Efficiency of Single Phage Suspensions and Phage Cocktail in the Inactivation of Escherichia coli and Salmonella Typhimurium: An In Vitro Preliminary Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Phage Preparation
2.3. Phage Host Range: Spot Test and Efficiency of Plating (EOP)
2.4. Killing Curves
2.5. Determination of the Frequency of Emergence of Phage Resistant Mutants
2.6. Fitness of Phage Resistant Mutants
2.7. Statistical Analysis
3. Results
3.1. Phage Host Range: Spot Test and Efficiency of Plating (EOP)
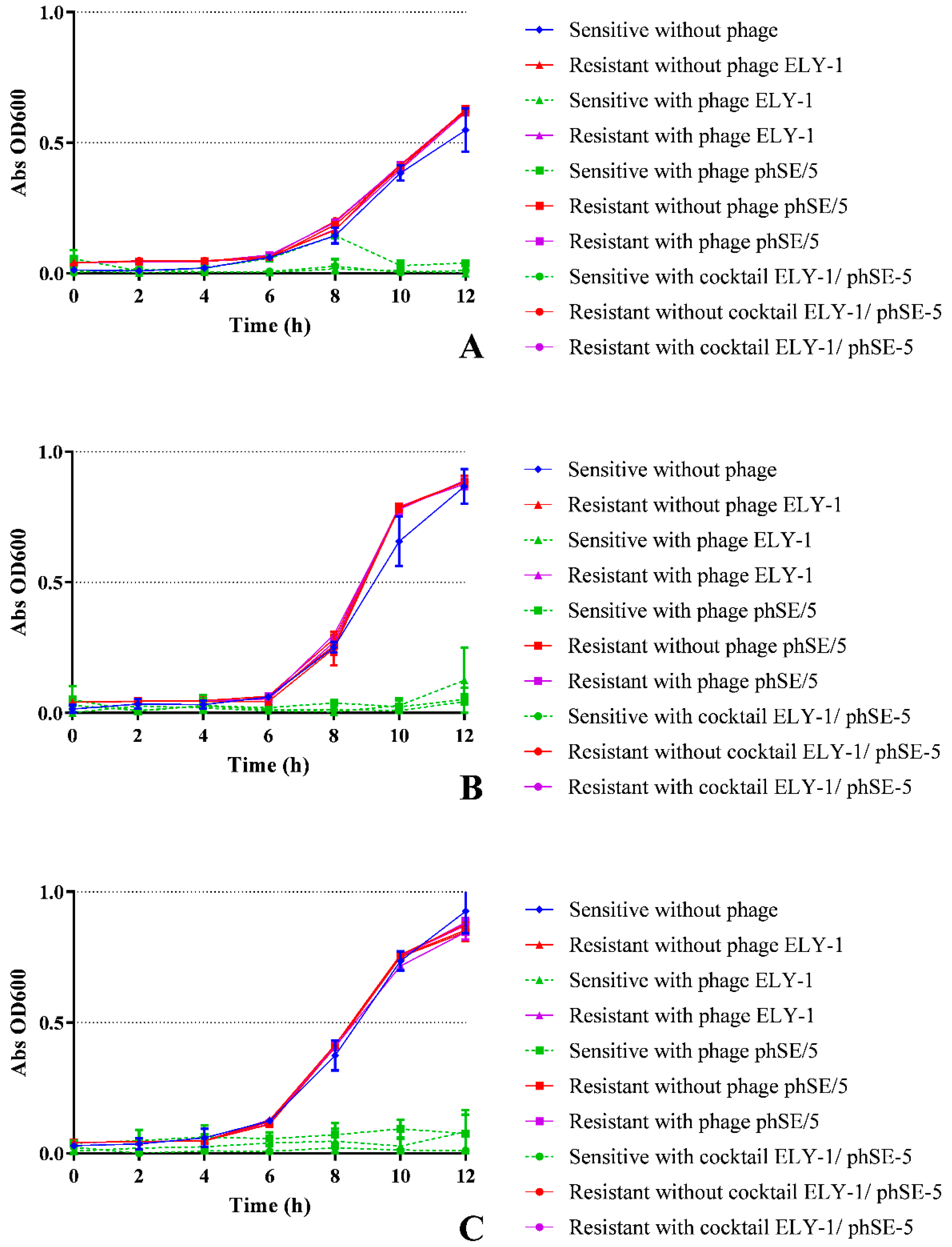
3.2. Effect of phage ELY-1, phSE-5, and cocktail ELY-1/phSE-5 on the inactivation of E. coli
3.3. Effect of Phage ELY-1, phSE-5 and Cocktail ELY-1/phSE-5 on the Inactivation of S. Typhimurium
3.4. Effect of Phages ELY-1, phSE-5 and Cocktail ELY-1/phSE-5 on the Inactivation of the Mixture of E. coli/S. Typhimurium
3.5. Determination of the Emergence Rate of Bacterial Mutants
3.6. Fitness of Phage Resistant Mutants
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Havelaar, A.H.; Kirk, M.D.; Torgerson, P.R.; Gibb, H.J.; Hald, T.; Lake, R.J.; Praet, N.; Bellinger, D.C.; de Silva, N.R.; Gargouri, N.; et al. World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 2015, 12, e1001923. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.Q.; Louie, J.W.; Feng, D.; Zhong, W.; Brandl, M.T. Curli fimbriae are conditionally required in Escherichia coli O157: H7 for initial attachment and biofilm formation. Food Microbiol. 2016, 57, 81–89. [Google Scholar] [CrossRef]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the food industry: Health aspects and control methods. Front. Microbiol. 2018, 9, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage applications for food production and processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef]
- Bolocan, A.S.; Callanan, J.; Forde, A.; Ross, P.; Hill, C. Phage therapy targeting Escherichia coli—A story with no end? FEMS Microbiol. Lett. 2016, 363, 1–5. [Google Scholar] [CrossRef]
- Brüssow, H. Targeting the gut to protect the bladder: Oral phage therapy approaches against urinary Escherichia coli infections? Environ. Microbiol. 2016, 18, 2084–2088. [Google Scholar] [CrossRef]
- Peng, Q.; Yuan, Y. Characterization of a newly isolated phage infecting pathogenic Escherichia coli and analysis of its mosaic structural genes. Sci. Rep. 2018, 8, 8086. [Google Scholar] [CrossRef]
- Mora, A.; Herrera, A.; López, C.; Dahbi, G.; Mamani, R.; Pita, J.M.; Alonso, M.P.; Llovo, J.; Bernárdez, M.I.; Blanco, J.E.; Blanco, M.; Blanco, J. Characteristics of the Shiga-toxin-producing enteroaggregative Escherichia coli O104:H4 German outbreak strain and of STEC strains isolated in Spain. Int. Microbiol. 2011, 14, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Massier, S.; Darfeuille-Michaud, A.; Billard, E.; Barnich, N. Understanding host-adherent-invasive Escherichia coli interaction in Crohn’s disease: Opening up new therapeutic strategies. Biomed Res. Int. 2014, 2014. [Google Scholar] [CrossRef]
