Changes in Mouse Gut Microbial Community in Response to the Different Types of Commonly Consumed Meat
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Feeding Schedule
2.2. DNA Extraction and Sequencing
2.3. Bioinformatics Analysis
2.4. Blood and Fecal Assays
2.5. Diversity and Statistical Analysis
3. Results
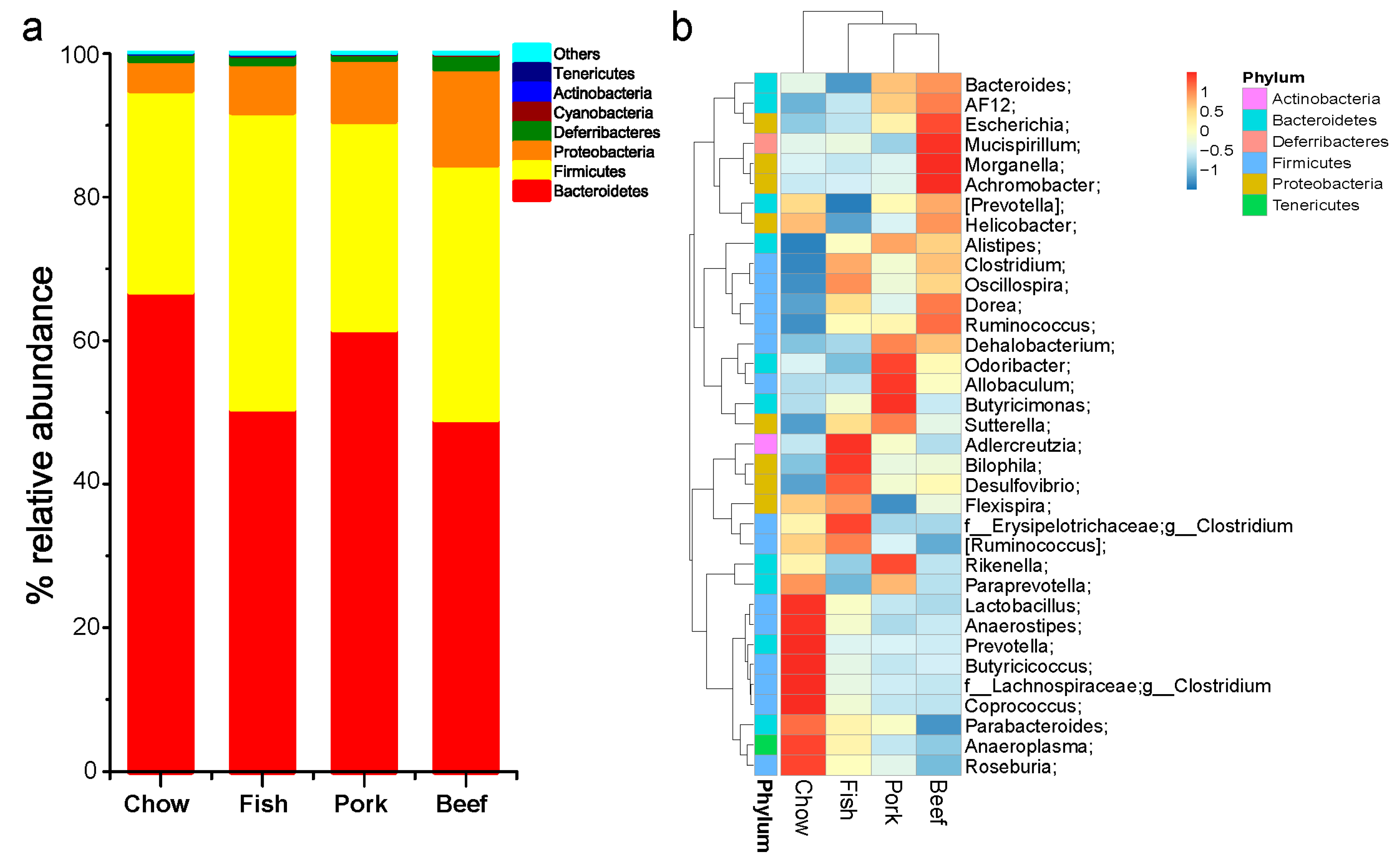
3.1. Taxonomic Structure and Characterization
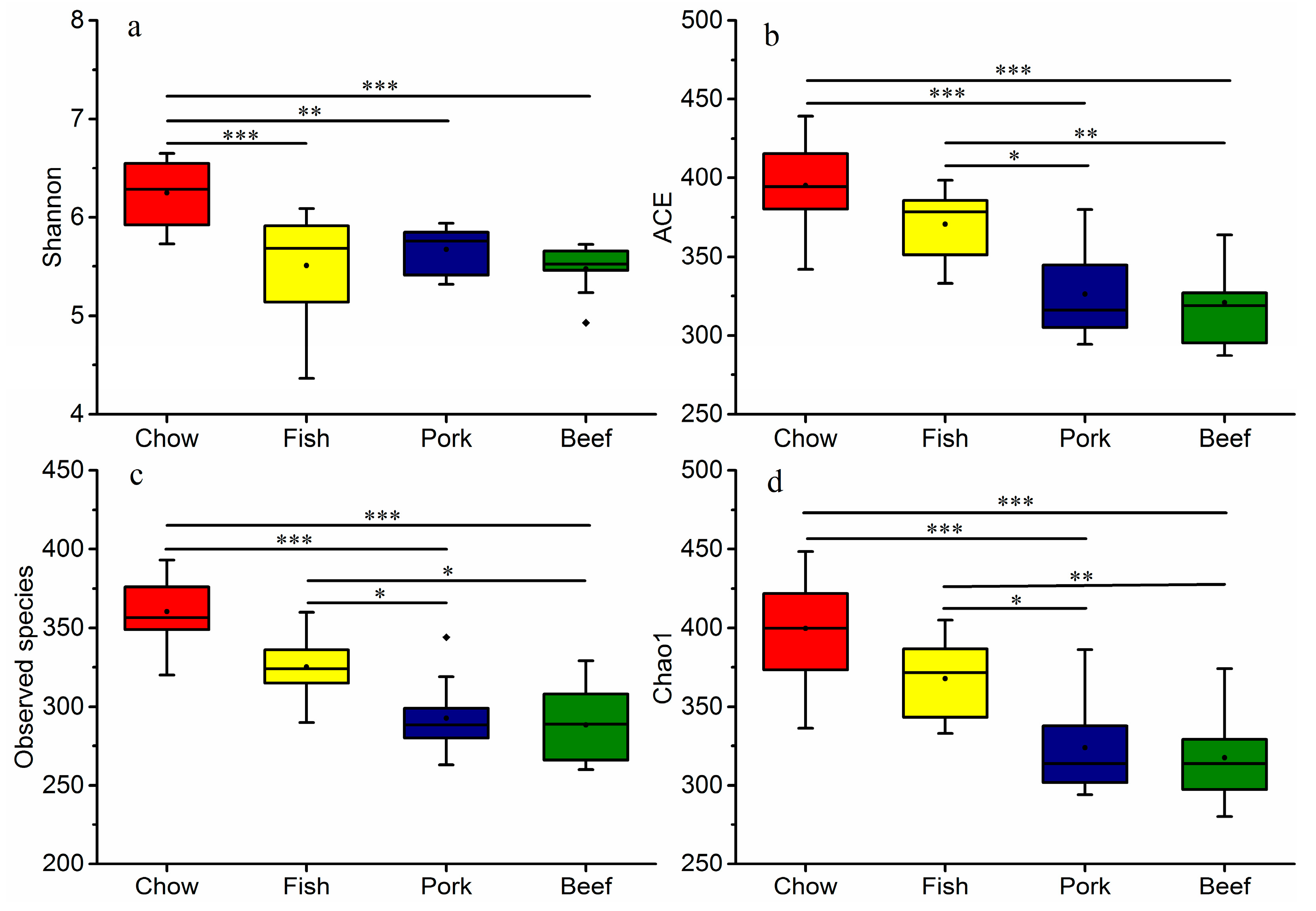
3.2. Meat Consumption Reduces Richness and Diversity of Gut Microbiota
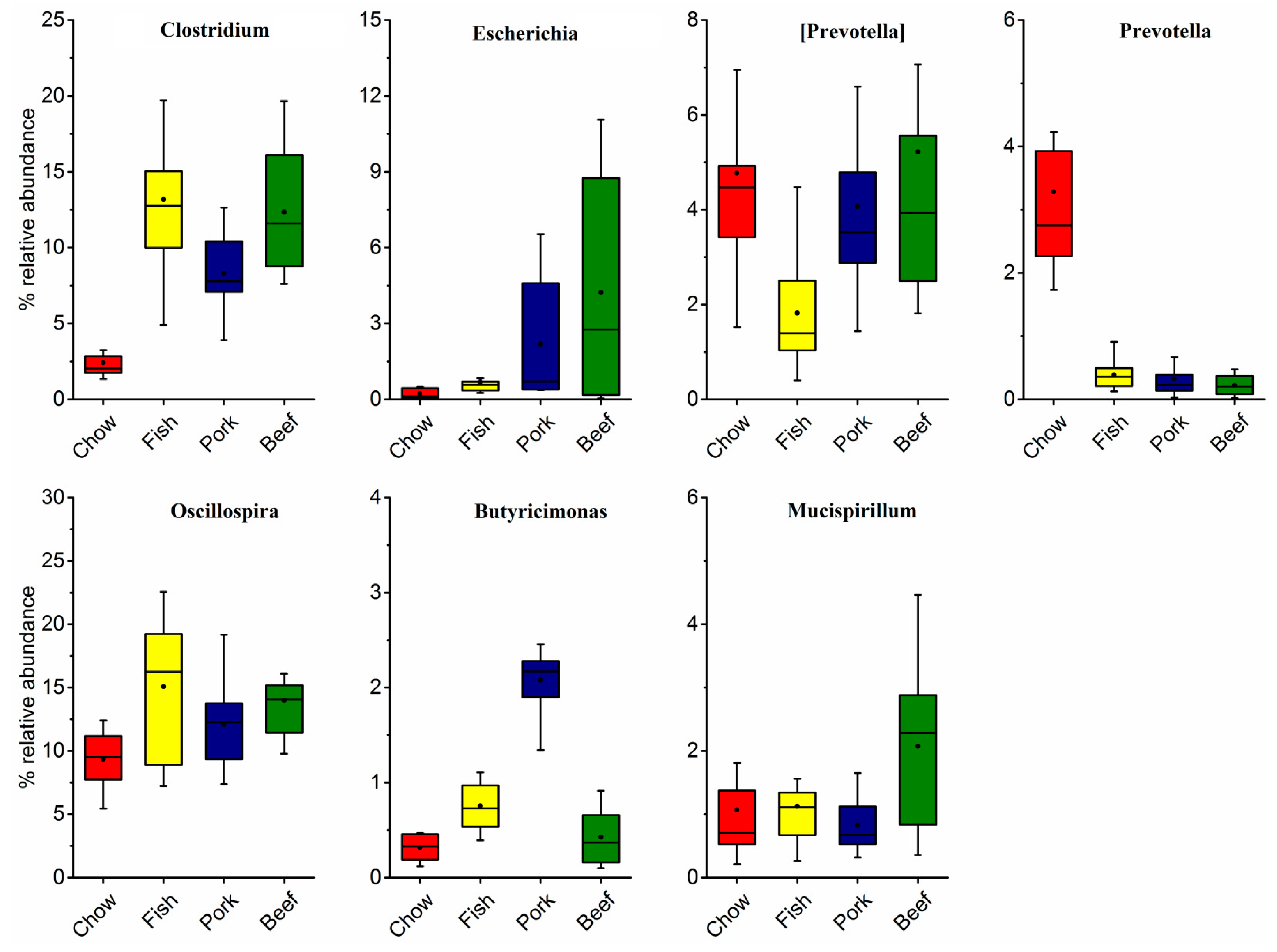
3.3. Multivariate Analysis of Gut Microbial Responses to Meat
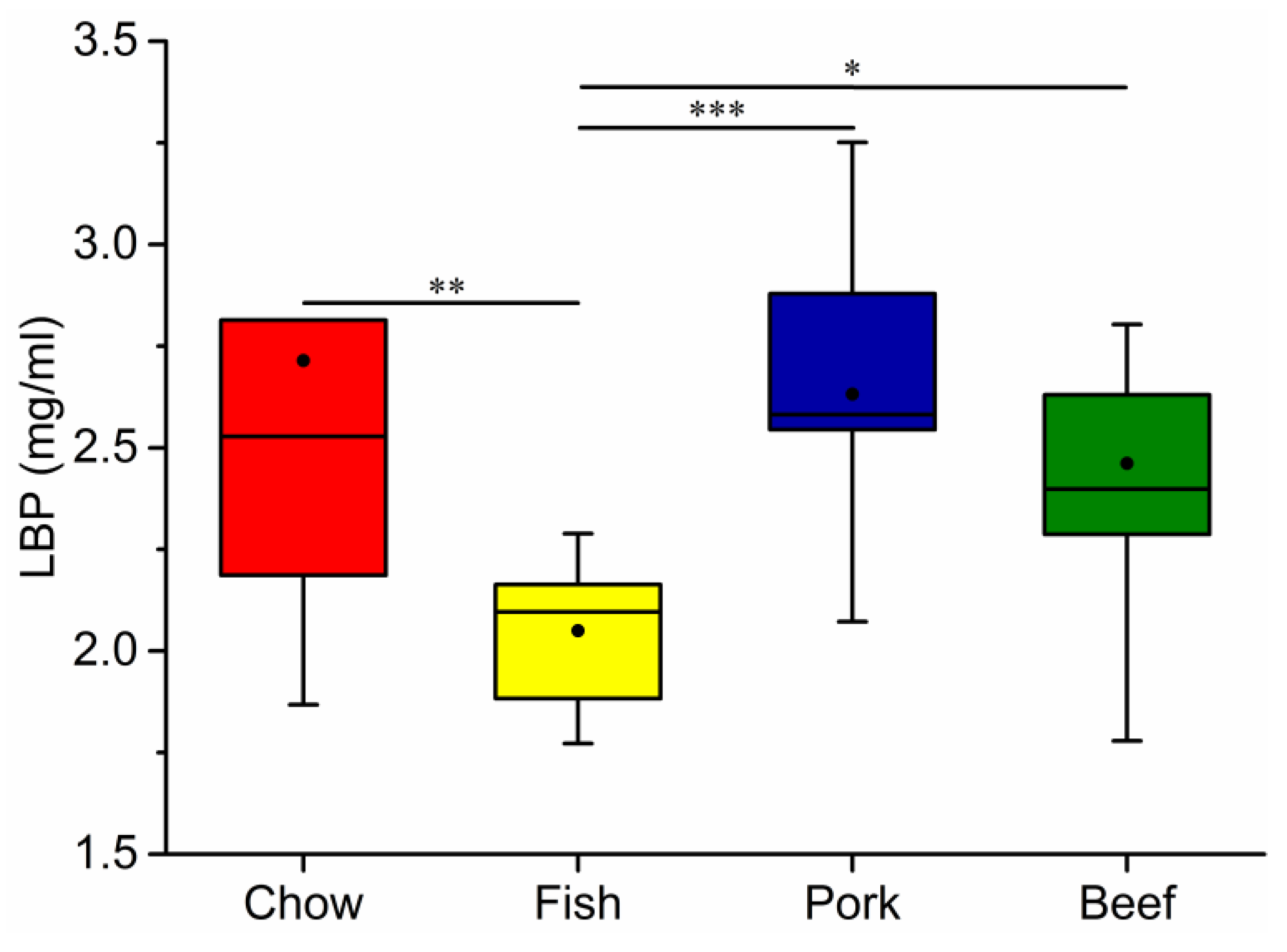
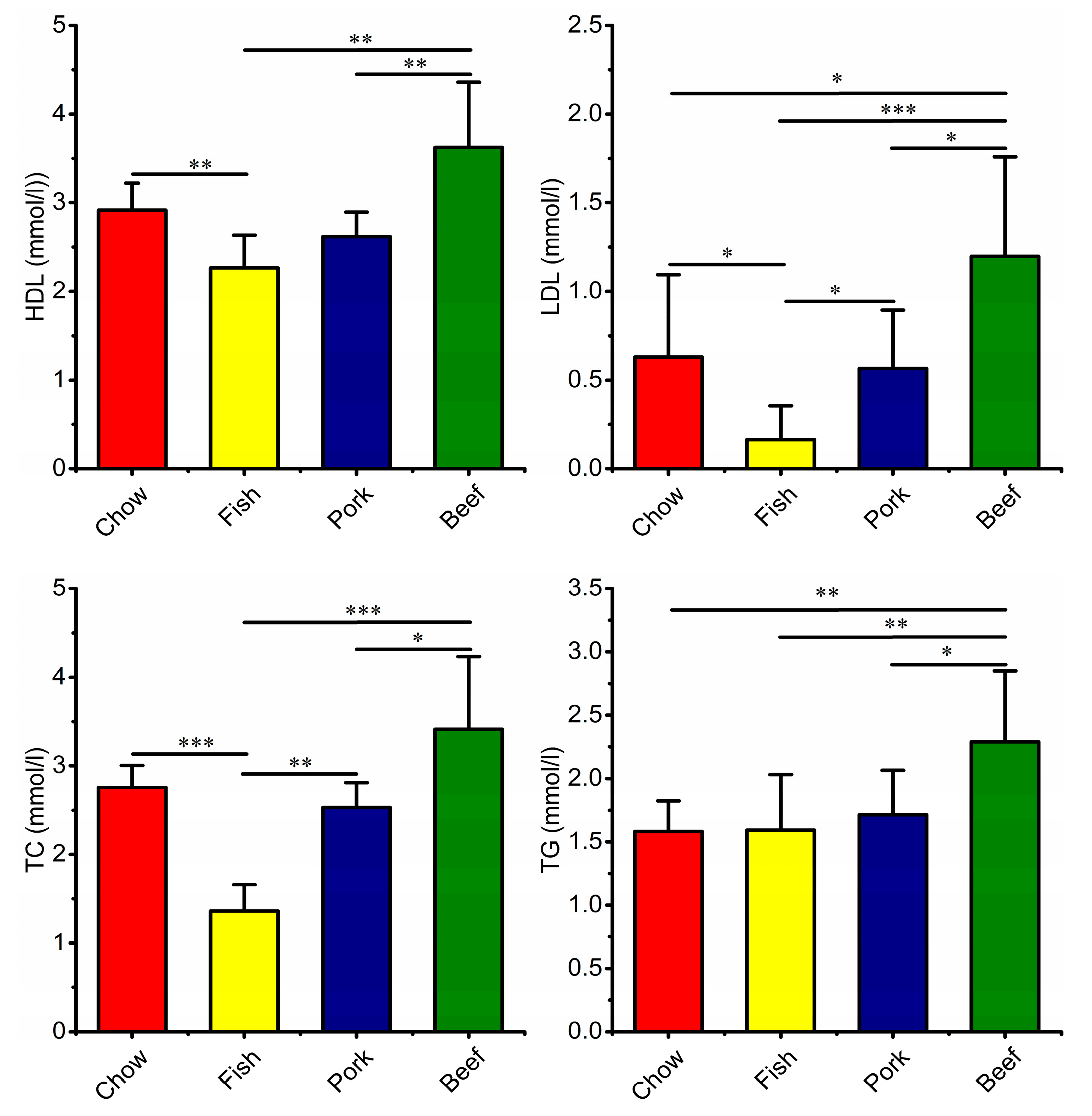
3.4. Growth, Blood Metabolic Indices, and Fecal Characteristics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed]
- Patricia, L.L.; Nicholas, R.F.; María, A.Z.; Jose, A.M.; Ian, D.C. The influence of mediterranean, carbohydrate and high protein diets on gut microbiota composition in the treatment of bbesity and associated inflammatory state. Asia Pac. J. Clin. Nutr. 2014, 23, 360–368. [Google Scholar]
- Schnorr, S.L.; Candela, M.; Rampelli, S.; Centanni, M.; Consolandi, C.; Basaglia, G.; Turroni, S.; Biagi, E.; Peano, C.; Severgnini, M.; et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014, 5, 3654. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.K.; Mazmanian, S.K. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 2010, 330, 1768–1773. [Google Scholar] [CrossRef]
