Comparison of Six Commercial Meat Starter Cultures for the Fermentation of Yellow Mealworm (Tenebrio molitor) Paste
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Materials
2.2. Experimental Design, Fermentation Procedure, and Online pH Measurement
2.3. Microbiological Plate Counts
2.4. Bacterial Community Analysis Yielding Relative Abundances
2.5. Quantitative Real-Time PCR Amplification to Determine Absolute Abundances
2.6. Metabolite Target Analysis
2.7. Statistical Analyses
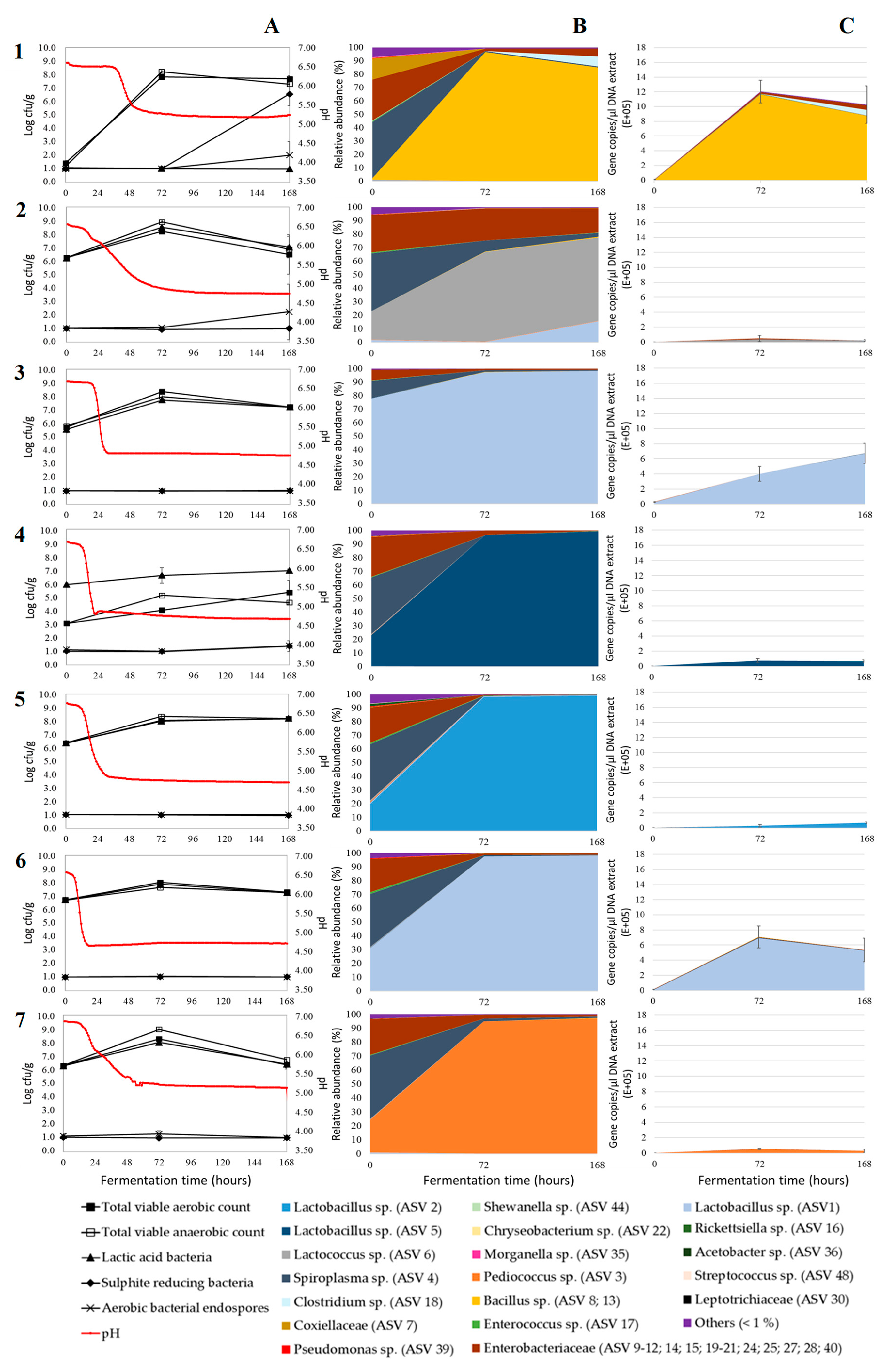
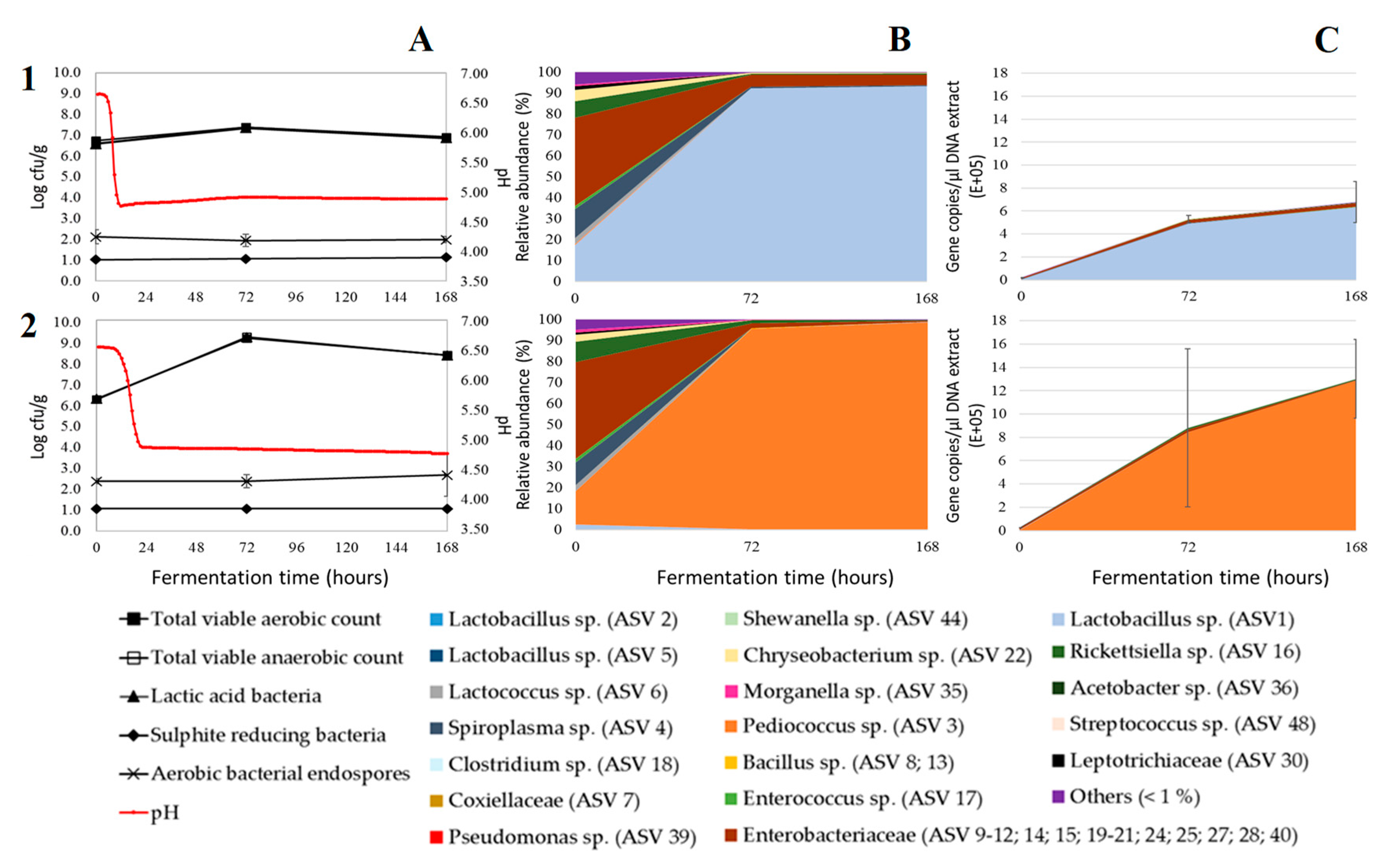
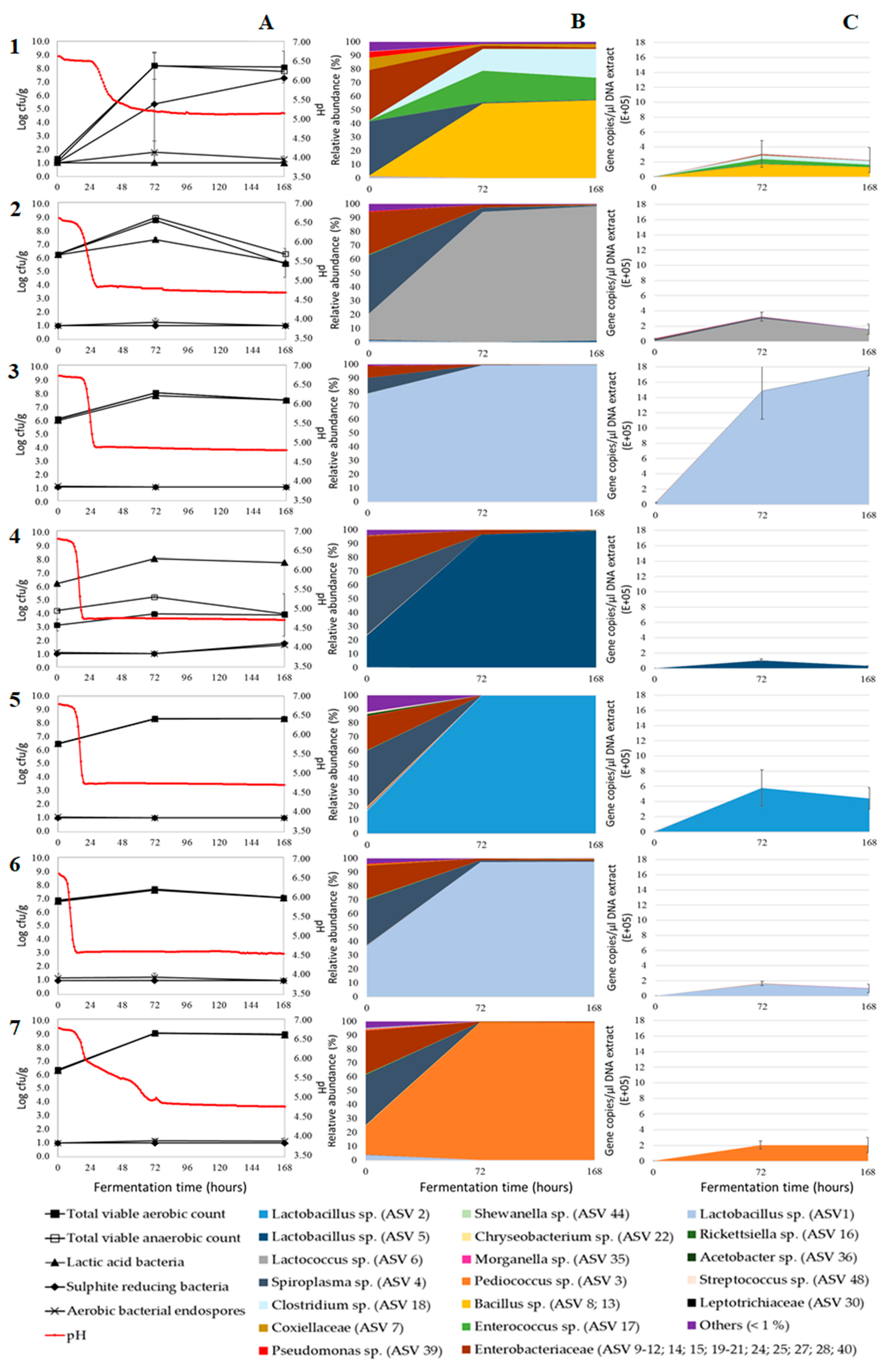
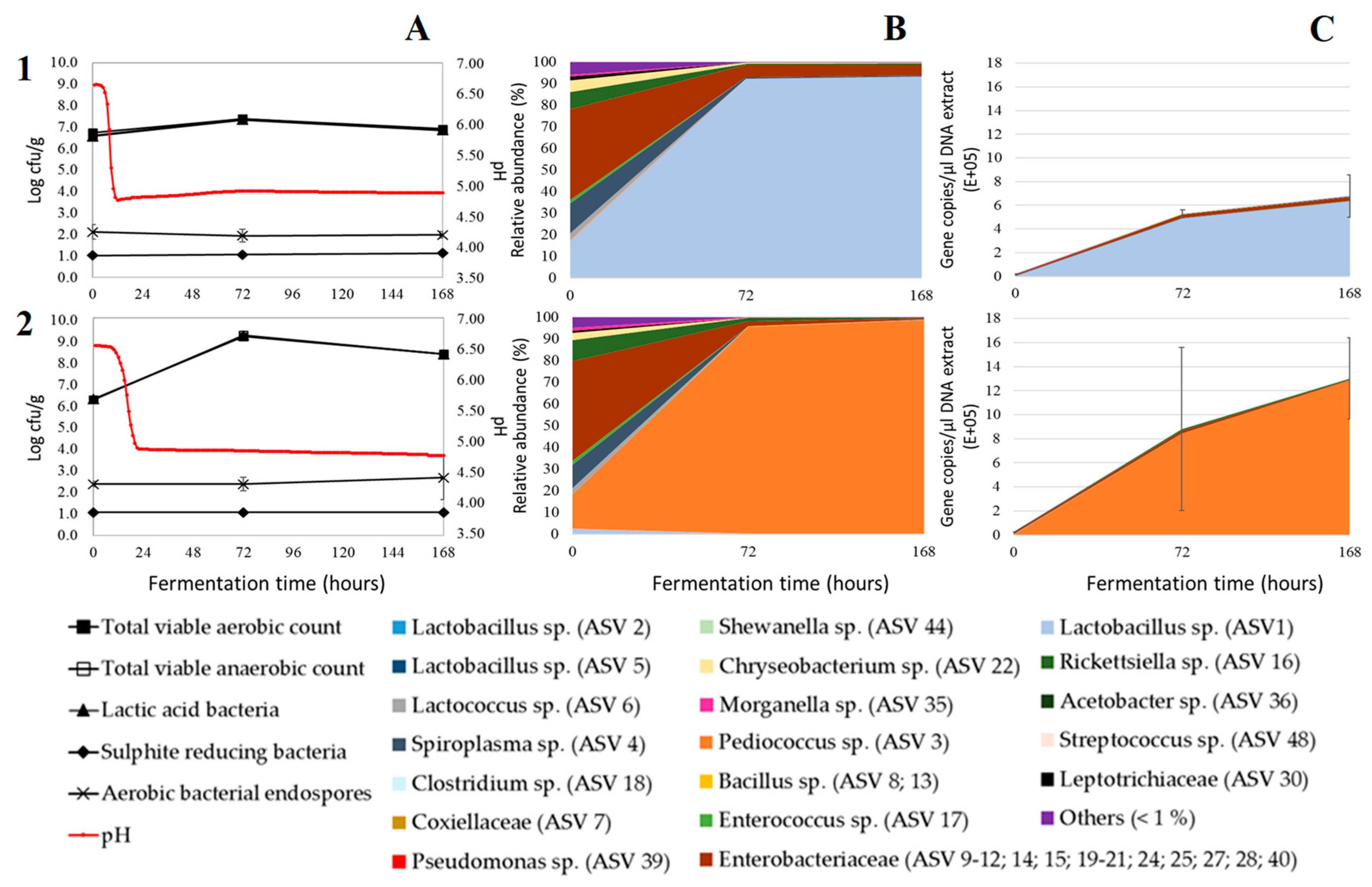
3. Results and Discussion
3.1. Acidification
3.2. Microbial Plate Counts
3.3. Relative Bacterial Abundances and Bacterial Diversity
3.4. Absolute Bacterial Abundances
3.5. Consumption of Glucose and Production of Specific Amino Acids
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Observed ASV 1 Richness | Shannon-Wiener 2 | ||||||
|---|---|---|---|---|---|---|---|
| Day 0 | Day 3 | Day 7 | Day 0 | Day 3 | Day 7 | ||
| Batch 1 | Control | 78.67 ± 2.52 a | 30.33 ± 2.31 b | 28.00 ± 4.00 b | 2.02 ± 0.17 a | 0.89 ± 0.05 b | 0.77 ± 0.19 b |
| Lactococcus lactis | 77.33 ± 7.09 a | 43.67 ± 8.08 b | 43.33 ± 23.09 b | 2.03 ± 0.11 a | 0.96 ± 0.17 b | 0.95 ± 0.21 b | |
| Lactobacillus curvatus | 47.00 ± 2.83 a | 22.33 ± 2.08 b | 20.00 ± 3.46 b | 0.89 ± 0.04 a | 0.17 ± 0.01 b | 0.11 ± 0.02 b | |
| Lactobacillus farciminis | 83.00 ± 10.00 a | 22.33 ± 5.03 b | 16.67 ± 4.04 b | 1.91 ± 0.05 a | 0.07 ± 0.02 b | 0.04 ± 0.01 b | |
| Lactobacillus plantarum | 87.33 ± 9.02 a | 30.67 ± 3.06 b | 20.33 ± 2.08 b | 2.18 ± 0.09 a | 0.14 ± 0.01 b | 0.06 ± 0.01 b | |
| Lactobacillus sakei | 73.33 ± 19.60 a,A | 24.00 ± 4.58 b,A | 20.33 ± 2.08 b,A | 1.87 ± 0.07 a,A | 0.16 ± 0.02 b,A | 0.13 ± 0.01 b,A | |
| Pediococcus acidilactici | 71.33 ± 3.21 a,C | 43.67 ± 5.51 b,C | 33.33 ± 4.51 b,C | 1.83 ± 0.05 a,C | 0.31 ± 0.07 b,C | 0.19 ± 0.03 b,C | |
| Batch 2 | Lactobacillus sakei | 116.00 ± 12.77 a,B | 51.67 ± 8.33 b,B | 56.67 ± 10.60 b,B | 2.83 ± 0.06 a,B | 0.88 ± 0. 41 b,B | 1.16 ± 0.32 b,B |
