MALDI-TOF Mass Spectrometry and Specific Biomarkers: Potential New Key for Swift Identification of Antimicrobial Resistance in Foodborne Pathogens
Abstract
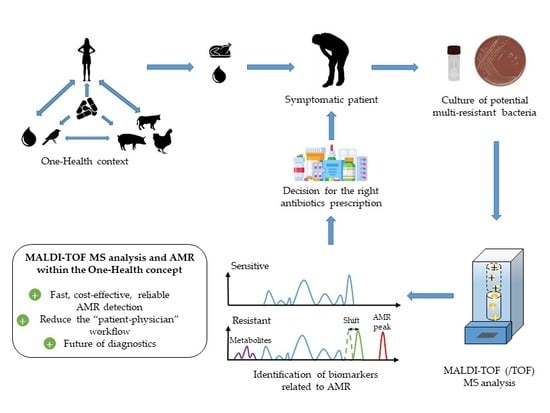
1. The Burden of Antimicrobial Resistances Worldwide: The Case of Foodborne Pathogens
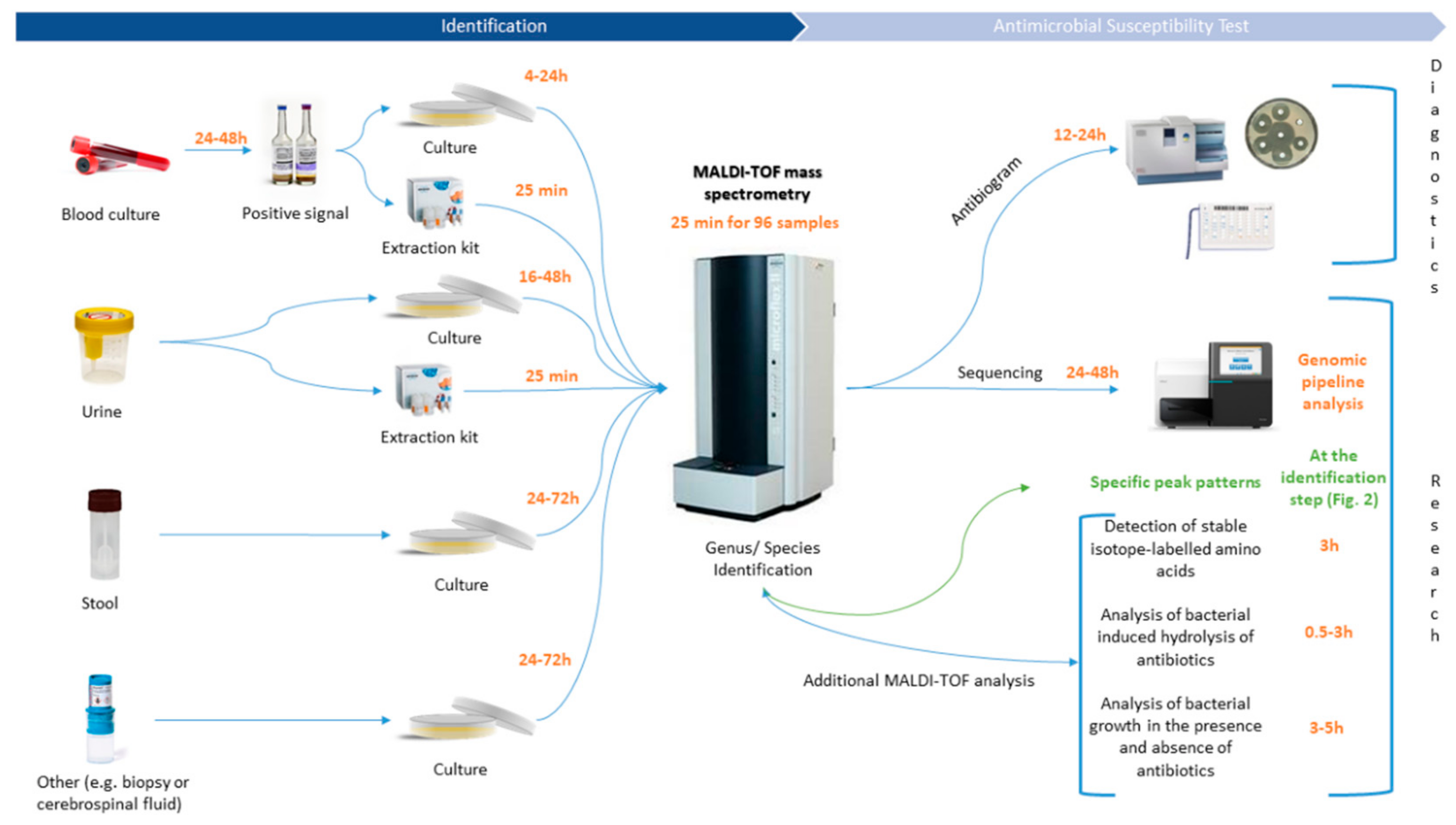
2. MALDI-TOF MS: A New Era for the Diagnostic Field
3. Specific Biomarkers as a Future Key for the Detection of AMR
4. Bioinformatics: A Powerful Tool to Reinforce Diagnostics
5. MALDI-TOF/TOF Tandem Mass Spectrometry: To Infinity and Beyond
6. Outlook and Future Challenges for MALDI-TOF MS and AMR in the Diagnostic Field
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Antibiotic Use in the United States, 2017: Progress and Opportunities. Available online: https://www.cdc.gov/antibiotic-use/stewardship-report/index.html (accessed on 9 October 2019).
