar]
- Baker, A.C. Reef corals bleach to survive change. Nature 2001, 411, 765. [Google Scholar] [CrossRef]
- Hoegh-Guldberg, O. Climate change, coral bleaching and the future of the world’s coral reefs. Mar. Freshw. Res. 1999, 50, 839–866. [Google Scholar] [CrossRef]
- Rosset, S.; Wiedenmann, J.; Reed, A.J.; D’Angelo, C. Phosphate deficiency promotes coral bleaching and is reflected by the ultrastructure of symbiotic dinoflagellates. Mar. Poll. Bull. 2017, 118, 180–187. [Google Scholar] [CrossRef]
- Takahashi, S.; Yoshioka-Nishimura, M.; Nanba, D.; Badger, M.R. Thermal acclimation of the symbiotic alga Symbiodinium spp. alleviates photobleaching under heat stress. Plant Physiol. 2013, 161, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Barshis, D.J.; Ladner, J.T.; Oliver, T.A.; Palumbi, S.R. Lineage-Specific Transcriptional Profiles of Symbiodinium spp. unaltered by heat stress in a coral host. Mol. Biol. Evol. 2014, 31, 1343–1352. [Google Scholar] [CrossRef]
- Karim, W.; Nakaema, S.; Hidaka, M. Temperature Effects on the growth rates and photosynthetic activities of Symbiodinium cells. J. Mar. Sci. Eng. 2015, 3, 368. [Google Scholar] [CrossRef]
- Gierz, S.L.; Forêt, S.; Leggat, W. Transcriptomic Analysis of Thermally stressed Symbiodinium reveals differential expression of stress and metabolism genes. Front. Plant Sci. 2017, 8, 271. [Google Scholar] [CrossRef] [PubMed]
- Karl, D.M. Microbially Mediated Transformations of phosphorus in the sea: New views of an old cycle. Annu. Rev. Mar. Sci. 2014, 6, 279–337. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Litaker Richard, W.; Sunda William, G.; Wood, M. Phosphorus physiological ecology and molecular mechanisms in marine phytoplankton. J. Phycol. 2016, 52, 10–36. [Google Scholar] [CrossRef] [PubMed]
- Dyhrman, S.N.T.; Ammerman, J.W.; van Mooy, B.A.S. Microbes and the marine phosphorus cycle. Oceanography 2007, 20, 110–116. [Google Scholar] [CrossRef]
- Trench, R.K.; Blank, R.J. Symbiodinium microadriaticum freudenthal, S. goreauii sp. nov., S. kawagutii sp. nov. and S. pilosum sp. nov.: Gymnodinioid dinoflagellate symbionts of marine invertebrates. J. Phycol. 1987, 23, 469–481. [Google Scholar] [CrossRef]
- Suggett, D.J.; Goyen, S.; Evenhuis, C.; Szabo, M.; Pettay, D.T.; Warner, M.E.; Ralph, P.J. Functional diversity of photobiological traits within the genus Symbiodinium appears to be governed by the interaction of cell size with cladal designation. New Phytol. 2015, 208, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.; Berkelmans, R. Potential costs of acclimatization to a warmer climate: Growth of a reef coral with heat tolerant vs. sensitive symbiont types. PLoS ONE 2010, 5, e10437. [Google Scholar] [CrossRef] [PubMed]
- Cunning, R.; Silverstein, R.N.; Baker, A.C. Investigating the causes and consequences of symbiont shuffling in a multi-partner reef coral symbiosis under environmental change. Proc. Biol. Sci. 2015, 282, 20141725. [Google Scholar] [CrossRef]
- Jones, A.M.; Berkelmans, R. Tradeoffs to Thermal acclimation: Energetics and reproduction of a reef coral with heat tolerant Symbiodinium Type-D. J. Mar. Biol. 2011, 2011, 12. [Google Scholar] [CrossRef]
- Lin, S.; Cheng, S.; Song, B.; Zhong, X.; Lin, X.; Li, W.; Li, L.; Zhang, Y.; Zhang, H.; Ji, Z.; et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 2015, 350, 691–694. [Google Scholar] [CrossRef]
- Zhang, H.; Zhuang, Y.; Gill, J.; Lin, S. Proof that dinoflagellate spliced leader (DinoSL) is a useful hook for fishing dinoflagellate transcripts from mixed microbial samples: Symbiodinium kawagutii as a case study. Protist 2013, 164, 510–527. [Google Scholar] [CrossRef]
- Goldstone, J.V.; Hamdoun, A.; Cole, B.J.; Howard-Ashby, M.; Nebert, D.W.; Scally, M.; Dean, M.; Epel, D.; Hahn, M.E.; Stegeman, J.J. The chemical defensome: Environmental sensing and response genes in the Strongylocentrotus purpuratus genome. Dev. Biol. 2006, 300, 366–384. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 1. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- García-Alcalde, F.; Okonechnikov, K.; Carbonell, J.; Cruz, L.M.; Götz, S.; Tarazona, S.; Dopazo, J.; Meyer, T.F.; Conesa, A. Qualimap: Evaluating next-generation sequencing alignment data. Bioinformatics 2012, 28, 2678–2679. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; García, F.; Ferrer, A.; Dopazo, J.; Conesa, A. NOIseq: A RNA-seq differential expression method robust for sequencing depth biases. EMBnet J. 2012, 17, 18–19. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Enrichplot: Visualization of Functional Enrichment Result, R package version 1.1.2; 2018; Available online: https://github.com/GuangchuangYu/enrichplot.