| Pediococcus acidilactici | 89.00 ± 17.35 a,D | 42.67 ± 11.37 b,C | 39.67 ± 0.58 b,D | 2.82 ± 0.05 a,D | 0.46 ± 0.29 b,D | 0.40 ± 0.08 b,D | |
| Batch 1 | Batch 2 | ||||
|---|---|---|---|---|---|
| Asp 2 | Glu 2 | Asp 2 | Glu 2 | ||
| Day 0 | 4.32 ± 0.89 | 18.06 ± 2.97 | 4.05 ± 0.46 | 21.08 ± 3.55 | |
| Day 7 | Control | 2.64 ± 0.23 | 16.43 ± 1.49 | ||
| Lactococcus lactis | 14.83 ± 1.77 | 21.08 ± 0.90 | |||
| Lactobacillus curvatus | 1.85 ± 0.20 | 26.11 ± 3.65 | |||
| Lactobacillus farciminis | 9.72 ± 0.42 | 29.31 ± 0.82 | |||
| Lactobacillus plantarum | 4.84 ± 0.42 | 19.04 ± 0.36 | |||
| Lactobacillus sakei | 1.23 ± 0.09 | 32.66 ± 1.83 | 2.68 ± 0.32 | 36.70 ± 2.76 | |
| Pediococcus acidilactici | 1.80 ± 0.14 | 37.12 ± 0.09 | 2.91 ± 0.27 | 32.80 ± 0.81 | |
References
- De Smet, J.; Lenaerts, S.; Borremans, A.; Scholliers, J.; Van Der Borght, M.; Van Campenhout, L. Stability assessment and laboratory scale fermentation of pastes produced on a pilot scale from mealworms (Tenebrio molitor). LWT 2019, 102, 113–121. [Google Scholar] [CrossRef]
- Deroy, O.; Reade, B.; Spence, C. The insectivore’s dilemma, and how to take the West out of it. Food Qual. Prefer. 2015, 44, 44–55. [Google Scholar] [CrossRef]
- Hartmann, C.; Shi, J.; Giusto, A.; Siegrist, M. The psychology of eating insects: A cross-cultural comparison between Germany and China. Food Qual. Prefer. 2015, 44, 148–156. [Google Scholar] [CrossRef]
- Schouteten, J.J.; De Steur, H.; De Pelsmaeker, S.; Lagast, S.; Juvinal, J.G.; De Bourdeaudhuij, I.; Verbeke, W.; Gellynck, X. Emotional and sensory profiling of insect-, plant-, and meat-based burgers under blind, expected and informed conditions. Food Qual. Prefer. 2016, 52, 27–31. [Google Scholar] [CrossRef]
- Tan, H.S.G.; Fischer, A.R.H.; Tinchan, P.; Stieger, M.; Steenbekkers, L.P.A.; van Trijp, H.C.M. Insects as food: Exploring cultural exposure and individual experience as determinants of acceptance. Food Qual. Prefer. 2015, 42, 78–89. [Google Scholar] [CrossRef]