- Schwarz, S.; Kehrenberg, C.; Walsh, T.R. Use of antimicrobial agents in veterinary medicine and food animal production. Int. J. Antimicrob. Agents 2001, 17, 431–437. [Google Scholar] [CrossRef]
- Ventola, C.L. The Antibiotic Resistance: Part 1: Causes and Threats. P T a Peer Rev. J. Formul. Manag. 2015, 40, 277–283. [Google Scholar]
- Orruño, M.; Garaizabal, I.; Arana, I.; Barcina, I. Persistence and dissemination of antimicrobial resistances in aquatic systems. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education; Formatex Research Center: Badajoz, Spain, 2013; pp. 624–629. [Google Scholar]
- The Organisation for Economic Co-operation Development. Stemming the Superbug Tide; OECD Health Policy Studies: Paris, France, 2018. [Google Scholar]
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015. Available online: https://apps.who.int/iris/handle/10665/199350 (accessed on 9 October 2019).
- Priyanka, B.; Patil, R.K.; Dwarakanath, S. A review on detection methods used for foodborne pathogens. Indian J. Med. Res. 2016, 144, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Lammie, S.L.; Hughes, J.M. Antimicrobial Resistance, Food Safety, and One Health: The Need for Convergence. Annu. Rev. Food Sci. Technol. 2016, 7, 287–312. [Google Scholar] [CrossRef] [PubMed]
- Destoumieux-Garzón, D.; Mavingui, P.; Boetsch, G.; Boissier, J.; Darriet, F.; Duboz, P.; Fritsch, C.; Giraudoux, P.; Le Roux, F.; Morand, S.; et al. The one Health Concept: 10 years old and a long road ahead. Front. Vet. Sci. 2018, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). The European union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA J. 2013, 11, 3129. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 2017, 15, 5077. [Google Scholar]
- Allos, B.M. Campylobacter jejuni Infections: Update on Emerging Issues and Trends. Clin. Infect. Dis. 2002, 32, 1201–1206. [Google Scholar]
- Chlebicz, A.; Śliżewska, K. Campylobacteriosis, Salmonellosis, Yersiniosis, and Listeriosis as Zoonotic Foodborne Diseases: A Review. Int. J. Environ. Res. Public Health 2018, 15, 863. [Google Scholar] [CrossRef]
- Szczepanska, B.; Andrzejewska, M.; Spica, D.; Klawe, J.J. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from children and environmental sources in urban and suburban areas. BMC Microbiol. 2017, 17, 80. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, A.; Dionisi, A.M.; Arena, S.; Iglesias-Torrens, Y.; Carattoli, A.; Luzzi, I. Human campylobacteriosis in Italy: Emergence of multi-drug resistance to ciprofloxacin, tetracycline, and erythromycin. Front. Microbiol. 2018, 9, 1906. [Google Scholar] [CrossRef] [PubMed]
- Karikari, A.B.; Obiri-Danso, K.; Frimpong, E.H.; Krogfelt, K.A. Antibiotic resistance of Campylobacter recovered from faeces and carcasses of healthy livestock. Biomed. Res. Int. 2017, 2017, 4091856. [Google Scholar] [CrossRef] [PubMed]
- Maktabi, S.; Ghorbanpoor, M.; Hossaini, M.; Motavalibashi, A.; Dvm, S.M. Detection of multi-antibiotic resistant Campylobacter coli and Campylobacter jejuni in beef, mutton, chicken and water buffalo meat in Ahvaz, Iran. Artic. Vet. Res. Forum 2019, 10, 37–42. [Google Scholar]
- Gölz, G.; Schielke, A.; Josenhans, C.; Rosner, B.; Löwenstein, A.; Alter, T.; Wieler, L.H.; Kreienbrock, L.; Stark, K.; Suerbaum, S.; et al. Relevance of Campylobacter to public health—The need for a One Health approach. Int. J. Med. Microbiol. 2014, 304, 817–823. [Google Scholar] [CrossRef]
- Mangioni, D.; Viaggi, B.; Giani, T.; Arena, F.; D’Arienzo, S.; Forni, S.; Tulli, G.; Rossolini, G.M. Diagnostic stewardship for sepsis: The need for risk stratification to triage patients for fast microbiology workflows. Future Microbiol. 2019, 14, 169–174. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Munk, P.; Njage, P.; van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J.; et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef]
- World Health Organization. Global Antimicrobial Resistance Surveillance System. Manual for Early Implementation. Available online: https://www.who.int/antimicrobial-resistance/publications/surveillance-system-manual/en/ (accessed on 9 October 2019).