- Chinen, T.; Rudensky, A.Y. The effects of commensal microbiota on immune cell subsets and inflammatory responses. Immunol. Rev. 2012, 245, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Knight, D.J.W.; Girling, K.J. Gut flora in health and disease. Lancet 2003, 361, 1831. [Google Scholar] [CrossRef]
- Kiecolt-Glaser, J.K.; Belury, M.A.; Andridge, R.; Malarkey, W.B.; Hwang, B.S.; Glaser, R. Omega-3 supplementation lowers inflammation in healthy middle-aged and older adults: A randomized controlled trial. Brain Behav. Immun. 2012, 26, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.; Fleming, T.; Robinson, M.; Thomson, B.; Graetz, N.; Margono, C.; Mullany, E.C.; Biryukov, S.; Abbafati, C.; Abera, S.F.; et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014, 384, 766–781. [Google Scholar] [CrossRef]
- de La Serre, C.B.; Ellis, C.L.; Lee, J.; Hartman, A.L.; Rutledge, J.C.; Raybould, H.E. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am. J. Physiol-Gastrointest. Liver Physiol. 2010, 299, G440–G448. [Google Scholar] [CrossRef] [PubMed]
- Lam, Y.Y.; Ha, C.W.; Hoffmann, J.M.; Oscarsson, J.; Dinudom, A.; Mather, T.J.; Cook, D.I.; Hunt, N.H.; Caterson, I.D.; Holmes, A.J.; et al. Effects of dietary fat profile on gut permeability and microbiota and their relationships with metabolic changes in mice. Obesity 2015, 23, 1429–1439. [Google Scholar] [CrossRef]