- Laranjo, M.; Potes, M.E.; Elias, M. Role of starter cultures on the safety of fermented meat products. Front. Microbiol. 2019, 10, 853. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.J.; Schieber, A.; Gänzle, M.G. Formation of taste-active amino acids, amino acid derivatives and peptides in food fermentation—A review. Food Res. Int. 2016, 89, 39–47. [Google Scholar] [CrossRef]
- Klunder, H.C.; Wolkers-Rooijackers, J.; Korpela, J.M.; Nout, M.J.R. Microbiological aspects of processing and storage of edible insects. Food Control 2012, 26, 628–631. [Google Scholar] [CrossRef]
- Borremans, A.; Lenaerts, S.; Crauwels, S.; Lievens, B.; Van Campenhout, L. Marination and fermentation of yellow mealworm larvae (Tenebrio molitor). Food Control 2018, 92, 47–52. [Google Scholar] [CrossRef]
- Hugas, M.; Monfort, J.M. Bacterial starter cultures for meat fermentation. Food Chem. 1997, 59, 547–554. [Google Scholar] [CrossRef]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Fadda, S.; López, C.; Vignolo, G. Role of lactic acid bacteria during meat conditioning and fermentation: Peptides generated as sensorial and hygienic biomarkers. Meat Sci. 2010, 86, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Katla, A.K.; Kruse, H.; Johnsen, G.; Herikstad, H. Antimicrobial susceptibility of starter culture bacteria used in Norwegian dairy products. Int. J. Food Microbiol. 2001, 67, 147–152. [Google Scholar] [CrossRef]
- Castellano, P.; Belfiore, C.; Fadda, S.; Vignolo, G. A review of bacteriocinogenic lactic acid bacteria used as bioprotective cultures in fresh meat produced in Argentina. Meat Sci. 2008, 79, 483–499. [Google Scholar] [CrossRef]
- Verluyten, J.; Messens, W.; de Vuyst, L. The curing agent sodium nitrite, used in the production of fermented sausages, is less inhibiting to the bacteriocin-producing meat starter culture Lactobacillus curvatus LTH 1174 under anaerobic conditions. Appl. Environ. Microbiol. 2003, 69, 3833–3839. [Google Scholar] [CrossRef]
- Nowak, V.; Persijn, D.; Rittenschober, D.; Charrondiere, U.R. Review of food composition data for edible insects. Food Chem. 2016, 193, 39–46. [Google Scholar] [CrossRef]