- Whitehouse, C.A.; Young, S.; Li, C.; Hsu, C.H.; Martin, G.; Zhao, S. Use of whole-genome sequencing for Campylobacter surveillance from NARMS retail poultry in the United States in 2015. Food Microbiol. 2018, 73, 122–128. [Google Scholar] [CrossRef]
- Anjum, M.F.; Zankari, E.; Hasman, H. Molecular methods for detection of antimicrobial resistance. Microbiol. Spectr. 2017, 5, 1–17. [Google Scholar] [CrossRef]
- Vestal, M.L. Evolution of quantitative MALDI-TOF mass spectrometry for clinical applications. Clin. Chem. 2016, 62, 20–23. [Google Scholar] [CrossRef]
- Anhalt, J.P.; Fenselau, C. Identification of bacteria using mass spectrometry. Anal. Chem. 1975, 47, 219–225. [Google Scholar] [CrossRef]
- Karas, M.; Bachmann, D.; Bahr, U.; Hillenkamp, F. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int. J. Mass Spectrom. Ion Process. 1987, 78, 53–68. [Google Scholar] [CrossRef]
- Tanaka, K.; Waki, H.; Ido, Y.; Akita, S.; Yoshida, Y.; Yoshida, T.; Matsuo, T. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1988, 2, 151–153. [Google Scholar] [CrossRef]
- Clark, A.E.; Kaleta, E.J.; Arora, A.; Wolk, D.M. Matrix-Assisted laser desorption ionization-time of flight mass spectrometry: A fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 2013, 26, 547–603. [Google Scholar] [CrossRef] [PubMed]
- Pusch, W.; Kostrzewa, M. Application of MALDI-TOF mass spectrometry in screening and diagnostic research. Curr. Pharm. Des. 2005, 11, 2577–2591. [Google Scholar] [CrossRef]
- Hou, T.Y.; Chiang-Ni, C.; Teng, S.H. Current status of MALDI-TOF mass spectrometry in clinical microbiology. J. Food Drug Anal. 2019, 27, 404–414. [Google Scholar] [CrossRef]
- Chabriere, E.; Bassène, H.; Drancourt, M.; Sokhna, C. MALDI-TOF-MS and point of care are disruptive diagnostic tools in Africa. New Microbes New Infect. 2018, 26, S83–S88. [Google Scholar] [CrossRef]
- Carbonnelle, E.; Mesquita, C.; Bille, E.; Day, N.; Dauphin, B.; Beretti, J.L.; Ferroni, A.; Gutmann, L.; Nassif, X. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin. Biochem. 2011, 44, 104–109. [Google Scholar] [CrossRef]
- Rotcheewaphan, S.; Lemon, J.K.; Desai, U.U.; Henderson, C.M.; Zelazny, A.M. Rapid one-step protein extraction method for the identification of mycobacteria using MALDI-TOF MS. Diagn. Microbiol. Infect. Dis. 2019, 94, 355–360. [Google Scholar] [CrossRef]
- L’Ollivier, C.; Ranque, S. MALDI-TOF-Based Dermatophyte Identification. Mycopathologia 2017, 182, 183–192. [Google Scholar] [CrossRef]
- Sjöholm, M.I.L.; Dillner, J.; Carlson, J. Multiplex detection of human herpesviruses from archival specimens by using matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2008, 46, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Boucheikhchoukh, M.; Laroche, M.; Aouadi, A.; Dib, L.; Benakhla, A.; Raoult, D.; Parola, P. MALDI-TOF MS identification of ticks of domestic and wild animals in Algeria and molecular detection of associated microorganisms. Comp. Immunol. Microbiol. Infect. Dis. 2018, 57, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, A.; Buttrini, M.; Montecchini, S.; Rossi, S.; Piccolo, G.; Arcangeletti, M.C.; Medici, M.C.; Chezzi, C.; De Conto, F. MALDI-TOF MS as a new tool for the identification of Dientamoeba fragilis. Parasites Vectors 2018, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Mayer-Scholl, A.; Murugaiyan, J.; Neumann, J.; Bahn, P.; Reckinger, S.; Nöckler, K. Rapid identification of the foodborne pathogen Trichinella spp. by matrix-assisted laser desorption/ionization mass spectrometry. PLoS ONE 2016, 11, e0152062. [Google Scholar] [CrossRef]
- Feucherolles, M.; Poppert, S.; Utzinger, J.; Becker, S.L. MALDI-TOF mass spectrometry as a diagnostic tool in human and veterinary helminthology: A systematic review. Parasite Vectors 2019, 12, 245. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef]
- Rodríguez-Sánchez, B.; Cercenado, E.; Coste, A.T.; Greub, G. Review of the impact of MALDI-TOF MS in public health and hospital hygiene, 2018. Eurosurveillance 2019, 24, 1800193. [Google Scholar] [CrossRef]
- Pliakos, E.E.; Andreatos, N.; Shehadeh, F.; Ziakas, P.D.; Mylonakis, E. The cost-effectiveness of rapid diagnostic testing for the diagnosis of bloodstream infections with or without antimicrobial stewardship. Clin. Microbiol. Rev. 2018, 31, e00095-17. [Google Scholar] [CrossRef]