- Zhang, C.; Yin, A.; Li, H.; Wang, R.; Wu, G.; Shen, J.; Zhang, M.; Wang, L.; Hou, Y.; Ouyang, H.; et al. Dietary Modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. EBioMedicine 2015, 2, 968–984. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Lin, X.; Zhao, F.; Shi, X.; Li, H.; Li, Y.; Zhu, W.; Xu, X.; Li, C.; Zhou, G. Meat, dairy and plant proteins alter bacterial composition of rat gut bacteria. Sci. Rep. 2015, 5, 15220. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Frese, S.A.; Parker, K.; Calvert, C.C.; Mills, D.A. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome 2015, 3, 28. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Beydoun, M.A. Meat consumption is associated with obesity and central obesity among US adults. Int. J. Obes. 2009, 33, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Fang, Y.-J.; Chen, Y.-M.; Lu, M.-S.; Pan, Z.-Z.; Yan, B.; Zhong, X.; Zhang, C.X. Higher freshwater fish and sea fish intake is inversely associated with colorectal cancer risk among Chinese population: A case-control study. Sci. Rep. 2015, 5, 12976. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, A.M.; Sun, Q.; Hu, F.B.; Stampfer, M.J.; Manson, J.E.; Willett, W.C. Major dietary protein sources and the risk of coronary heart disease in women. Circulation 2010, 122, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Takata, Y.; Zhang, X.; Li, H.; Gao, Y.T.; Yang, G.; Gao, J. Fish intake and risks of total and cause-specific mortality in 2 population-based cohort studies of 134,296 men and women. Am. J. Epidemiol. 2013, 178, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Liisberg, U.; Fauske, K.R.; Kuda, O.; Fjære, E.; Myrmel, L.S.; Norberg, N.; Frøyland, L.; Graff, I.E.; Liaset, B.; Kristiansen, K.; et al. Intake of a Western diet containing cod instead of pork alters fatty acid composition in tissue phospholipids and attenuates obesity and hepatic lipid accumulation in mice. J. Nutr. Biochem. 2016, 33, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Chen, Y.A.; Tuohy, K.M. A comparative in vitro investigation into the effects of cooked meats on the human faecal microbiota. Anaerobe 2010, 16, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Tamaki, H.; Wright, C.L.; Li, X.Z.; Lin, Q.; Hwang, C.; Wang, S.; Thimmapuram, J.; Kamagata, Y.; Liu, W.-T. Analysis of 16S rRNA amplicon sequencing options on the roche/454 next-generation titanium sequencing platform. PLoS ONE 2011, 6, e25263. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, D141–D145. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimerachecked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microb. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Wang, Y.; Yang, T.; Ao, H.; Chen, S.; Xing, K.; Zhang, F.; Zhao, X.; Liu, J.; Wang, C. Differences in gut microbiota composition in finishing Landrace pigs with low and high feed conversion ratios. Antonie Van Leeuwenhoek 2018, 111, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Lozupone, C.A.; Hamady, M.; Knight, R.; Gordon, J.I. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008, 6, 776–788. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.B.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Schiraldi, A.; Fessas, D.; Signorelli, M. Water activity in biological systems—A review. Pol. J. Food Nutr. Sci. 2012, 62, 5–13. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, D.; Falony, G.; Vieira-Silva, S.; Tito, R.Y.; Joossens, M.; Raes, J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 2016, 65, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.; Ley, R. The human gut microbiome: Ecology and recent evolutionary changes. Annu. Rev. Microbiol. 2011, 65, 411–429. [Google Scholar] [CrossRef] [PubMed]
- Diether, N.E.; Willing, B.P. Microbial fermentation of dietary protein: An important factor in diet-microbe-host interaction. Microorganisms 2019, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Allison, C.; Gibson, S.A.; Cummings, J.H. Contribution of the microflora to proteolysis in the human large intestine. J. Appl. Bacteriol. 