- Hutkins, R.W. Microbiology and Technology of Fermented Foods; Blackwell Publishing Ltd.: Chicago, IL, USA, 2006. [Google Scholar] [CrossRef]
- Dijk, R.; van den Berg, D.; Beumer, R.; de Boer, E.; Dijkstra, A.; Mout, L.; Stegeman, H.; Uyttendaele, M.; In’t Veld, S. Microbiologie van Voedingsmiddelen: Methoden, Principes en Criteria, 5th ed.; MYbusinessmedia: Capelle aan den Ijssel, The Netherlands, 2015; pp. 403–516. [Google Scholar]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analysing amplicon sequence data on the MiSeq Illumina Sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. Genome analysis QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS 581 amplicon sequencing. bioRxiv 2016, 081257. [Google Scholar] [CrossRef]
- Edgar, R.C. SINTAX: A simple non-Bayesian taxonomy classifier for 16S and ITS583 sequences. bioRxiv 2016, 074161. [Google Scholar] [CrossRef]
- Altschul, S.; Gish, G.; Miller, W.; Myers, E.W.; Lipman, D. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Leroy, F.; Verluyten, J.; De Vuyst, L. Functional meat starter cultures for improved sausage fermentation. Int. J. Food Microbiol. 2006, 106, 270–285. [Google Scholar] [CrossRef]
- Vandeweyer, D.; Crauwels, S.; Lievens, B.; Van Campenhout, L. Microbial counts of mealworms (Tenebrio molitor) and crickets (Acheta domesticus and Gryllodes sigillatus) from different rearing companies and different production batches. Int. J. Food Microbiol. 2017, 242, 13–18. [Google Scholar] [CrossRef]
- Korkeala, H.; Alanko, T.; Tiusanen, T. Effect of sodium nitrite and sodium chloride on growth of lactic acid bacteria. Acta Vet. Scand. 1992, 33, 27–32. [Google Scholar]
- Verluyten, J.; Leroy, F.; de Vuyst, L. Effects of different spices used in production of fermented sausages on growth of and curvacin A production of Lactobacillus curvatus LTH 1174. Appl. Environ. Microbiol. 2004, 70, 4807–4813. [Google Scholar] [CrossRef]