- Hill, R.; Hargrove, M.; Arredondo-Peter, R. Phytoglobin: A novel nomenclature for plant globins accepted by the globin community at the 2014 XVIII conference on oxygen-binding and sensing proteins. F1000Research 2016, 5, 212. [Google Scholar] [CrossRef] [PubMed]
- Rosic, N.N.; Leggat, W.; Kaniewska, P.; Dove, S.; Hoegh-Guldberg, O. New-old hemoglobin-like proteins of symbiotic dinoflagellates. Ecol. Evol. 2013, 3, 822–834. [Google Scholar] [CrossRef]
- Lee, S.K.; Eom, J.S.; Voll, L.M.; Prasch, C.M.; Park, Y.I.; Hahn, T.R.; Ha, S.H.; An, G.; Jeon, J.S. Analysis of a triose phosphate/phosphate translocator-deficient mutant reveals a limited capacity for starch synthesis in rice leaves. Mol. Plant 2014, 7, 1705–1708. [Google Scholar] [CrossRef]
- Li, J.; Guo, J.; Ou, X.; Zhang, M.; Li, Y.; Liu, Z. Mechanical coupling of the multiple structural elements of the large-conductance mechanosensitive channel during expansion. Proc. Natl. Acad. Sci. USA 2015, 112, 10726. [Google Scholar] [CrossRef] [PubMed]
- Roche, J.; Geider, R.J.; Graziano, L.M.; Murray, H.; Lewis, K. Induction of specific proteins in eukaryotic algae grown under iron-deficient, phosphorus-deficient, or nitrogen-deficient conditions. J. Phycol. 1993, 29, 767–777. [Google Scholar] [CrossRef]
- McKay, R.M.L.; Geider, R.J.; LaRoche, J. Physiological and biochemical response of the photosynthetic apparatus of two marine diatoms to Fe stress. Plant Physiol. 1997, 114, 615. [Google Scholar] [CrossRef] [PubMed]
- McKay, R.; Laroche, J.; Yakunin, A.; Durnford, D.; Geider, R. Accumulation of ferredoxin and flavodoxin in a marine diatom in response to Fe. J. Phycol. 2002, 35, 510–519. [Google Scholar] [CrossRef]
- Chappell, P.D.; Whitney, L.P.; Wallace, J.R.; Darer, A.I.; Jean-Charles, S.; Jenkins, B.D. Genetic indicators of iron limitation in wild populations of Thalassiosira oceanica from the northeast Pacific ocean. ISME J. 2015, 9, 592–602. [Google Scholar] [CrossRef]
- Kochian, L.V. Rooting for more phosphorus. Nature 2012, 488, 466. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-A.; Li, Q.; Ge, X.-Y.; Yang, C.-L.; Luo, X.-L.; Zhang, A.-H.; Xiao, J.-L.; Tian, Y.-C.; Xia, G.-X.; Chen, X.-Y.; et al. The mitochondrial malate dehydrogenase 1 gene GhmMDH1 is involved in plant and root growth under phosphorus deficiency conditions in cotton. Sci. Rep. 2015, 5, 10343. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Burki, F.; Wilcox, H.M.; Allam, B.; Allen, E.E.; Amaral-Zettler, L.A.; Armbrust, E.V.; Archibald, J.M.; Bharti, A.K.; Bell, C.J.; et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014, 12, e1001889. [Google Scholar] [CrossRef]
- Nikolayeva, O.; Robinson, M.D. edgeR for differential rna-seq and ChIP-seq analysis: An application to stem cell biology. In Stem Cell Transcriptional Networks: Methods and Protocols; Kidder, B.L., Ed.; Springer: New York, NY, USA, 2014; pp. 45–79. [Google Scholar]
- Gong, W.; Browne, J.; Hall, N.; Schruth, D.; Paerl, H.; Marchetti, A. Molecular insights into a dinoflagellate bloom. ISME J. 2017, 11, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Lin, X.; Li, L.; Li, M.; Palenik, B.; Lin, S. Transcriptomic and microRNAomic profiling reveals multi-faceted mechanisms to cope with phosphate stress in a dinoflagellate. ISME J. 2017, 11, 2209. [Google Scholar] [CrossRef]
- Harke, M.J.; Juhl, A.R.; Haley, S.T.; Alexander, H.; Dyhrman, S.T. Conserved Transcriptional Responses to Nutrient Stress in Bloom-Forming Algae. Front. Microbiol. 2017, 8, 1279. [Google Scholar] [CrossRef]
- Consortium, E.P. The ENCODE (ENCyclopedia of DNA Elements) Project. Science 2004, 306, 636–640. [Google Scholar] [CrossRef]
- Tarazona, S.; García-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Prieto, R.; Matta, J.L.; Robins, W.A.; Trench, R.K. Photosynthetic response to elevated temperature in the symbiotic dinoflagellate Symbiodinium microadriaticum in culture. Proc. Natl. Acad. Sci. USA 1992, 89, 10302. [Google Scholar] [CrossRef] [PubMed]
- Leggat, W.; Hoegh-Guldberg, O.; Dove, S.; Yellowlees, D. Analysis of an EST library from the dinoflagellate (Symbiodinium sp.) symbiont of reef-building corals1. J. Phycol. 2007, 43, 1010–1021. [Google Scholar] [CrossRef]