- Conte, M.P.; Longhi, C.; Marazzato, M.; Conte, A.L.; Aleandri, M.; Lepanto, M.S.; Zagaglia, C.; Nicoletti, M.; Aloi, M.; Totino, V.; et al. Adherent-invasive Escherichia coli (AIEC) in pediatric Crohn’s disease patients: Phenotypic and genetic pathogenic features. BMC Res. Notes 2014, 7, 1–12. [Google Scholar] [CrossRef]
- Clements, A.; Young, J.C.; Constantinou, N.; Frankel, G. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes 2012, 3, 71–87. [Google Scholar] [CrossRef]
- Bell, C.; Kyriakides, A. Salmonella: A Practical Approach to the Organism and Its Control in Foods; Practical Food Microbiology Series; Blackwell Science Ltd.: Oxford, UK, 2002. [Google Scholar]
- Wang, R.; Bono, J.L.; Kalchayanand, N.; Shackelford, S.; Harhay, D.M. Biofilm formation by Shiga toxin–producing Escherichia coli O157:H7 and non-O157 strains and their tolerance to sanitizers commonly used in the food processing environment. J. Food Prot. 2012, 75, 1418–1428. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Moreirinha, C.; Lewicka, M.; Almeida, P.; Clemente, C.; Cunha, Â.; Delgadillo, I.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Bacteriophages with potential to inactivate Salmonella Typhimurium: Use of single phage suspensions and phage cocktails. Virus Res. 2016, 220. [Google Scholar] [CrossRef] [PubMed]
- Allocati, N.; Masulli, M.; Alexeyev, M.F.; Di Ilio, C. Escherichia coli in Europe: An overview. Int. J. Environ. Res. Public Health 2013, 10, 6235–6254. [Google Scholar] [CrossRef] [PubMed]
- Iredell, J.; Brown, J.; Tagg, K. Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. BMJ 2016, 356, 1–19. [Google Scholar] [CrossRef]
- Fair, R.J.; Tor, Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Medicin. Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef]
- WHO. Antimicrobial resistance. Bull. World Health Organ. 2014, 61, 383–394. [Google Scholar] [CrossRef]
- Rong, R.; Lin, H.; Wang, J.; Khan, M.N.; Li, M. Reductions of Vibrio parahaemolyticus in oysters after bacteriophage application during depuration. Aquaculture 2014, 418–419, 171–176. [Google Scholar] [CrossRef]
- Pereira, C.; Moreirinha, C.; Lewicka, M.; Almeida, P.; Clemente, C.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Characterization and in vitro evaluation of new bacteriophages for the biocontrol of Escherichia coli. Virus Res. 2017, 227. [Google Scholar] [CrossRef]
- Golkar, Z. Experimental phage therapy on multiple drug resistant Pseudomonas aeruginosa infection in mice. J. Antivir. Antiretrovir. 2013, 05. [Google Scholar] [CrossRef]