- Vandeweyer, D.; Lenaerts, S.; Callens, A.; Van Campenhout, L. Effect of blanching followed by refrigerated storage or industrial microwave drying on the microbial load of yellow mealworms (Tenebrio molitor). Food Control 2017, 71, 311–314. [Google Scholar] [CrossRef]
- Leroy, F.; De Vuyst, L. Temperature and pH conditions that prevail during fermentation of sausages are optimal for production of the antilisterial bacteriocin sakacin K. Appl. Environ. Microbiol. 1999, 65, 974–981. [Google Scholar] [PubMed]
- Leroy, F.; Lievens, K.; de Vuyst, L. Interactions of meat-associated bacteriocin-producing Lactobacilli with Listeria innocula under stringent sausage fermentation conditions. J. Food Protect. 2005, 68, 2078–2084. [Google Scholar] [CrossRef] [PubMed]
- Sriphochanart, W.; Skolpap, W. The use of selected lactic acid bacteria starter cultures for improved Thai sausage fermentation. J. Food Process. Preserv. 2010, 35, 291–298. [Google Scholar] [CrossRef]
- Shannon, C. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Farrelly, V.; Rainley, F.A.; Stackebrandt, E. Effect of genome size and rrn gene copy number in PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 1995, 61, 2798–2801. [Google Scholar]
- Garofalo, C.; Osimani, A.; Milanovic, V.; Taccari, M.; Cardinali, F.; Aquilanti, L.; Riolo, P.; Ruschioni, S.; Isidoro, N.; Clementi, F. The microbiota of marketed processed edible insects as revealed by high-throughput sequencing. Food Microbiol. 2017, 62, 15–22. [Google Scholar] [CrossRef]
- Jung, J.; Heo, A.; Park, Y.W.; Kim, Y.J.; Koh, H.; Park, W. Gut microbiota of Tenebrio molitor and their response to environmental change. J. Microbiol. Biotechnol. 2014, 24, 888–897. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y. Investigation of gut-associated bacteria in Tenebrio molitor (Coleoptera: Tenebrionidae) larvae using culture-dependent and DGGE methods. Ann. Entomol. Soc. Am. 2015, 108, 941–949. [Google Scholar] [CrossRef]
- Vandeweyer, D.; Crauwels, S.; Lievens, B.; Van Campenhout, L. Metagenetic analysis of the bacterial communities of edible insects from diverse production cycles at industrial rearing companies. Int. J. Food Microbiol. 2017, 261, 11–18. [Google Scholar] [CrossRef]