- Florio, W.; Tavanti, A.; Ghelardi, E.; Lupetti, A. MALDI-TOF MS applications to the detection of antifungal resistance: State of the art and future perspectives. Front. Microbiol. 2018, 9, 2577. [Google Scholar] [CrossRef]
- Florio, W.; Tavanti, A.; Barnini, S.; Ghelardi, E.; Lupetti, A. Recent advances and ongoing challenges in the diagnosis of microbial infections by MALDI-TOF mass spectrometry. Front. Microbiol. 2018, 9, 1097. [Google Scholar] [CrossRef]
- Vrioni, G.; Tsiamis, C.; Oikonomidis, G.; Theodoridou, K.; Kapsimali, V.; Tsakris, A. MALDI-TOF mass spectrometry technology for detecting biomarkers of antimicrobial resistance: Current achievements and future perspectives. Ann. Transl. Med. 2018, 6, 240. [Google Scholar] [CrossRef] [PubMed]
- Oviaño, M.; Bou, G. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the rapid detection of antimicrobial resistance mechanisms and beyond. Clin. Microbiol. Rev. 2019, 32, e00037-18. [Google Scholar] [CrossRef] [PubMed]
- Sparbier, K.; Schubert, S.; Kostrzewa, M. MBT-ASTRA: A suitable tool for fast antibiotic susceptibility testing? Methods 2016, 104, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Kostrzewa, M.; Sparbier, K.; Maier, T.; Schubert, S. MALDI-TOF MS: An upcoming tool for rapid detection of antibiotic resistance in microorganisms. Proteomics-Clin. Appl. 2013, 7, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Ballesté, R.N. Proteomics: Technology and Applications. In The Use of Mass Spectrometry Technology (MALDI-TOF) in Clinical Microbiology; Academic Press: London, UK, 2018; pp. 1–17. [Google Scholar]
- Edwards-Jones, V.; Claydon, M.A.; Evason, D.J.; Walker, J.; Fox, A.J.; Gordon, D.B. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J. Med. Microbiol. 2000, 49, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Song, Y.; Wang, J.; Yang, R.; Guo, Z. Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 2002, 74, 5487–5491. [Google Scholar] [CrossRef]
- Hindre, T.; Didelot, S.; Le Pennec, J.-P.; Haras, D.; Dufour, A.; Vallee-Rehel, K. Bacteriocin detection from whole bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 2003, 69, 1051–1058. [Google Scholar] [CrossRef][Green Version]
- Camara, J.E.; Hays, F.A. Discrimination between wild-type and ampicillin-resistant Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Bioanal. Chem. 2007, 389, 1633–1638. [Google Scholar] [CrossRef]
- Nagy, E.; Becker, S.; Sóki, J.; Urbán, E.; Kostrzewa, M. Differentiation of division I (cfiA-negative) and division II (cfiA-positive) Bacteroides fragilis strains by matrix-assisted laser desorption/ionization time of-flight mass spectrometry. J. Med. Microbiol. 2011, 60, 1584–1590. [Google Scholar] [CrossRef]
- Cai, J.C.; Hu, Y.Y.; Zhang, R.; Zhou, H.W.; Chen, G.-X. Detection of OmpK36 porin loss in Klebsiella spp. by matrix-assisted laser desorption ionization time of flight mass spectrometry. J. Clin. Microbiol. 2012, 50, 2179–2182. [Google Scholar] [CrossRef]
- Griffin, P.M.; Price, G.R.; Schooneveldt, J.M.; Schlebusch, S.; Tilse, M.H.; Urbanski, T.; Hamilton, B.; Venter, D. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J. Clin. Microbiol. 2012, 50, 2918–2931. [Google Scholar] [CrossRef] [PubMed]
- Griffin, P.M.; Price, G.R.; Schooneveldt, J.M.; Schlebusch, S.; Urbanski, T.; Hamilton, B.; Venter, D. Rapid identification of VRE with MALDI-TOF MS. Pathology 2013, 45, S57. [Google Scholar] [CrossRef]
- Lau, A.F.; Wang, H.; Weingarten, R.A.; Drake, S.K.; Suffredini, A.F.; Garfield, M.K.; Chen, Y.; Gucek, M.; Youn, J.H.; Stock, F.; et al. A rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for single-plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 2014, 52, 2804–2812. [Google Scholar] [CrossRef] [PubMed]
- Penny, C.; Grothendick, B.; Zhang, L.; Borror, C.M.; Barbano, D.; Cornelius, A.J.; Gilpin, B.J.; Fagerquist, C.K.; Zaragoza, W.J.; Jay-Russell, M.T. A designed experiments approach to optimizing MALDI-TOF MS spectrum processing parameters enhances detection of antibiotic resistance in Campylobacter jejuni. Front. Microbiol. 2016, 7, 818. [Google Scholar] [CrossRef]
- Rhoads, D.D.; Wang, H.; Karichu, J.; Richter, S.S. The presence of a single MALDI-TOF mass spectral peak predicts methicillin resistance in staphylococci. Diagn. Microbiol. Infect. Dis. 2016, 86, 257–261. [Google Scholar] [CrossRef]