- Jun, J.W.; Shin, T.H.; Kim, J.H.; Shin, S.P.; Han, J.E.; Heo, G.J.; De Zoysa, M.; Shin, G.W.; Chai, J.Y.; Park, S.C. Bacteriophage therapy of a Vibrio parahaemolyticus infection caused by a multiple-antibiotic–resistant O3:K6 pandemic clinical strain. J. Infect. Dis. 2014, 210, 72–78. [Google Scholar] [CrossRef]
- Fish, R.; Kutter, E.; Wheat, G.; Blasdel, B.; Kutateladze, M.; Kuhl, S. Bacteriophage treatment of intransigent diabetic toe ulcers: a case series. J. Wound Care 2016, 25, S27–S33. [Google Scholar] [CrossRef]
- Adebayo, O.S.; Gabriel-Ajobiewe, R.A.O.G.; Taiwo, M.O.; Kayode, S. Phage therapy: A potential alternative in the treatment of multi-drug resistant bacterial infections. J. Microbiol. Exp. 2017, 5. [Google Scholar] [CrossRef]
- LaVergne, S.; Hamilton, T.; Biswas, B.; Kumaraswamy, M.; Schooley, R.T.; Wooten, D. Phage therapy for a multidrug-resistant Acinetobacter baumannii craniectomy site infection. Open Forum Infect. Dis. 2018, 5, ofy064. [Google Scholar] [CrossRef]
- Park, S.; Nakai, T. Bacteriophage control of Pseudomonas plecoglossicida infection in ayu. Dis. Aquat. Organ. 2003, 53, 33–39. [Google Scholar] [CrossRef]
- Pereira, C.; Silva, Y.J.; Santos, A.L.; Cunha, Â.; Gomes, N.C.M.; Almeida, A. Bacteriophages with potential for inactivation of fish pathogenic bacteria: Survival, host specificity and effect on bacterial community structure. Mar. Drugs 2011, 9, 2236–2255. [Google Scholar] [CrossRef]
- Nilsson, A.S. Phage therapy-constraints and possibilities. Ups. J. Med. Sci. 2014, 119, 192–198. [Google Scholar] [CrossRef]
- Kutter, E.M.; Kuhl, S.J.; Abedon, S.T. Re-establishing a place for phage therapy in western medicine. Future Microbiol. 2015, 10, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Cooper, C.J.; Mirzaei, M.K.; Nilsson, A.S. Adapting drug approval pathways for bacteriophage-based therapeutics. Front. Microbiol. 2016, 7, 1–15. [Google Scholar] [CrossRef]
- Flynn, G.O.; Ross, R.P.; Fitzgerald, G.F.; Coffey, A. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 2004, 70, 3417–3424. [Google Scholar] [CrossRef]
- Sharma, M.; Patel, J.R.; Conway, W.S.; Ferguson, S.; Sulakvelidze, A. Effectiveness of bacteriophages in reducing Escherichia coli O157:H7 on fresh-cut cantaloupes and lettucet. J. Food Prot. 2009, 72, 1481–1485. [Google Scholar] [CrossRef]
- Rozema, E.A.; Stephens, T.P.; Bach, S.J.; Okine, E.K.; Johnson, R.P.; Stanford, K.; McAllister, T.A. Oral and rectal administration of bacteriophages for control of Escherichia coli O157:H7 in feedlot cattle. J. Food Prot. 2009, 72, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Rivas, L.; Coffey, B.; McAuliffe, O.; McDonnell, M.J.; Burgess, C.M.; Coffey, A.; Ross, R.P.; Duffy, G. In vivo and ex vivo evaluations of bacteriophages e11/2 and e4/1c for use in the control of Escherichia coli O157:H7. Appl. Environ. Microbiol. 2010, 76, 7210–7216. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Sharma, M.; Millner, P.; Calaway, T.; Singh, M. Inactivation of Escherichia coli O157:H7 attached to spinach harvester blade using bacteriophage. Foodborne Pathog. Dis. 2011, 8, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Tomat, D.; Migliore, L.; Aquili, V.; Quiberoni, A.; Balagué, C. Phage biocontrol of enteropathogenic and Shiga toxin-producing Escherichia coli during milk fermentation. Front. Cell. Infect. Microbiol. 2013, 3, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Moreirinha, C.; Teles, L.; Rocha, R.; Calado, R.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Application of phage therapy during bivalve depuration improves Escherichia coli decontamination. Food Microbiol. 2017, 61, 102–112. [Google Scholar] [CrossRef]
- Berchieri, A.; Lovell, M.A.; Barrow, P.A. The activity in the chicken alimentary tract of bacteriophages lytic for Salmonella Typhimurium. Res. Microbiol. 1991, 142, 541–549. [Google Scholar] [CrossRef]
- Bigwood, T.; Hudson, J.A.; Billington, C. Influence of host and bacteriophage concentrations on the inactivation of food-borne pathogenic bacteria by two phages. FEMS Microbiol. Lett. 2009, 291, 59–64. [Google Scholar] [CrossRef]
- Wall, S.K.; Zhang, J.; Rostagno, M.H.; Ebner, P.D. Phage therapy to reduce preprocessing Salmonella infections in market-weight swine. Appl. Environ. Microbiol. 2010, 76, 48–53. [Google Scholar] [CrossRef]
- Callaway, T.R.; Edrington, T.S.; Brabban, A.; Kutter, B.; Karriker, L.; Stahl, C.; Wagstrom, E.; Anderson, R.; Poole, T.L.; Genovese, K.; et al. Evaluation of phage treatment as a strategy to reduce Salmonella populations in growing swine. Foodborne Pathog. Dis. 2011, 8, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Shi, J.; Ma, W.; Li, Z.; Wang, J.; Li, J.; Wang, X. Isolation, characterization, and application of a novel specific Salmonella bacteriophage in different food matrices. Food Res. Int. 2018, 111, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Moreirinha, C.; Rocha, R.J.M.; Calado, R.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Application of bacteriophages during depuration reduces the load of Salmonella Typhimurium in cockles. Food Res. Int. 2016, 90, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.J.; Hyman, P. Phage choice, isolation, and preparation for phage therapy. Curr. Pharm. Biotechnol. 2010, 11, 2–14. [Google Scholar] [CrossRef]
- Deveau, H.; Garneau, J.E.; Moineau, S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010, 64, 475–493. [Google Scholar] [CrossRef]
- Bull, J.J.; Vegge, C.S.; Schmerer, M.; Chaudhry, W.N.; Levin, B.R. Phenotypic resistance and the dynamics of bacterial escape from phage control. PLoS ONE 2014, 9, e94690. [Google Scholar] [CrossRef]
- Vieira, A.; Silva, Y.J.; Cunha, Â.; Gomes, N.C.M.; Ackermann, H.W.; Almeida, A. Phage therapy to control multidrug-resistant Pseudomonas aeruginosa skin infections: In vitro and ex vivo experiments. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 3241–3249. [Google Scholar] [CrossRef]