- Madigan, M.T.; Martinko, J.M.; Dunlap, P.V.; Clark, D.P. Brock Biology of Microorganisms, 12th ed.; Pearson/Benjamin Cummings: San Francisco, CA, USA, 2009. [Google Scholar]
- Baylis, C.; Uyttendaele, M.; Joosten, H.; Davies, A. The Enterobacteriaceae and Their Significance to the Food Industry; ILSI Europe: Brussels, Belgium, 2011. [Google Scholar]
- Osimani, A.; Milanović, V.; Cardinali, F.; Garofalo, C.; Clementi, F.; Pasquini, M.; Riolo, P.; Ruschioni, S.; Isidoro, N.; Loreto, N.; et al. The bacterial biota of laboratory-reared edible mealworms (Tenebrio molitor L.): From feed to frass. Int. J. Food Microbiol. 2018, 272, 49–60. [Google Scholar] [CrossRef]
- Stoops, J.; Crauwels, S.; Waud, M.; Claes, J.; Lievens, B.; Van Campenhout, L. Microbial community assessment of mealworms (Tenebrio molitor) and grasshoppers (Locusta migratoria migratorioides) sold for human consumption. Food Microbiol. 2016, 53, 122–127. [Google Scholar] [CrossRef] [PubMed]
- M’hir, S.; Minervini, F.; Di Cagno, R.; Chammem, N.; Hamdi, M. Technological, functional and safety aspects of enterococci in fermented vegetable products: A mini-review. Ann. Microbiol. 2012, 62, 469–481. [Google Scholar] [CrossRef]
- Pot, B.; Felis, G.E.; De Bruyne, K.; Tsakalidou, E.; Papadimitriou, K.; Leisner, J.; Vandamme, P. The genus Lactobacillus. In Lactic Acid Bacteria: Biodiversity and Taxonomy; Holzapfel, W.P., Wood, B.J.B., Eds.; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2014; pp. 249–353. [Google Scholar]
- Zheng, J.; Ruan, L.; Sun, M.; Gänzle, M. A genomic view of lactobacilli and pediococci demonstrates that phylogeny matches ecology and physiology. Appl. Environ. Microbiol. 2015, 81, 7233–7243. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Harris, H.M.; McCann, A.; Guo, C.; Argimón, S.; Zhang, W.; Yang, X.; Jeffery, I.B.; Cooney, J.C.; Kagawa, T.F.; et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 2015, 6, 8322. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.T.; Lee, F.L.; Tai, C.J.; Kasai, H. Comparison of gyrB gene sequences, 16S rRNA gene sequences and DNA–DNA hybridization in the Bacillus subtilis group. Int. J. Syst. Evol. Microbiol. 2007, 57, 1846–1850. [Google Scholar] [CrossRef]