- Dortet, L.; Bonnin, R.A.; Pennisi, I.; Gauthier, L.; Jousset, A.B.; Dabos, L.; Furniss, R.C.D.; Mavridou, D.A.I.; Bogaerts, P.; Glupczynski, Y.; et al. Rapid detection and discrimination of chromosome- and MCR-plasmid-mediated resistance to polymyxins by MALDI-TOF MS in Escherichia coli: The MALDIxin test. J. Antimicrob. Chemother. 2018, 73, 3359–3367. [Google Scholar] [CrossRef]
- Figueroa-Espinosa, R.; Costa, A.; Cejas, D.; Barrios, R.; Vay, C.; Radice, M.; Gutkind, G.; Conza, J. Di MALDI-TOF MS based procedure to detect KPC-2 directly from positive blood culture bottles and colonies. J. Microbiol. Methods 2019, 159, 120–127. [Google Scholar] [CrossRef]
- Jeverica, S.; Sóki, J.; Premru, M.M.; Nagy, E.; Papst, L. High prevalence of division II (cfiA positive) isolates among blood stream Bacteroides fragilis in Slovenia as determined by MALDI-TOF MS. Anaerobe 2019, 58, 30–34. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.-H.; McDermott, P.F. Validating the NCBI AMRFinder tool and resistance gene database using antimicrobial resistance genotype-phenotype correlation in a collection of NARMS isolates. Antimicrob. Agents Chemother. 2019, 63, 550707. [Google Scholar] [CrossRef]
- Yang, Y.; Jiang, X.; Chai, B.; Ma, L.; Li, B.; Zhang, A.; Cole, J.R.; Tiedje, J.M.; Zhang, T. ARGs-OAP: Online analysis pipeline for antibiotic resistance genes detection from metagenomic data using an integrated structured ARG-database. Bioinformatics 2016, 32, 2346–2351. [Google Scholar] [CrossRef] [PubMed]
- Rowe, W.; Baker, K.S.; Verner-Jeffreys, D.; Baker-Austin, C.; Ryan, J.J.; Maskell, D.; Pearce, G. Search engine for antimicrobial resistance: A cloud compatible pipeline and web interface for rapidly detecting antimicrobial resistance genes directly from sequence data. PLoS ONE 2015, 10, e0133492. [Google Scholar] [CrossRef] [PubMed]
- Argoty, G.A.A.; Guron, G.K.; Garner, E.; Riquelme, M.V.; Heath, L.; Pruden, A.; Vikesland, P.; Zhang, L. ARG-miner: A web platform for crowdsourcing-based curation of antibiotic resistance genes. BioRxiv 2018, 274282. [Google Scholar] [CrossRef]
- Huang, Y.; Li, W.; Liu, K.; Xiong, C.; Cao, P.; Tao, J. New detection method in experimental mice for schistosomiasis: ClinProTool and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Parasitol. Res. 2016, 115, 4173–4181. [Google Scholar] [CrossRef]
- Hoyos-Mallecot, Y.; Riazzo, C.; Miranda-Casas, C.; Rojo-Martín, M.D.; Gutiérrez-Fernández, J.; Navarro-Marí, J.M. Rapid detection and identification of strains carrying carbapenemases directly from positive blood cultures using MALDI-TOF MS. J. Microbiol. Methods 2014, 105, 98–101. [Google Scholar] [CrossRef]
- Vranckx, K.; De Bruyne, K.; Pot, B. Analysis of MALDI-TOF MS Spectra using the BioNumerics Software. In MALDI-TOF and Tandem MS for Clinical Microbiology, First Edition; Gharbia, H.N.S.S.E., Ed.; John Wiley & Sons Ltd.: Oxford, UK, 2017; pp. 539–562. [Google Scholar]
- Tracz, D.M.; Tyler, A.D.; Cunningham, I.; Antonation, K.S.; Corbett, C.R. Custom database development and biomarker discovery methods for MALDI-TOF mass spectrometry-based identification of high-consequence bacterial pathogens. J. Microbiol. Methods 2017, 134, 54–57. [Google Scholar] [CrossRef]
- Rodrigues, C.; Passet, V.; Brisse, S.; Brisse, S. Identification of Klebsiella pneumoniae complex members using MALDI-TOF mass spectrometry. Front. Microbiol. 2018. [Google Scholar] [CrossRef]
- Pinto, T.C.A.; Costa, N.S.; Castro, L.F.S.; Ribeiro, R.L.; Botelho, A.C.N.; Neves, F.P.G.; Peralta, J.M.; Teixeira, L.M. Potential of MALDI-TOF MS as an alternative approach for capsular typing Streptococcus pneumoniae isolates. Sci. Rep. 2017, 7, 45572. [Google Scholar] [CrossRef]
- Huber, C.A.; Pflüger, V.; Reed, S.; Cottrell, K.; Sidjabat, H.E.; Ranasinghe, A.; Zowawi, H.M.; Harris, P.; Paterson, D.L. Bacterial identification using a SCIEX 5800 TOF/TOF MALDI research instrument and an external database. J. Microbiol. Methods 2019, 164, 105685. [Google Scholar] [CrossRef]
- Karlsson, R.; Gonzales-Siles, L.; Boulund, F.; Lindgren, Å.; Svensson-Stadler, L.; Karlsson, A.; Kristiansson, E.; Moore, E.R.B. Proteotyping: Tandem mass spectrometry shotgun proteomic characterization and typing of pathogenic microorganisms. In MALDI-TOF and Tandem MS for Clinical Microbiology; John Wiley & Sons Ltd.: Oxford, UK, 2017; pp. 419–450. [Google Scholar]