- Takahashi, S.; Whitney, S.; Itoh, S.; Maruyama, T.; Badger, M. Heat stress causes inhibition of the de novo synthesis of antenna proteins and photobleaching in cultured Symbiodinium. Proc. Natl. Acad. Sci. USA 2008, 105, 4203–4208. [Google Scholar] [CrossRef] [PubMed]
- Abrego, D.; Ulstrup, K.E.; Willis, B.L.; van Oppen, M.J.H. Species–specific interactions between algal endosymbionts and coral hosts define their bleaching response to heat and light stress. Proc. R. Soc. B Biol. Sci. 2008, 275, 2273–2282. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, J.N.; Yamasaki, H. Heat Stress Stimulates Nitric Oxide Production in Symbiodinium microadriaticum: A possible linkage between nitric oxide and the coral bleaching phenomenon. Plant Cell Physiol. 2008, 49, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Whitney, S.M.; Badger, M.R. Different thermal sensitivity of the repair of photodamaged photosynthetic machinery in cultured Symbiodinium species. Proc. Natl. Acad. Sci. USA 2009, 106, 3237–3242. [Google Scholar] [CrossRef] [PubMed]
- Leggat, W.; Seneca, F.; Wasmund, K.; Ukani, L.; Yellowlees, D.; Ainsworth, T.D. Differential responses of the coral host and their algal symbiont to thermal stress. PLoS ONE 2011, 6, e26687. [Google Scholar] [CrossRef]
- Bayer, T.; Aranda, M.; Sunagawa, S.; Yum, L.K.; DeSalvo, M.K.; Lindquist, E.; Coffroth, M.A.; Voolstra, C.R.; Medina, M. Symbiodinium Transcriptomes: Genome insights into the dinoflagellate symbionts of reef-building corals. PLoS ONE 2012, 7, e35269. [Google Scholar] [CrossRef]
- Krueger, T.; Becker, S.; Pontasch, S.; Dove, S.; Hoegh-Guldberg, O.; Leggat, W.; Fisher, P.L.; Davy, S.K. Antioxidant plasticity and thermal sensitivity in four types of Symbiodinium sp. J. Phycol. 2014, 50, 1035–1047. [Google Scholar] [CrossRef]
- Krueger, T.; Hawkins, T.D.; Becker, S.; Pontasch, S.; Dove, S.; Hoegh-Guldberg, O.; Leggat, W.; Fisher, P.L.; Davy, S.K. Differential coral bleaching-contrasting the activity and response of enzymatic antioxidants in symbiotic partners under thermal stress. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2015, 190, 15–25. [Google Scholar] [CrossRef]
- Gierz, S.L.; Gordon, B.R.; Leggat, W. Integral light-harvesting complex expression in Symbiodinium within the coral Acropora aspera under thermal stress. Sci. Rep. 2016, 6, 25081. [Google Scholar] [CrossRef] [PubMed]
- Levin, R.A.; Beltran, V.H.; Hill, R.; Kjelleberg, S.; McDougald, D.; Steinberg, P.D.; van Oppen, M.J. Sex, Scavengers, and Chaperones: Transcriptome secrets of divergent Symbiodinium thermal tolerances. Mol. Biol. Evol. 2016, 33, 2201–2215. [Google Scholar] [CrossRef]
- Goyen, S.; Pernice, M.; Szabó, M.; Warner, M.E.; Ralph, P.J.; Suggett, D.J. A molecular physiology basis for functional diversity of hydrogen peroxide production amongst Symbiodinium spp. (Dinophyceae). Mar. Biol. 2017, 164, 46. [Google Scholar] [CrossRef]
- Grégoire, V.; Schmacka, F.; Coffroth, M.A.; Karsten, U. Photophysiological and thermal tolerance of various genotypes of the coral endosymbiont Symbiodinium sp. (Dinophyceae). J. Appl. Phycol. 2017, 29, 1893–1905. [Google Scholar] [CrossRef]
- Chen, J.E.; Cui, G.; Wang, X.; Liew, Y.J.; Aranda, M. Recent expansion of heat-activated retrotransposons in the coral symbiont Symbiodinium microadriaticum. ISME J. 2018, 12, 639–643. [Google Scholar] [CrossRef]
- Fujise, L.; Nitschke, M.R.; Frommlet, J.C.; Serodio, J.; Woodcock, S.; Ralph, P.J.; Suggett, D.J. Cell cycle dynamics of cultured coral endosymbiotic microalgae (Symbiodinium) across different types (species) under alternate light and temperature conditions. J. Eukaryot. Microb. 2018, 65, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Rosic, N.; Pernice, M.; Rodriguez-Lanetty, M.; Hoegh-Guldberg, O. Validation of Housekeeping Genes for Gene Expression Studies in Symbiodinium Exposed to Thermal and Light Stress. Mar. Biotechnol. 2011, 13, 355–365. [Google Scholar] [CrossRef]