1988, 64, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Sabja, D.; Sarker, M.R. Clostridium perfringens sporulation and its relevance to pathogenesis. Future Microbiol. 2009, 4, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Close, R.; Hughes, E.A.; Magee, S.; Bingham, S. Protein degradation in the large intestine: Relevance to colorectal cancer. Intest. Microbiol. 2000, 1, 51–58. [Google Scholar]
- Evers, E.G.; Pielaat, A.; Smid, J.H.; van Duijkeren, E.; Vennemann, F.B.; Wijnands, L.M.; Chardon, J.E. Comparative exposure assessment of ESBL-producing Escherichia coli through meat consumption. PLoS ONE 2017, 12, e0169589. [Google Scholar] [CrossRef] [PubMed]
- Hermes, R.G.; Molist, F.; Ywazaki, M.; Nofrarías, M.; Gomez de Segura, A.; Gasa, J.; Pérez, J.F. Effect of dietary level of protein and fiber on the productive performance and health status of piglets. J. Anim. Sci. 2009, 87, 3569–3577. [Google Scholar] [CrossRef] [PubMed]
- Tims, S.; Derom, C.; Jonkers, D.M.; Vlietinck, R.; Saris, W.H.; Kleerebezem, M.; de Vos, W.M.; Zoetendal, E.G. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. 2013, 7, 707–717. [Google Scholar] [CrossRef]
- Escobar, J.S.; Klotz, B.; Valdes, B.E.; Agudelo, G.M. The gut microbiota of colombians differs from that of Americans, Europeans and Asians. BMC Microbiol. 2014, 14, 311. [Google Scholar] [CrossRef] [PubMed]