- Jung, J.Y.; Lee, S.H.; Kim, J.M.; Park, M.S.; Bae, J.W.; Hahn, Y.; Madsen, E.L.; Jeon, C.O. Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl. Environ. Microbiol. 2011, 77, 2264–2274. [Google Scholar] [CrossRef]
- Park, E.J.; Chang, H.W.; Kim, K.H.; Nam, Y.D.; Roh, S.W.; Bae, J.W. Application of quantitative real-time PCR for enumeration of total bacterial, archaeal, and yeast populations in kimchi. J. Microbiol. 2009, 47, 682–685. [Google Scholar] [CrossRef]
- Acinas, S.G.; Marcelino, L.A.; Klepac-Ceraj, V.; Polz, M.F. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 2004, 186, 2629–2635. [Google Scholar] [CrossRef]
- Chevallier, B.; Hubert, J.C.; Kammerer, B. Determination of chromosome size and number of rrn loci in Lactobacillus plantarum by pulse-field gel electrophoresis. FEMS Microbiol. Lett. 1994, 120, 51–56. [Google Scholar] [CrossRef][Green Version]
- Kleerebezem, M.; Boekhorst, J.; van Kranenburg, R.; Molenaar, D.; Kuipers, O.P.; Leer, R.; Tarchini, R.; Peters, S.A.; Sandbrink, H.M.; Fiers, M.W.; et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 2003, 100, 1990–1995. [Google Scholar] [CrossRef]
- Dudez, A.M.; Chaillou, S.; Hissler, L.; Stentz, R.; Champomier-Vergès, M.C.; Alpert, C.A.; Zagorec, M. Physical and genetic map of the Lactobacillus sakei 23K chromosome. Microbiology 2002, 148, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Panicker, G.; Myers, M.L.; Bej, A.K. Rapid detection of Vibrio vulnificus in shellfish and gulf water by real-time PCR. Appl. Environ. Microbiol. 2004, 70, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Boncristiani, H.; Li, J.; Evans, J.D.; Pettis, J.; Chen, Y. Scientific note on PCR inhibitors in the compound eyes of honey bees, Apis Mellifera. Apidologie 2011, 42, 457–460. [Google Scholar] [CrossRef]
- Shamim, G.; Ranjan, S.K.; Pandey, D.M.; Ramani, R. Biochemistry and biosynthesis of insect pigments. Eur. J. Entomol. 2014, 111, 149–164. [Google Scholar] [CrossRef]