- Chong, G.; Kuo, F.-W.; Tsai, S.; Lin, C. Validation of reference genes for cryopreservation studies with the gorgonian coral endosymbiont Symbiodinium. Sci. Rep. 2017, 7, 39396. [Google Scholar] [CrossRef]
- Small, I.D.; Rackham, O.; Filipovska, A. Organelle transcriptomes: Products of a deconstructed genome. Curr. Opin. Microbiol. 2013, 16, 652–658. [Google Scholar] [CrossRef]
- Manna, S. An overview of pentatricopeptide repeat proteins and their applications. Biochimie 2015, 113, 93–99. [Google Scholar] [CrossRef]
- Kohl, A.; Binz, H.K.; Forrer, P.; Stumpp, M.T.; Plückthun, A.; Grütter, M.G. Designed to be stable: Crystal structure of a consensus ankyrin repeat protein. Proc. Natl. Acad. Sci. USA 2003, 100, 1700–1705. [Google Scholar] [CrossRef]
- Mosavi, L.K.; Cammett, T.J.; Desrosiers, D.C.; Peng, Z.y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004, 13, 1435–1448. [Google Scholar] [CrossRef]
- Levin, R.A.; Voolstra, C.R.; Weynberg, K.D.; van Oppen, M.J.H. Evidence for a role of viruses in the thermal sensitivity of coral photosymbionts. ISME J. 2016, 11, 808. [Google Scholar] [CrossRef]
- Ikura, M. Calcium binding and conformational response in EF-hand proteins. Trends Biochem. Sci. 1996, 21, 14–17. [Google Scholar] [CrossRef]
- Lewit-Bentley, A.; Réty, S. EF-hand calcium-binding proteins. Curr. Opin. Struct. Biol. 2000, 10, 637–643. [Google Scholar] [CrossRef]
- Skelton, N.J.; Kördel, J.; Akke, M.; Forsén, S.; Chazin, W. Signal transduction versus buffering activity in Ca(2+)-binding proteins. Nat. Struct. Biol. 1994, 1, 239–245. [Google Scholar] [CrossRef]
- Donato, R. Functional roles of S100 proteins, calcium-binding proteins of the EF-hand type. Biochim. Biophys. Acta 1999, 1450, 191–231. [Google Scholar] [CrossRef]
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef]
- Dasso, M. RCC1 in the cell cycle: The regulator of chromosome condensation takes on new roles. Trends Biochem. Sci. 1993, 18, 96–101. [Google Scholar] [CrossRef]
- Renault, L.; Kuhlmann, J.; Henkel, A.; Wittinghofer, A. Structural basis for guanine nucleotide exchange on ran by the regulator of chromosome condensation (RCC1). Cell 2001, 105, 245–255. [Google Scholar] [CrossRef]
- Shoguchi, E.; Shinzato, C.; Kawashima, T.; Gyoja, F.; Mungpakdee, S.; Koyanagi, R.; Takeuchi, T.; Hisata, K.; Tanaka, M.; Fujiwara, M.; et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 2013, 23, 1399–1408. [Google Scholar] [CrossRef]
- Shinzato, C.; Inoue, M.; Kusakabe, M. A snapshot of a coral “holobiont”: A transcriptome assembly of the scleractinian coral, porites, captures a wide variety of genes from both the host and symbiotic zooxanthellae. PLoS ONE 2014, 9, e85182. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Cao, P.; Jung, K.-H.; Sharma, M.K.; Ronald, P.C. Construction of a rice glycoside hydrolase phylogenomic database and identification of targets for biofuel research. Front. Plant Sci. 2013, 4, 330. [Google Scholar] [CrossRef]
- Pauchet, Y.; Kirsch, R.; Giraud, S.; Vogel, H.; Heckel, D.G. Identification and characterization of plant cell wall degrading enzymes from three glycoside hydrolase families in the cerambycid beetle Apriona japonica. Insect Biochem. Mol. Biol. 2014, 49, 1–13. [Google Scholar] [CrossRef]
- Saxena, I.M.; Brown, R.M.; Fevre, M.; Geremia, R.A.; Henrissat, B. Multidomain architecture of beta-glycosyl transferases: Implications for mechanism of action. J. Bacteriol. 1995, 177, 1419–1424. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Khurana, P. Characterization of a novel zinc finger transcription factor (TaZnF) from wheat conferring heat stress tolerance in Arabidopsis. Cell Stress Chaperones 2018, 23, 253–267. [Google Scholar] [CrossRef]
- Van den Brule, S.; Muller, A.; Fleming, A.J.; Smart, C.C. The ABC transporter SpTUR2 confers resistance to the antifungal diterpene sclareol. Plant J. 2002, 30, 649–662. [Google Scholar] [CrossRef]
- Kuromori, T.; Miyaji, T.; Yabuuchi, H.; Shimizu, H.; Sugimoto, E.; Kamiya, A.; Moriyama, Y.; Shinozaki, K. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. USA 2010, 107, 2361–2366. [Google Scholar] [CrossRef]