- Konikoff, T.; Gophna, U. Oscillospira: A central, enigmatic component of the human gut microbiota. Trends Microbiol. 2016, 24, 523–524. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Polyunsaturated fatty acids, inflammatory processes and inflammatory bowel diseases. Mol. Nutr. Food Res. 2008, 52, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Mujico, J.R.; Baccan, G.C.; Gheorghe, A.; Díaz, L.E.; Marcos, A. Changes in gut microbiota due to supplemented fatty acids in diet-induced obese mice. Br. J. Nutr. 2013, 110, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Rist, V.T.S.; Weiss, E.; Eklund, M.; Mosenthin, R. Impact of dietary protein on microbiota composition and activity in the gastrointestinal tract of piglets in relation to gut health: A review. Animal 2013, 7, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Frick, M.H.; Manninen, V.; Huttunen, J.K.; Heinonen, O.P.; Tenkanen, L.; Mänttäri, M. HDL-cholesterol as a risk factor in coronary heart disease. Drugs 1990, 40, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Després, J.P.; Lemieux, I.; Dagenais, G.R.; Cantin, B.; Lamarche, B.T. HDL-cholesterol as a marker of coronary heart disease risk: The Québec cardiovascular study. Atherosclerosis 2000, 153, 263–272. [Google Scholar] [CrossRef]
- Zanoni, P.; Khetarpal, S.A.; Larach, D.B.; Hancock-Cerutti, W.F.; Millar, J.S.; Cuchel, M.; DerOhannessian, S.; Kontush, A.; Surendran, P.; Saleheen, D.; et al. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science 2016, 351, 1166–1171. [Google Scholar] [CrossRef] [PubMed]