- Chan, B.K.; Abedon, S.T.; Loc-carrillo, C. Phage cocktail and the future of phage therapy. Future Med. 2013, 158, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.; Pereira, C.; Moreirinha, C.; Salvio, R.; Lopes, A.; Wang, D.; Almeida, A. New insights on phage efficacy to control Aeromonas salmonicida in aquaculture systems: An in vitro preliminary study. Aquaculture 2018, 495, 970–982. [Google Scholar] [CrossRef]
- Cairns, B.J.; Timms, A.R.; Jansen, V.A.A.; Connerton, I.F.; Payne, R.J.H. Quantitative models of in vitro bacteriophage–host dynamics and their application to phage therapy. PLoS Pathog. 2009, 5, e1000253. [Google Scholar] [CrossRef] [PubMed]
- Kunisaki, H.; Tanji, Y. Intercrossing of phage genomes in a phage cocktail and stable coexistence with Escherichia coli O157: H7 in anaerobic continuous culture. Appl. Genet. Mol. Biotechnol. 2010, 85, 1533–1540. [Google Scholar] [CrossRef]
- Merabishvili, M.; Pirnay, J.-P.; Verbeken, G.; Chanishvili, N.; Tediashvili, M.; Lashkhi, N.; Glonti, T.; Krylov, V.; Mast, J.; Van Parys, L.; et al. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS ONE 2009, 4, e4944. [Google Scholar] [CrossRef] [PubMed]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J.G. Bacteriophage therapy. Antimicrob. Agents Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef]
- Kutter, E.; De Vos, D.; Gvasalia, G.; Alavidze, Z.; Gogokhia, L.; Kuhl, S.; Abedon, S.T. Phage therapy in clinical practice: treatment of human infections. Curr. Pharm. Biotechnol. 2010, 11, 69–86. [Google Scholar] [CrossRef] [PubMed]
- McCallin, S.; Alam Sarker, S.; Barretto, C.; Sultana, S.; Berger, B.; Huq, S.; Krause, L.; Bibiloni, R.; Schmitt, B.; Reuteler, G.; et al. Safety analysis of a Russian phage cocktail: From MetaGenomic analysis to oral application in healthy human subjects. Virology 2013, 443, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Manohar, P.; Nachimuthu, R.; Lopes, B.S. The therapeutic potential of bacteriophages targeting gram-negative bacteria using Galleria mellonella infection model. BMC Microbiol. 2018, 18, 97. [Google Scholar] [CrossRef]
- Alves, E.; Carvalho, C.M.B.; Tomé, J.P.C.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Tomé, A.C.; Cavaleiro, J.A.S.; Cunha, Â.; Mendo, S.; Almeida, A. Photodynamic inactivation of recombinant bioluminescent Escherichia coli by cationic porphyrins under artificial and solar irradiation. J. Ind. Microbiol. Biotechnol. 2008, 35, 1447–1454. [Google Scholar] [CrossRef]
- Silva, I.; Tacão, M.; Tavares, R.D.S.; Miranda, R.; Araújo, S.; Manaia, C.M.; Henriques, I. Fate of cefotaxime-resistant Enterobacteriaceae and ESBL-producers over a full-scale wastewater treatment process with UV disinfection. Sci. Total Environ. 2018, 639, 1028–1037. [Google Scholar] [CrossRef]