- Matsuda, S.; Funabiki, A.; Furukawa, K.; Komori, N.; Koike, M.; Tokuji, Y.; Takamure, I.; Kato, K. Genome-wide analysis and expression profiling of half-size ABC protein subgroup G in rice in response to abiotic stress and phytohormone treatments. Mol. Genet. Genomics 2012, 287, 819–835. [Google Scholar] [CrossRef] [PubMed]
- Bienert, M.D.; Baijot, A.; Boutry, M. ABCG transporters and their role in the biotic stress response. In Plant ABC Transporters; Geisler, M., Ed.; Springer: Cham, Switzerland, 2014; pp. 137–162. [Google Scholar] [CrossRef]
- Maher, T.J.; Ren, Y.; Li, Q.; Braunlin, E.; Garry, M.G.; Sorrentino, B.P.; Martin, C.M. ATP-binding cassette transporter Abcg2 lineage contributes to the cardiac vasculature after oxidative stress. Am. J. Physiol. Heart Circ. Physiol. 2014, 306, H1610–H1618. [Google Scholar] [CrossRef]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, R.I. The heat shock response: Systems biology of proteotoxic stress in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 91–99. [Google Scholar] [CrossRef]
- Qu, A.L.; Ding, Y.F.; Jiang, Q.; Zhu, C. Molecular mechanisms of the plant heat stress response. Cold Spring Harb. Symp. Quant. Biol. 2013, 432, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Bachinski, N.; Koziol, C.; Batel, R.; Labura, Z.; Schröder, H.C.; Müller, W.E.G. Immediate early response of the marine sponge Suberites domuncula to heat stress: Reduction of trehalose and glutathione concentrations and glutathione S-transferase activity. J. Exp. Mar. Biol. Ecol. 1997, 210, 129–141. [Google Scholar] [CrossRef]
- Murata, N.; Takahashi, S.; Nishiyama, Y.; Allakhverdiev, S.I. Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta 2007, 1767, 414–421. [Google Scholar] [CrossRef]
- Marutani, Y.; Yamauchi, Y.; Kimura, Y.; Mizutani, M.; Sugimoto, Y. Damage to photosystem II due to heat stress without light-driven electron flow: Involvement of enhanced introduction of reducing power into thylakoid membranes. Planta 2012, 236, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, I.B.; Lin, S.; Ho, J.; Ho, T.-Y. Effects of trace metal concentrations on the growth of the coral endosymbiont Symbiodinium kawagutii. Front. Microbiol. 2016, 7, 82. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, I.B.; Ho, T.-Y. Trace metal requirements and interactions in Symbiodinium kawagutii. Front. Microbiol. 2018, 9, 142. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, L.; Tang, Y.; Tang, R.; Jing, Y.; Zhang, C.; Zhang, B.; Li, X.; Cui, Y.; Zhang, C.; et al. Arabidopsis choline transporter-like 1 (CTL1) regulates secretory trafficking of auxin transporters to control seedling growth. PLoS Biol. 2017, 15, e2004310. [Google Scholar] [CrossRef] [PubMed]
- Willecke, M.; Hamaratoglu, F.; Kango-Singh, M.; Udan, R.; Chen, C.-l.; Tao, C.; Zhang, X.; Halder, G. The Fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr. Biol. 2006, 16, 2090–2100. [Google Scholar] [CrossRef]
- Bosch, J.A.; Sumabat, T.M.; Hafezi, Y.; Pellock, B.J.; Gandhi, K.D.; Hariharan, I.K. The Drosophila F-box protein Fbxl7 binds to the protocadherin Fat and regulates Dachs localization and Hippo signaling. eLife 2014, 3, e03383. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, M.; Yuyama, I.; Shimizu, H.; Nozawa, M.; Ikeo, K.; Gojobori, T. Different endosymbiotic interactions in two hydra species reflect the evolutionary history of endosymbiosis. Genome Biol. Evol. 2016, 8, 2155–2163. [Google Scholar] [CrossRef]
- Sahar, T.; Reddy, K.S.; Bharadwaj, M.; Pandey, A.K.; Singh, S.; Chitnis, C.E.; Gaur, D. Plasmodium falciparum reticulocyte binding-like homologue protein 2 (PfRH2) is a key adhesive molecule involved in erythrocyte invasion. PLoS ONE 2011, 6, e17102. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Wang, L.; Shi, X.; Lin, S. Rapidly diverging evolution of an atypical alkaline phosphatase (PhoAaty) in marine phytoplankton: Insights from dinoflagellate alkaline phosphatases. Front. Microbiol. 2015, 6, 868. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.-E.; Lomeda, R.-A.R.; Ryu, S.-H.; Sohn, H.-Y.; Shin, H.-I.; Beattie, J.H.; Kwun, I.-S. Zinc deficiency negatively affects alkaline phosphatase and the concentration of Ca, Mg and P in rats. Nutr. Res. Pract. 2007, 1, 113–119. [Google Scholar] [CrossRef]