- Welker, M.; Moore, E.R.B. Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst. Appl. Microbiol. 2011, 34, 2–11. [Google Scholar] [CrossRef]
- Greaves, J.; Roboz, J.; Roboz, J. Mass Spectrometry for the Novice, First Edition; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Yergey, A.L.; Coorssen, J.R.; Backlund, P.S.; Blank, P.S.; Humphrey, G.A.; Zimmerberg, J.; Campbell, J.M.; Vestal, M.L. De novo sequencing of peptides using MALDI/TOF-TOF. J. Am. Soc. Mass Spectrom. 2002, 13, 784–791. [Google Scholar] [CrossRef]
- Pusch, W.; Flocco, M.T.; Leung, S.-M.; Thiele, H.; Kostrzewa, M. Mass spectrometry-based clinical proteomics. Pharmacogenomics 2003, 4, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Pieper, R.; Gatlin-Bunai, C.L.; Mongodin, E.F.; Parmar, P.P.; Huang, S.T.; Clark, D.J.; Fleischmann, R.D.; Gill, S.R.; Peterson, S.N. Comparative proteomic analysis of Staphylococcus aureus strains with differences in resistance to the cell wall-targeting antibiotic vancomycin. Proteomics 2006, 6, 4246–4258. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.J.; Mkrtchyan, H.V.; Xu, Z.; Serafim, V.; Shah, H.N. Determination of antimicrobial resistance using tandem mass spectrometry. In MALDI-TOF and Tandem MS for Clinical Microbiology; John Wiley & Sons Ltd.: Oxford, UK, 2017; pp. 399–418. [Google Scholar]
- Welker, M.; van Belkum, A.; Girard, V.; Charrier, J.-P.; Pincus, D. An Update on the Routine Application of MALDI-TOF MS in Clinical Microbiology. Expert Rev Proteomics 2019, 8, 695–710. [Google Scholar] [CrossRef]
- Bruker. Bruker Launches MALDI Biotyper Sirius at ASM Microbe Conference. Available online: https://www.prnewswire.com/news-releases/bruker-launches-maldi-biotyper-sirius-at-asm-microbe-conference-300871836.html (accessed on 22 June 2019).
- Bruker. Innovation with Integrity Changing Microbiology MALDI Biotyper ® Subtyping Module. Available online: https://www.bruker.com/fileadmin/user_upload/8-PDF-Docs/Separations_MassSpectrometry/Literature/Brochures/1851663_MBT_Subtyping_brochure_04-2017_ebook.pdf (accessed on 10 September 2019).
- Cordovana, M.; Pranada, A.B.; Ambretti, S.; Kostrzewa, M. MALDI-TOF bacterial subtyping to detect antibiotic resistance. Clin. Mass Spectrom. 2019, in press. [Google Scholar] [CrossRef]
- Youn, J.-H.; Drake, S.K.; Weingarten, R.A.; Frank, K.M.; Dekker, J.P.; Lau, A.F. Clinical performance of a matrix-assisted laser desorption ionization time of flight mass spectrometry method for detection of certain bla KPC -containing plasmids. J. Clin. Microbiol. 2016, 54, 35–42. [Google Scholar] [CrossRef]
- Sydenham, T.V.; Sóki, J.; Hasman, H.; Wang, M.; Justesen, U.S. Identification of antimicrobial resistance genes in multidrug-resistant clinical Bacteroides fragilis isolates by whole genome shotgun sequencing. Anaerobe 2015, 31, 59–64. [Google Scholar] [CrossRef]
- Metcalf, B.J.; Chochua, S.; Gertz, R.E.; Li, Z.; Walker, H.; Tran, T.; Hawkins, P.A.; Glennen, A.; Lynfield, R.; Li, Y.; et al. Using whole genome sequencing to identify resistance determinants and predict antimicrobial resistance phenotypes for year 2015 invasive pneumococcal disease isolates recovered in the United States. Clin. Microbiol. Infect. 2016, 22, 1002.e1–1002.e8. [Google Scholar] [CrossRef]
- Tyson, G.H.; McDermott, P.F.; Li, C.; Chen, Y.; Tadesse, D.A.; Mukherjee, S.; Bodeis-Jones, S.; Kabera, C.; Gaines, S.A.; Loneragan, G.H.; et al. WGS accurately predicts antimicrobial resistance in Escherichia coli. J. Antimicrob. Chemother. 2015, 70, 2763–2769. [Google Scholar] [CrossRef]
- Schlebusch, S.; Price, G.R.; Gallagher, R.L.; Horton-Szar, V.; Elbourne, L.D.H.; Griffin, P.; Venter, D.J.; Jensen, S.O.; Van Hal, S.J. MALDI-TOF MS meets WGS in a VRE outbreak investigation. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 495–499. [Google Scholar] [CrossRef]
- Eriksen, H.B.; Gumpert, H.; Faurholt, C.H.; Westh, H. Determination of Elizabethkingia diversity by MALDI-TOF mass spectrometry and whole-genome sequencing. Emerg. Infect. Dis. 2017, 23, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Batz, M.B.; Hoffmann, S.A.; Morris, J.G. Ranking the Risks: The 10 Pathogen-Food Combinations with the Greatest Burden on Public Health. Available online: https://folio.iupui.edu/bitstream/handle/10244/1022/72267report.pdf?sequence=1 (accessed on 10 September 2019).