- Leaf, D.A.; Hatcher, L. The effect of lean fish consumption on triglyceride levels. Phys. Sportsmed. 2009, 37, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Mocellin, M.C.; Camargo, C.Q.; Nunes, E.A.; Fiates, G.M.R.; Trindade, E.B.S.M. A systematic review and meta-analysis of the n-3 polyunsaturated fatty acids effects on inflammatory markers in colorectal cancer. Clin. Nutr. 2016, 35, 359–369. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Li, D.; Tang, R. Changes in Mouse Gut Microbial Community in Response to the Different Types of Commonly Consumed Meat. Microorganisms 2019, 7, 76. https://doi.org/10.3390/microorganisms7030076
Zhang Z, Li D, Tang R. Changes in Mouse Gut Microbial Community in Response to the Different Types of Commonly Consumed Meat. Microorganisms. 2019; 7(3):76. https://doi.org/10.3390/microorganisms7030076
Chicago/Turabian StyleZhang, Zhimin, Dapeng Li, and Rong Tang. 2019. "Changes in Mouse Gut Microbial Community in Response to the Different Types of Commonly Consumed Meat" Microorganisms 7, no. 3: 76. https://doi.org/10.3390/microorganisms7030076
APA StyleZhang, Z., Li, D., & Tang, R. (2019). Changes in Mouse Gut Microbial Community in Response to the Different Types of Commonly Consumed Meat. Microorganisms, 7(3), 76. https://doi.org/10.3390/microorganisms7030076