- Pereira, S.; Pereira, C.; Santos, L.; Klumpp, J.; Almeida, A. Potential of phage cocktails in the inactivation of Enterobacter cloacae—An in vitro study in a buffer solution and in urine samples. Virus Res. 2016, 211, 199–208. [Google Scholar] [CrossRef]
- Silva, Y.J.; Costa, L.; Pereira, C.; Cunha, A.; Calado, R.; Gomes, N.C.M.; Almeida, A. Influence of environmental variables in the efficiency of phage therapy in aquaculture. Microb. Biotechnol. 2014, 7, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H. Bacteriophages; Interscience Publishers, Inc.: New York, NY, USA, 1959. [Google Scholar]
- Filippov, A.; Sergueev, K.V.; He, Y.; Huang, X.Z.; Gnade, B.T.; Mueller, A.J.; Fernandez-Prada, C.; Nikolich, M.P. Bacteriophage-resistant mutants in Yersinia pestis: Identification of phage receptors and attenuation for mice. PLoS ONE 2011, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.; Cunha, Â.; Gomes, N.C.M.; Alves, E.; Costa, L.; Faustino, M.A.F. Phage therapy and photodynamic therapy: Low environmental impact approaches to inactivate microorganisms in fish farming plants. Mar. Drugs 2009, 7, 268–313. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, K.M.; Nilsson, A.S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Nakai, T. Application of bacteriophages for control of infectious diseases in aquaculture. In Bacteriophages in the Control of Food and Waterborne Pathogens; Sabour, P., Griffiths, M., Eds.; USA ASM Press: Washington, DC, USA, 2010; pp. 257–272. [Google Scholar]
- Brown, C.M.; Bidle, K.D. Attenuation of virus production at high multiplicities of infection in Aureococcus anophagefferens. Virology 2014, 466–467, 71–81. [Google Scholar] [CrossRef]
- Arisaka, F.; Kanamaru, S.; Leiman, P.; Rossmann, M.G. The tail lysozyme complex of bacteriophage T4. Int. J. Biochem. Cell Biol. 2003, 35, 16–21. [Google Scholar] [CrossRef]
- Kao, S.H.; McClain, W.H. Baseplate protein of bacteriophage T4 with both structural and lytic functions. J. Virol. 1980, 34, 95–103. [Google Scholar] [PubMed]
- Essoh, C.; Blouin, Y.; Loukou, G.; Cablanmian, A.; Lathro, S.; Kutter, E.; Thien, H.V.; Vergnaud, G.; Pourcel, C. The susceptibility of Pseudomonas aeruginosa strains from cystic fibrosis atients to bacteriophages. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Wagner, P.L.; Waldor, M.K. Bacteriophage control of bacterial virulence. Infect. Immun. 2002, 70, 3985–3993. [Google Scholar] [CrossRef] [PubMed]
- O’Flynn, G.; Coffey, A.; Fitzgerald, G.; Ross, R. The newly isolated lytic bacteriophages st104a and st104b are highly virulent against Salmonella enterica. J. Appl. Microbiol. 2006, 101, 251–259. [Google Scholar] [CrossRef]
- Mateus, L.; Costa, L.; Silva, Y.J.; Pereira, C.; Cunha, A.; Almeida, A. Efficiency of phage cocktails in the inactivation of Vibrio in aquaculture. Aquaculture 2014, 424–425. [Google Scholar] [CrossRef]