- Twining, B.S.; Baines, S.B. The trace metal composition of marine phytoplankton. Ann. Rev. Mar. Sci. 2013, 5, 191–215. [Google Scholar] [CrossRef]
- Wang, D.-Z.; Zhang, Y.-J.; Zhang, S.-F.; Lin, L.; Hong, H.-S. Quantitative proteomic analysis of cell cycle of the dinoflagellate Prorocentrum donghaiense (Dinophyceae). PLoS ONE 2013, 8, e63659. [Google Scholar] [CrossRef] [PubMed]
- Dinneny, J.R.; Long, T.A.; Wang, J.Y.; Jung, J.W.; Mace, D.; Pointer, S.; Barron, C.; Brady, S.M.; Schiefelbein, J.; Benfey, P.N. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 2008, 320, 942. [Google Scholar] [CrossRef]
- Xing, D.; Wu, Y. Effect of phosphorus deficiency on photosynthetic inorganic carbon assimilation of three climber plant species. Bot. Stud. 2014, 55, 60. [Google Scholar] [CrossRef]
- Shimoda, C.; Uehira, M.; Kishida, M.; Fujioka, H.; Iino, Y.; Watanabe, Y.; Yamamoto, M. Cloning and analysis of transcription of the mei2 gene responsible for initiation of meiosis in the fission yeast Schizosaccharomyces pombe. J. Bacteriol 1987, 169, 93–96. [Google Scholar] [CrossRef]
- Watanabe, Y.; Shinozaki-Yabana, S.; Chikashige, Y.; Hiraoka, Y.; Yamamoto, M. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 1997, 386, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Lino, Y.; Furuhata, K.; Shimoda, C.; Yamamoto, M. The S.pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J. 1988, 7, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Triglia, T.; Tham, W.-H.; Hodder, A.; Cowman, A.F. Reticulocyte binding protein homologues are key adhesins during erythrocyte invasion by Plasmodium falciparum. Cell. Microbiol. 2009, 11, 1671–1687. [Google Scholar] [CrossRef]
- Grüber, A.; Gunalan, K.; Ramalingam, J.K.; Manimekalai, M.S.S.; Grüber, G.; Preiser, P.R. Structural characterization of the erythrocyte binding domain of the reticulocyte binding protein homologue family of Plasmodium falciparum. Infect. Immun. 2011, 79, 2880–2888. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Koonin, E.V. A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol ligase activity. Protein Sci. 1997, 6, 2639–2643. [Google Scholar] [CrossRef] [PubMed]
- Shilton, B.H. Active transporters as enzymes: An energetic framework applied to major facilitator superfamily and ABC importer systems. Biochem. J. 2015, 467, 193. [Google Scholar] [CrossRef]
- Qiu, X.B.; Shao, Y.M.; Miao, S.; Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef]
- Lau, S.; Shao, N.; Bock, R.; Jurgens, G.; De Smet, I. Auxin signaling in algal lineages: Fact or myth? Trends Plant Sci. 2009, 14, 182–188. [Google Scholar] [CrossRef]
- Michniewicz, M.; Brewer, P.B.; Friml, J.I. Polar auxin transport and asymmetric auxin distribution. Arabidopsis Book 2007, 5, e0108. [Google Scholar] [PubMed]
- Žádníková, P.; Petrášek, J.; Marhavý, P.; Raz, V.; Vandenbussche, F.; Ding, Z.; Schwarzerová, K.; Morita, M.T.; Tasaka, M.; Hejátko, J.; et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 2010, 137, 607. [Google Scholar] [CrossRef]
- Forestan, C.; Varotto, S. The role of pin auxin efflux carriers in polar auxin transport and accumulation and their effect on shaping maize development. Mol. Plant 2012, 5, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, J.; Zhang, W.; Yan, S.; Wang, R.; Zhao, J.; Li, Y.; Qi, Z.; Sun, Z.; Zhu, Z. The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. Plant J. 2012, 72, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Mekhalfi, M.; Puppo, C.; Avilan, L.; Lebrun, R.; Mansuelle, P.; Maberly, S.C.; Gontero, B. Glyceraldehyde-3-phosphate dehydrogenase is regulated by ferredoxin-NADP reductase in the diatom Asterionella formosa. New Phytol. 2014, 203, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Chong, I.K.; Ho, W.S. Glyceraldehyde-3-phosphate dehydrogenase from Chironomidae showed differential activity towards metals. Protein Pept. Lett. 2013, 20, 970–976. [Google Scholar] [CrossRef] [PubMed]