- Zautner, A.E.; Masanta, W.O.; Tareen, A.M.; Weig, M.; Lugert, R.; Groß, U.; Bader, O. Discrimination of multilocus sequence typing-based Campylobacter jejuni subgroups by MALDI-TOF mass spectrometry. BMC Microbiol. 2013, 13, 247. [Google Scholar] [CrossRef] [PubMed]
- Emele, M.F.; Karg, M.; Hotzel, H.; Graaf-van Bloois, L.; Groß, U.; Bader, O.; Zautner, A.E. Differentiation of Campylobacter fetus subspecies by proteotyping. Eur. J. Microbiol. Immunol. 2019, 9, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Emele, M.F.; Možina, S.S.; Lugert, R.; Bohne, W.; Masanta, W.O.; Riedel, T.; Groß, U.; Bader, O.; Zautner, A.E. Proteotyping as alternate typing method to differentiate Campylobacter coli clades. Sci. Rep. 2019, 9, 4244. [Google Scholar] [CrossRef] [PubMed]
- Zautner, A.E.; Lugert, R.; Masanta, W.O.; Weig, M.; Groß, U.; Bader, O. Subtyping of Campylobacter jejuni ssp. doylei isolates using mass spectrometry-based phyloproteomics (MSPP). J. Vis. Exp. 2016, 116, e54165. [Google Scholar]
- Guarino, A.; Bruzzese, E.; Giannattasio, A. Antibiotic treatment of acute gastroenteritis in children. F1000Research 2018, 7, 193. [Google Scholar]
- World Health Organization. Critically Important Antimicrobials for Human Medicine 6th Revision 2018. Ranking of Medically Important Antimicrobials for Risk Management of Antimicrobial Resistance Due to Non-Human Use. Available online: https://www.who.int/foodsafety/publications/antimicrobials-sixth/en/ (accessed on 5 September 2019).
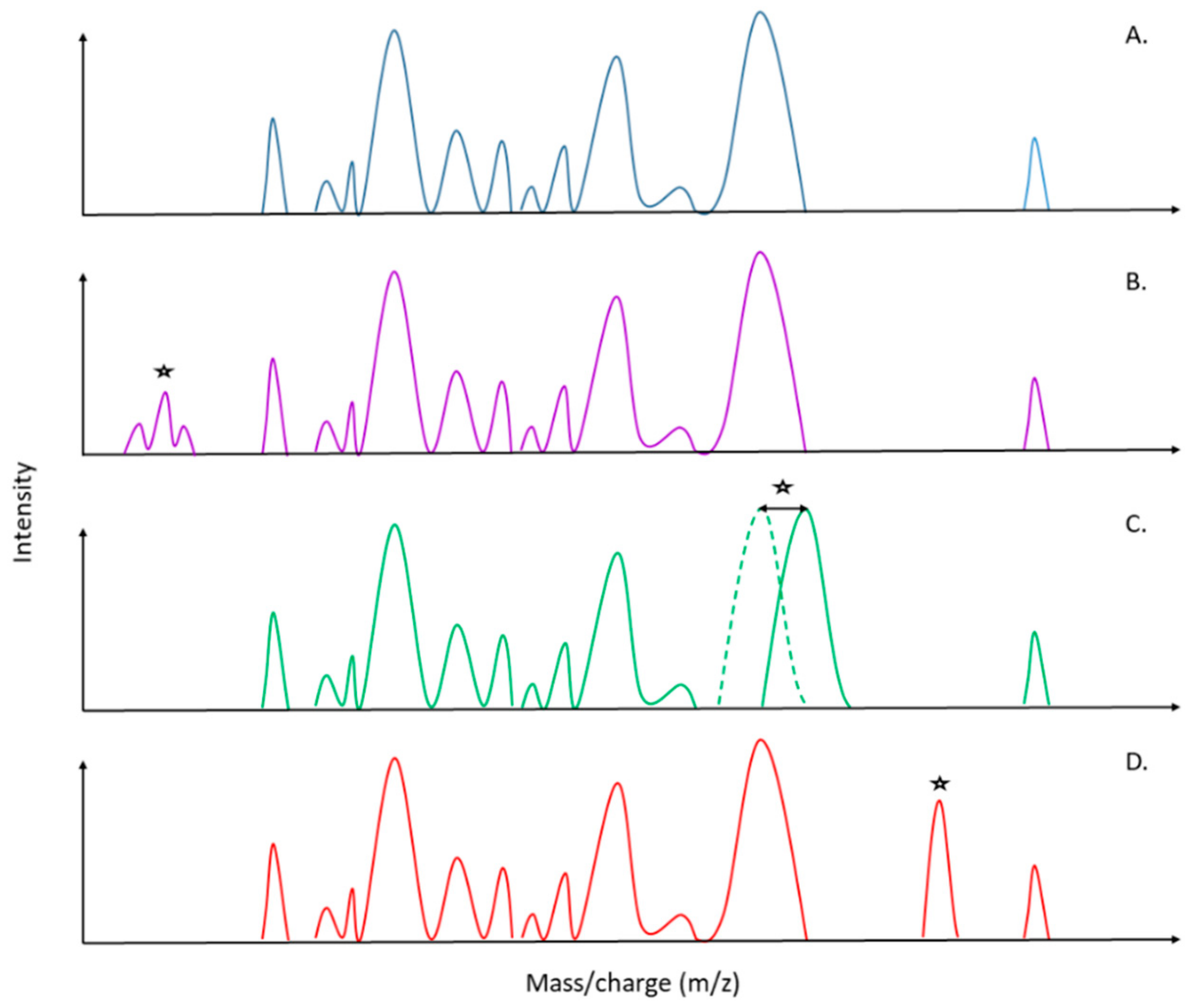
| Organism | Antibiotic Classes Tested | Biomarkers | Year | Reference |
|---|---|---|---|---|
| Staphylococcus aureus | β-lactams | MRSA: 891, 1140, 1165, 1229 and 2127 m/z MSSA: 2548 and 2647 m/z | 2000 | [51] |
| Staphylococcus aureus | β-lactams | Variation between in the spectral profiles in the mass range of m/z 500–3500 Da | 2002 | [52] |
| Lactococcus lactis Bacillus coagulans Escherichia coli | Bacteriocins (lantibiotic) | Lacticin 481: 2902, 2924,2940 m/z Nisin A: 3392 m/z Coagulin: 4650 m/z | 2003 | [53] |
| Escherichia coli | β-lactams | Ampicillin: 29.000 m/z | 2007 | [54] |