- Fischer, C.; Yoichi, M.; Unno, H.; Y, T. The coexistence of Escherichia coli serotype O157:H7 and its specific bacteriophage in continuous culture. FEMS Microbiol. Lett. 2004, 241, 171–177. [Google Scholar] [CrossRef]
- Bohannan, B.J.M.; Travisano, M.; Lenski, R.E. Epistatic interactions can lower the cost of resistance to multiple consumers. Evolution 1999, 53, 292–295. [Google Scholar] [CrossRef]
- Brockhurst, M.A.; Buckling, A.; Rainey, P.B. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc. Biol. Sci. 2005, 272, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Lennon, J.T.; Khatana, S.A.M.; Marston, M.F.; Martiny, J.B.H. Is there a cost of virus resistance in marine cyanobacteria? ISME J. 2007, 1, 300–312. [Google Scholar] [CrossRef]
- Quance, M.A.; Travisano, M. Effects of temperature on the fitness cost of resistance to bacteriophage T4 in Escherichia coli. Evolution 2009, 63, 1406–1416. [Google Scholar] [CrossRef] [PubMed]
- Silva, Y.J.; Costa, L.; Pereira, C.; Mateus, C.; Cunha, Â.; Calado, R.; Gomes, N.C.M.; Pardo, M.A.; Hernandez, I.; Almeida, A. Phage therapy as an approach to prevent Vibrio anguillarum infections in fish larvae production. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Silva, Y.; Moreirinha, C.; Pereira, C.; Costa, L.; Rocha, R.J.M.; Cunha, Â.; Gomes, N.C.M.; Calado, R.; Almeida, A. Biological control of Aeromonas salmonicida infection in juvenile Senegalese sole (Solea senegalensis) with phage AS-A. Aquaculture 2016, 450, 225–233. [Google Scholar] [CrossRef]
- Ly-Chatain, M.H. The factors affecting effectiveness of treatment in phages therapy. Front. Microbiol. 2014, 5, 1–7. [Google Scholar] [CrossRef]
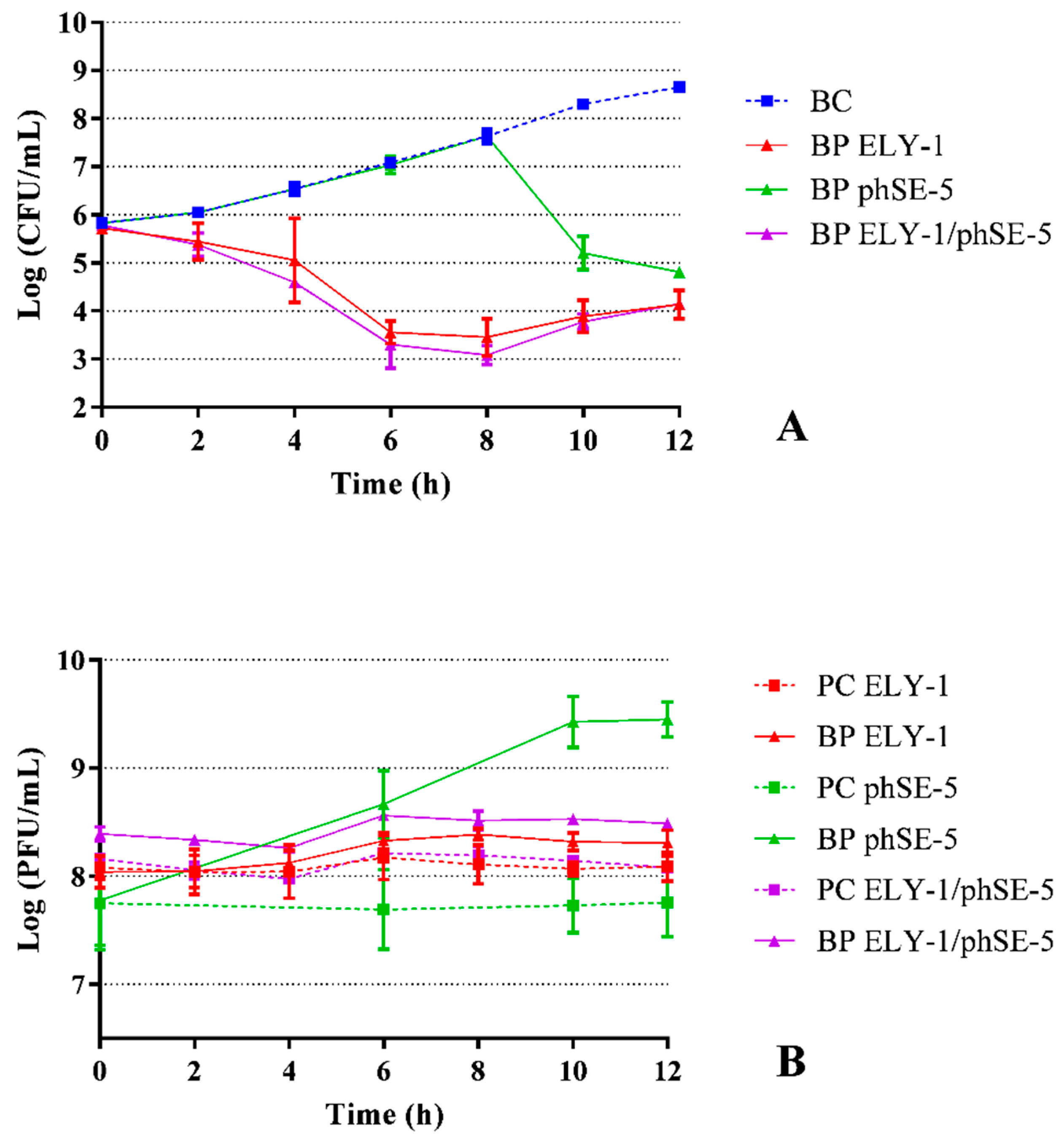
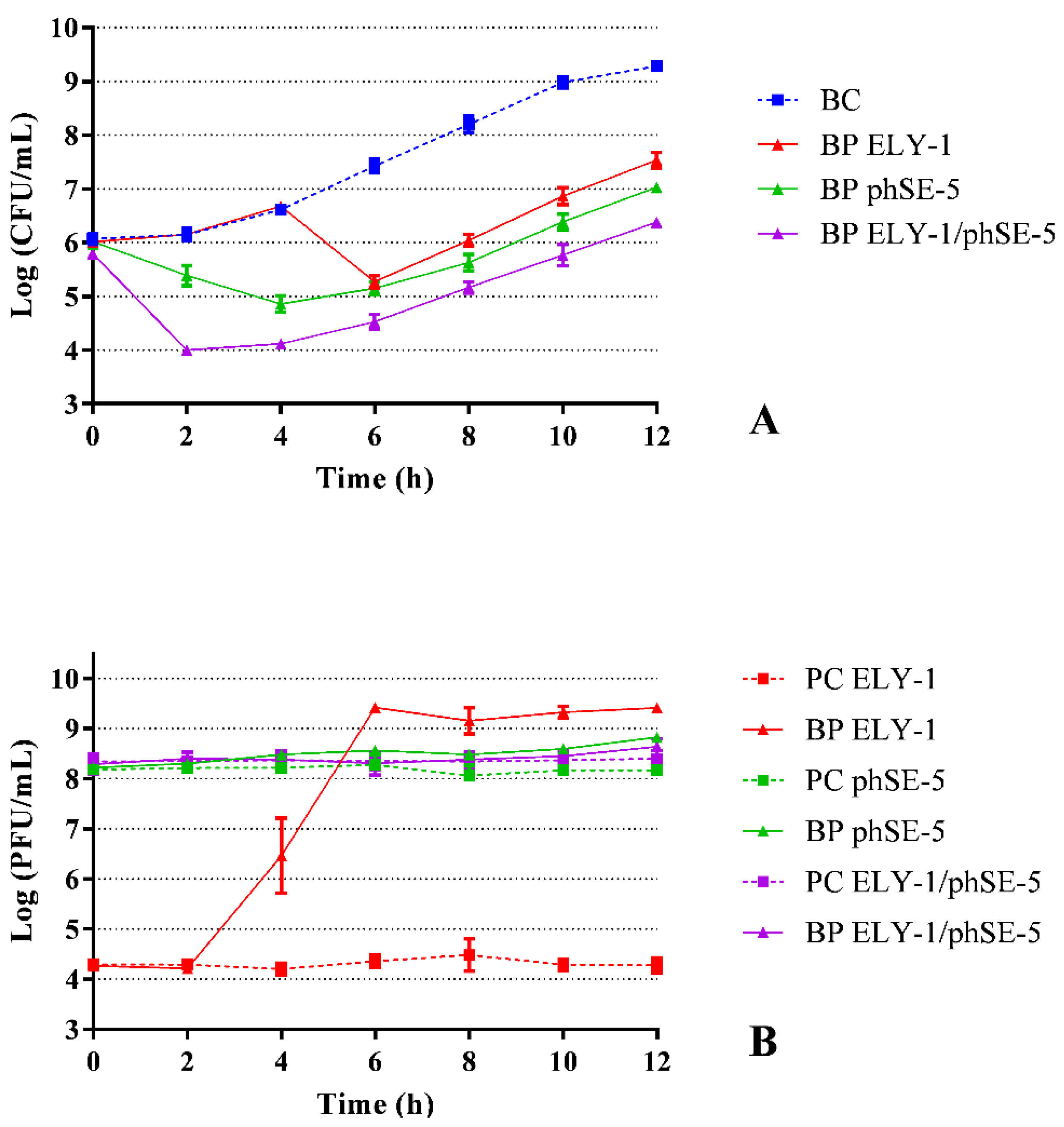
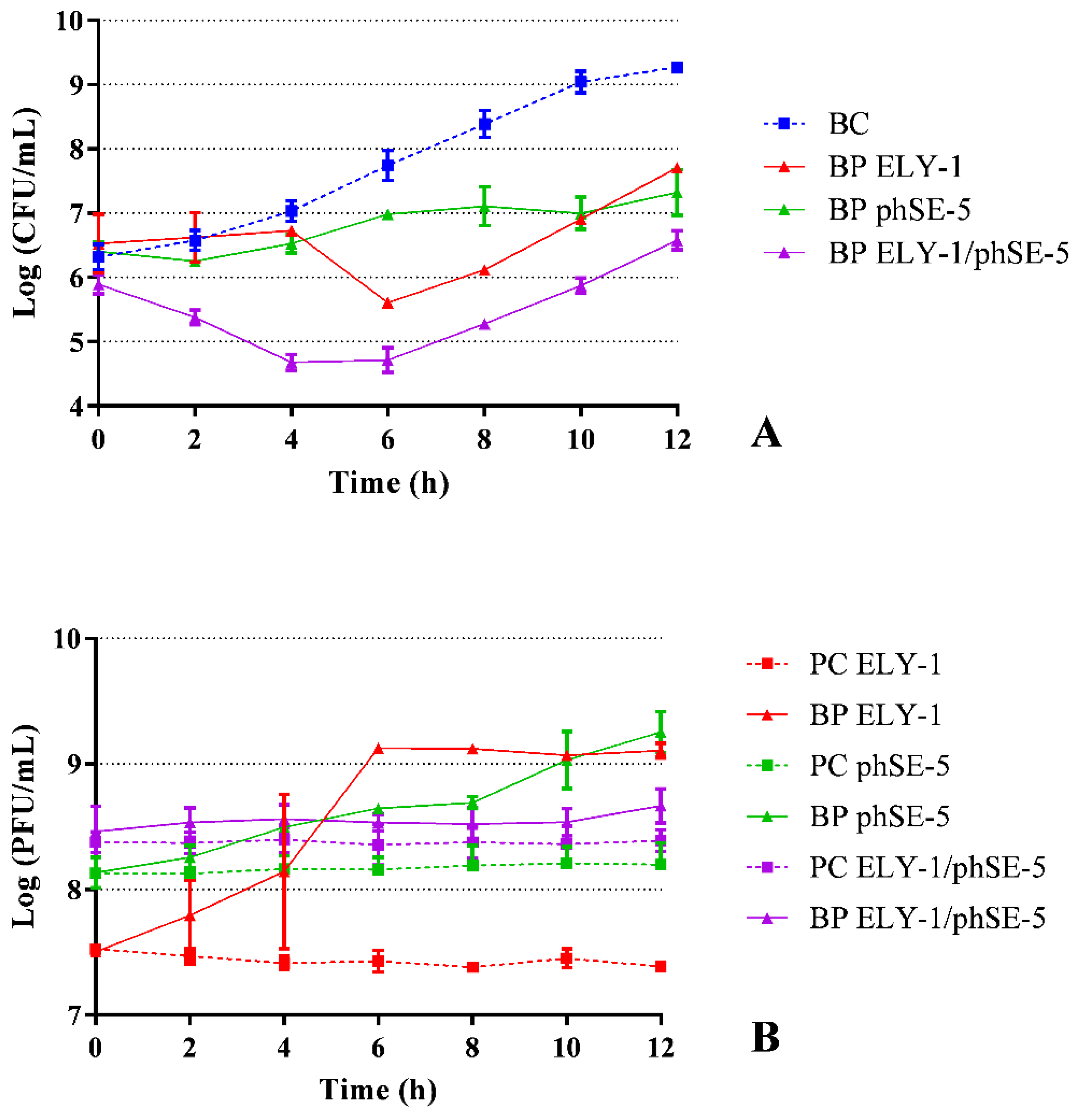
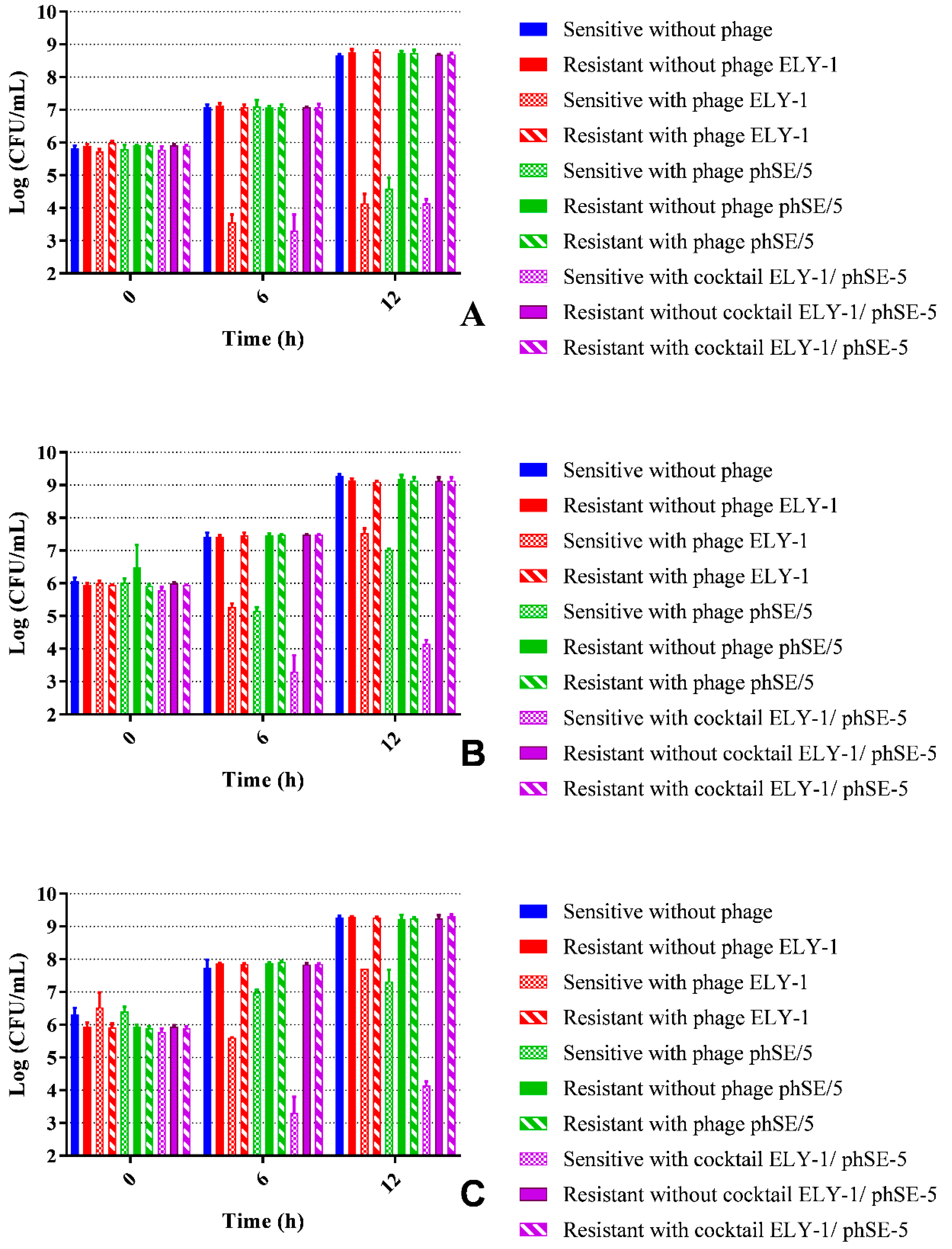
| Strains | ELY-1 | phSE-5 | ||
|---|---|---|---|---|
| Spot Test | EOP | Spot Test | EOP | |
| S. Typhimurium ATCC13311 | + | 5.7 × 10−4 ± 2.5 × 10−5 | + | 1 (host) |
| Bioluminescent E. coli | + | 1 (host) | + | 2.2 × 10−1 ± 1.5 × 10−2 |
| Citrobacter freundii 6F | − | 0 | + | 0 |
| Enterobacter cloacae | − | 0 | + | 0 |
| E. coli AC5 | − | 0 | + | 0 |
| E. coli AD6 | − | 0 | + | 0 |
| E. coli AE11 | − | 0 | + | 1.7 × 10−8 ± 5.6 × 10−9 |
| E. coli AF15 | − | 0 | − | 0 |
| E. coli AJ23 | − | 0 | + | 0 |
| E. coli AN19 | − | 0 | + | 0 |
| E. coli ATCC 13706 | − | 0 | + | 0 |
| E. coli ATCC 25922 | + | 22.7 ± 1.02 | + | 0 |
| E. coli BC30 | + | 0 | + | 0 |
| E. coli BM62 | − | 0 | + | 0 |
| E. coli BN65 | − | 0 | + | 0 |
| E. coli Scc 09 | − | 0 | − | 0 |
| E. coli Scc 33 | − | 0 | − | 0 |
| E. coli Scc 34 | + | 5.5 × 10−5 ± 5.0 × 10−6 | + | 1.8 × 10−5 ± 1.5 × 10−6 |
| E. coli Scc 35 | − | 0 | − | 0 |
| E. coli Scc 36 | − | 0 | − | 0 |
| E. coli Scc 37 | + | 0 | − | 0 |