- Mráček, T.; Drahota, Z.; Houštěk, J. The function and the role of the mitochondrial glycerol-3-phosphate dehydrogenase in mammalian tissues. Biochim. Biophys. Acta 2013, 1827, 401–410. [Google Scholar] [CrossRef] [PubMed]
| Sample ID | MMETSP ID | SRA ID | Condition | Clean Data Size (Mbp) | Mapping Rate |
|---|---|---|---|---|---|
| SymkaSL1 | MMETSP0132 | SRR1300302 | heat stress | 660 | 59.21% |
| SymkaSL2 | MMETSP0133 | SRR1300303 | normal | 550 | 61.29% |
| SymkaSL3 | MMETSP0134 | SRR1300304 | P deprivation | 695 | 67.43% |
| SymkaSL4 | MMETSP0135 | SRR1300305 | Gro3P replacement | 1150 | 62.34% |
| Group | AEG | NOIseq | edgeR | NOIseq + edgeR |
|---|---|---|---|---|
| HS | 8081 | 1108 (13.71%) | 450 (5.57%) | 357 (4.42%, 249↑+108↓) |
| P- | 8364 | 1535 (18.35%) | 557 (6.66%) | 396 (4.73%, 332↑+64↓) |
| DOP | 8335 | 4707 (56.47%) | 1111 (13.33%) | 671 (8.05%, 580↑+ 91↓) |
| Total union | 10,857 | 5397 | 1601 | 1091 |
| Clade/Type | Conditions (control; stress) | Major Findings | Reference |
|---|---|---|---|
| Symbiodinium. microadriaticum | Heat stress (26 °C; 20–36 °C) | Photosynthesis was impaired at temperatures above 30 °C and ceases completely at 34–36 °C. | Iglesias-Prieto et al. 1992 [53] |
| Symbiodiniaceae Clade C3 | Warming (21.1 °C; 28.7 °C); Eutrophication (ammounium); increasing CO2 levels | Identified 1456 unique ESTs, among which 561 (44%) were functionally annotated. Most of them were related to posttranslational modification, protein turnover, and chaperones; energy production and conversion. | Leggat et al. 2007 [54] |
| Symbiodiniaceae OTcH-1 (Clade A) CS-7 (Clade A) | Heat stress (25–34 °C) | Inhibition of de novo synthesis of intrinsic light-harvesting antennae [chlorophyll a– chlorophyll c2–peridinin–protein complexes (acpPC); photoinhibition of photosystem II observed in CS-7 at 34 °C, but not in OTcH-1. | Takahashi et al. 2008 [55] |
| Symbiodiniaceae Type C1 Clade D | bleaching (28 °C; 30, 31, and 32 °C) heat stress (26 °C; 29, and 32 °C) | Lower metabolic costs and enhanced physiological tolerance of Acropora tenuis juveniles when hosting Symbiodiniaceae type C1 compared with type D. | Abrego et al. 2008 [56] |
| Symbiodiniaceae CCMP829 (Clade A) | Heat stress (27 °C; 34 °C) | Enhanced nitric oxide (NO) production at high temperatures. | Bouchard et al. 2008 [57] |
| Symbiodiniaceae OTcH-1 (Clade A) CS-73 (Clade A) | Heat stress (25°C; ~34 °C) | Thermal resistance is not associated with de novo synthesis of D1 protein. | Takahashi et al. 2009 [58] |
| Symbiodiniaceae Type C3 | Heat stress (27 °C; 34 °C) | Expression of stress responsive and carbon metabolism genes were up-regulated in coral host, but seldom and with smaller fold changes in the symbiont, during the experimental bleaching event. | Leggat et al. 2011 [59] |
| Symbiodiniaceae CassKB8 (Clade A) Mf1.05b (Clade B) | Heat (27 °C; 30-31 °C); cold (27 °C; 19 °C); light (120 µmoL photons/m2/s); dark (darkness for 6 days) | Generated 56,000 assembled sequences per species; found a complete set of core histones, a low number of transcription factors (cold shock domain was predominant), and a high number of antioxidative genes. | Bayer et al. 2012 [60] |
| Symbiodiniaceae CCMP827 (Clade A) CCMP831 (Clade A) CCMP830 (Clade B) CCMP421 (Clade E) | Heat stress (25 °C; 30 °C, 35 °C) | Enhanced thermal tolerance of PSII at elevated temperatures. | Takahashi et al. 2013 [11] |
| Symbiodiniaceae Type D2 Type C3K | Heat stress (26.8–34.5 °C; 27–37.6 °C for 3 days) | No DEGs after heat stress within each type; Hundreds of DEGs after heat stress between the two types. | Barshis et al. 2014 [12] |
| Symbiodiniaceae Ap1(Clade B1) CCMP2466 (Clade C1) CCMP421 (Clade E) Mv (Clade F1) | Heat stress (25 °C; 29 °C, 33 °C) | In Symbiodiniaceae clades B1, C1, and E, declining photochemical efficiency (Fv /Fm) and death at 33 °C were generally associated with elevated superoxide dismutase (SOD) activity and a more oxidized glutathione pool. Clade F1 exhibited no decline in Fv /Fm or growth, but showed proportionally larger increases in ascorbate peroxidase (APX) activity and glutathione content (GSx), while maintaining GSx in a reduced state. | Krueger et al. 2014 [61] |