| Bacteroides fragilis | Carbapenems | cfiA negative: 4711, 4817, 5017, 5204, 5268 m/z cfiA positive: 4688, 4826, 5002, 5189, 5282 m/z | 2011 | [55] |
| Klebsiella spp. | Carbapenems | OmpK36 porin: 38000, 19000 m/z | 2012 | [56] |
| Enterococcus faecium | Glycopeptides | VanA/B: 6603 m/z | 2012 | [57,58] |
| Enterobacteriaceae | Carbapenems | blaKPC: 11109 m/z | 2014 | [59] |
| Campylobacter jejuni | β-lactams Tetracyclines Glycopeptides | Spectrum processing parameters increased the resistance detection | 2016 | [60] |
| Staphylococcus aureus Staphylococcus epidermidis | β-lactams | mecA: 2415 m/z | 2016 | [61] |
| Escherichia coli | Polymyxin | Lipid A modification: 1919 m/z | 2018 | [62] |
| Klebsiella pneumonia Enterobacter cloacae Escherichia coli Serratia marcescens Citrobacter braakii, Pseudomonas aeruginosa | Carbapenems | KPC-2: 28544 m/z | 2019 | [63] |
| Bacteroides fragilis | Carbapenems | Identification of B. fragilis with the validated “cfiA library” [55] | 2019 | [64] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feucherolles, M.; Cauchie, H.-M.; Penny, C. MALDI-TOF Mass Spectrometry and Specific Biomarkers: Potential New Key for Swift Identification of Antimicrobial Resistance in Foodborne Pathogens. Microorganisms 2019, 7, 593. https://doi.org/10.3390/microorganisms7120593
Feucherolles M, Cauchie H-M, Penny C. MALDI-TOF Mass Spectrometry and Specific Biomarkers: Potential New Key for Swift Identification of Antimicrobial Resistance in Foodborne Pathogens. Microorganisms. 2019; 7(12):593. https://doi.org/10.3390/microorganisms7120593
Chicago/Turabian StyleFeucherolles, Maureen, Henry-Michel Cauchie, and Christian Penny. 2019. "MALDI-TOF Mass Spectrometry and Specific Biomarkers: Potential New Key for Swift Identification of Antimicrobial Resistance in Foodborne Pathogens" Microorganisms 7, no. 12: 593. https://doi.org/10.3390/microorganisms7120593
APA StyleFeucherolles, M., Cauchie, H.-M., & Penny, C. (2019). MALDI-TOF Mass Spectrometry and Specific Biomarkers: Potential New Key for Swift Identification of Antimicrobial Resistance in Foodborne Pathogens. Microorganisms, 7(12), 593. https://doi.org/10.3390/microorganisms7120593