| E. coli Scc 38 | − | 0 | − | 0 |
| E. coli Scc 40 | − | 0 | − | 0 |
| E. coli Scc 41 | − | 0 | − | 0 |
| E. coli Scc 43 | − | 0 | − | 0 |
| E. coli Scc 45 | − | 0 | − | 0 |
| E. coli Scc 47 | − | 0 | − | 0 |
| E. coli Scc 48 | − | 0 | − | 0 |
| E. coli Scc 49 | − | 0 | − | 0 |
| E. coli Scc 50 | − | 0 | − | 0 |
| E. coli Scc 51 | + | 0 | − | 0 |
| E. coli Scc 52 | − | 0 | − | 0 |
| E. coli Scc 53 | − | 0 | − | 0 |
| E. coli Scc 55 | − | 0 | − | 0 |
| E. coli Scc 56 | − | 0 | − | 0 |
| E. coli Scc 58 | − | 0 | − | 0 |
| E. coli Scc 69 | + | 2.8 × 10−1 ± 8.7 × 10−2 | + | 2.5 × 10−8 ± 4.8 × 10−9 |
| E. coli Scc 77 | − | 0 | − | 0 |
| E. coli Scc 78 | − | 0 | − | 0 |
| E. coli Scc 91 | − | 0 | − | 0 |
| Proteus mirabilis | − | 0 | + | 0 |
| Providencia sp. | − | 0 | + | 0 |
| S. Enteriditis CVA | − | 0 | + | 0 |
| S. Enteriditis CVB | − | 0 | + | 0 |
| S. Enteriditis CVC | − | 0 | + | 0 |
| S. Enteriditis CVD | + | 0 | + | 0 |
| S. Enteriditis CVE | − | 0 | + | 0 |
| S. Typhimurium ATCC 14028 | − | 0 | + | 0 |
| S. Typhimurium WG49 | − | 0 | + | 0 |
| Shigella flexneri DSM 4782 | − | 0 | − | 0 |
| Bacteria | Phage | Frequency of Phage-Mutants |
|---|---|---|
| E. coli | phSE-5 | 1.29 × 10−3 ± 3.15 × 10−5 |
| ELY-1 | 5.06 × 10−5 ± 6.55 × 10−6 | |
| ELY-1/phSE-5 | 1.22 × 10−3 ± 6.53 × 10−5 | |
| S. Typhimurium | phSE-5 | 2.80 × 10−4 ± 3.44 × 10−5 |
| ELY-1 | 5.11 × 10−5 ± 6.60 × 10−6 | |
| ELY-1/phSE-5 | 2.75 × 10−4 ± 4.10 × 10−5 | |
| Mixture of E. coli/S. Typhimurium | phSE-5 | 5.09 × 10−4 ± 6.99 × 10−5 |
| ELY-1 | 4.66 × 10−4 ± 8.44 × 10−5 | |
| ELY-1/phSE-5 | 6.45 × 10−5 ± 1.11 × 10−5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, P.; Pereira, C.; Gomes, A.T.P.C.; Almeida, A. Efficiency of Single Phage Suspensions and Phage Cocktail in the Inactivation of Escherichia coli and Salmonella Typhimurium: An In Vitro Preliminary Study. Microorganisms 2019, 7, 94. https://doi.org/10.3390/microorganisms7040094
Costa P, Pereira C, Gomes ATPC, Almeida A. Efficiency of Single Phage Suspensions and Phage Cocktail in the Inactivation of Escherichia coli and Salmonella Typhimurium: An In Vitro Preliminary Study. Microorganisms. 2019; 7(4):94. https://doi.org/10.3390/microorganisms7040094
Chicago/Turabian StyleCosta, Pedro, Carla Pereira, Ana T. P. C. Gomes, and Adelaide Almeida. 2019. "Efficiency of Single Phage Suspensions and Phage Cocktail in the Inactivation of Escherichia coli and Salmonella Typhimurium: An In Vitro Preliminary Study" Microorganisms 7, no. 4: 94. https://doi.org/10.3390/microorganisms7040094
APA StyleCosta, P., Pereira, C., Gomes, A. T. P. C., & Almeida, A. (2019). Efficiency of Single Phage Suspensions and Phage Cocktail in the Inactivation of Escherichia coli and Salmonella Typhimurium: An In Vitro Preliminary Study. Microorganisms, 7(4), 94. https://doi.org/10.3390/microorganisms7040094