| Symbiodiniaceae Y106 (Clade A) K100 (Clade B) Y103 (Clade C) K111 (Clade D) K102 (Clade F) | Heat stress (25 °C; 33 °C) | Decreased growth rate and photosynthesis at elevated temperature in clades A and B, but not in clades D and F. | Karim et al. 2015 [13] |
| Symbiodiniaceae Type C3 Type C15 | Heat stress (28 °C; 33 °C) | No significant changes in enzymatic antioxidant defense detected in the symbiont. Preceded significant declines in PSII photochemical efficiencies. | Krueger T et al. 2015 [62] |
| Symbiodiniaceae Clade C3 | Heat stress (increasing daily from 25 °C to 34 °C) | At day 8, photochemical efficiency was decreased. On day 16, symbiont density was significantly lower. Three acpPC genes were up-regulated when temperatures above 31.5 °C. | Gierz et al. 2016 [63] |
| Symbiodiniaceae thermos-sensitive SM (Type C1) thermos-tolerant MI (Type C1) | Heat stress (27 °C; 32 °C) | After 9 days at 32 °C, the two populations showed no physiological stress, but the enhanced meiosis genes. After 13 days at 32 °C, SM population showed decreasing photochemical efficiency and increasing ROS, MI exhibited no physiological stress and enhanced expression of genes of ROS scavenging and molecular chaperone. | Levin et al. 2016 [64] |
| Symbiodiniaceae Clades A, B, D, F | Heat stress (25 °C; 32 °C) | Sixteen Symbiodiniaceae isolates were clustered into three novel functional groups based on their physiological response to heat stress: thermally tolerant, thermally susceptible and thermally. | Goyen et al. (2017) [65] |
| Symbiodiniaceae Clade F | Heat stress (24.5 °C; 31 °C for 28 days) | 37.01% DEGs of the transcriptome (∼23,654 unique genes found at FDR < 0.05), with 92.49% DEGs at ≤2-fold change. The DEGs encoded stress response components, glyoxylate cycle enzymes, and altered metabolic processes. | Gierz et al. 2017 [14] |
| Symbiodiniaceae Type A3 Type B1 Type B2 Type C2 Type D1a Type F | Heat stress (26 °C; 20–33 °C) | Six Symbiodiniaceae genotypes showed significant differences in the response patterns under heat stress. While some types photosynthesized, respired, and grew at 33 °C, others showed a partial or complete inhibition. | Gregoire et al. 2017 [66] |
| Symbiodiniaceae CCMP2467 (Clade A) | Heat stress (26 °C; 36 °C) Cold stress (26 °C; 16 °C) Dark stress (no daybreak) | Verified the existence of heat stress-activated Ty1-copia-type LTR retrotransposons and its recent expansion events in the S. microadriaticum. | Chen et al. 2018 [67] |
| Breviolum. minutum (Clade B) Cladocopium goreaui (Clade C) Durusdinium trenchii (Clade D) | Heat stress (26 °C; 32 °C) | Heat stress inhibited cell cycle progression and arrested all strains in G1 phase. | Fujise et al. 2018 [68] |
| Fugacium kawagutii CCMP2468 (Clade F) | Heat stress (25 °C; 30 °C) P deprivation (25 °C; P-) DOP utilization (25 °C; DOP) | Documented 357 (4.42%) DEGs under heat stress putatively involved in molecular interaction, cell wall modulation and transport, in addition to heat shock proteins reported previously. Documented 396 (4.73%) DEGs under P deprivation, and 671 (8.05%) DEGs under DOP utilization, which have not been studied previously, and both groups of DEGs putatively function in photosystem and defensome. | This study |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.; Yu, L.; Zhang, H. Transcriptomic Responses to Thermal Stress and Varied Phosphorus Conditions in Fugacium kawagutii. Microorganisms 2019, 7, 96. https://doi.org/10.3390/microorganisms7040096
Lin S, Yu L, Zhang H. Transcriptomic Responses to Thermal Stress and Varied Phosphorus Conditions in Fugacium kawagutii. Microorganisms. 2019; 7(4):96. https://doi.org/10.3390/microorganisms7040096
Chicago/Turabian StyleLin, Senjie, Liying Yu, and Huan Zhang. 2019. "Transcriptomic Responses to Thermal Stress and Varied Phosphorus Conditions in Fugacium kawagutii" Microorganisms 7, no. 4: 96. https://doi.org/10.3390/microorganisms7040096
APA StyleLin, S., Yu, L., & Zhang, H. (2019). Transcriptomic Responses to Thermal Stress and Varied Phosphorus Conditions in Fugacium kawagutii. Microorganisms, 7(4), 96. https://doi.org/10.3390/microorganisms7040096