- Morales-Ramos, J.A.; Guadalupe Rojas, M.; Shelby, K.; Coudron, T.A. Nutritional value of pupae versus larvae of Tenebrio molitor (Coleoptera: Tenebrionidae) as food for rearing Podisus maculiventris (Heteroptera: Pentatomidae). J. Econ. Entomol. 2016, 109, 564–571. [Google Scholar] [CrossRef]
- Limsuwan, S.; Visessanguan, W.; Kongkiattikajorn, J. The effects of starter cultures on biogenic amine and free amino acid contents in Nham during fermentation. Nat. Sci. 2007, 41, 363–372. [Google Scholar]
- Triki, M.; Herrero, A.M.; Jiménez-Colmenero, F.; Ruiz-Capillas, C. Quality assessment of fresh meat from several species based on free amino acid and biogenic amine contents during chilled storage. Foods 2018, 7, 132. [Google Scholar] [CrossRef]
| Observed ASV 1 Richness | Shannon-Wiener 2 | ||||||
|---|---|---|---|---|---|---|---|
| Day 0 | Day 3 | Day 7 | Day 0 | Day 3 | Day 7 | ||
| Batch 1 | Control | 92.33 ± 10.21 a | 36.00 ± 4.58 b | 40.00 ± 12.17 b | 2.15 ± 0.18 a | 1.23 ± 0.14 b | 1.26 ± 0.26 b |
| Lactococcus lactis | 68.33 ± 12.58 a | 34.00 ± 5.20 b | 27.00 ± 6.08 b | 2.07 ± 0.13 a | 0.33 ± 0.13 b | 0.17 ± 0.05 b | |
| Lactobacillus curvatus | 63.00 ± 9.17 a | 15.33 ± 4.62 b | 11.33 ± 0.58 b | 0.92 ± 0.01 a | 0.04 ± 0.00 b | 0.03 ± 0.00 b | |
| Lactobacillus farciminis | 78.33 ± 9.24 a | 19.00 ± 4.00 b | 21.00 ± 1.73 b | 1.95 ± 0.04 a | 0.17 ± 0.10 b | 0.06 ± 0.01 b | |
| Lactobacillus plantarum | 100.00 ± 21.70 a | 10.67 ± 1.53 b | 8.33 ± 1.53 b | 2.35 ± 0.34 a | 0.02 ± 0.00 b | 0.02 ± 0.00 b | |
| Lactobacillus sakei | 70.33 ± 3.79 a,A | 23.00 ± 3.61 b,A | 29.00 ± 6.08 b,A | 1.89 ± 0.02 a,A | 0.18 ± 0.02 b,A | 0.18 ± 0.05 b,A | |
| Pediococcus acidilactici | 74.00 ± 3.61 a,C | 17.67 ± 0.58 b,C | 16.00 ± 4.36 b,C | 2.15 ± 0.05 a,C | 0.07 ± 0.03 b,C | 0.07 ± 0.05 b,C | |
| Batch 2 | Lactobacillus sakei | 101.67 ± 21.22 a,B | 45.00 ± 2.65 b,B | 42.33 ± 3.51 b,B | 2.87 ± 0.07 a,B | 0.48 ± 0.05 b,B | 0.42 ± 0.06 b,B |
| Pediococcus acidilactici | 97.33 ± 23.44 a,C | 36.00 ± 11.36 b,D | 30.00 ± 1.00 b,D | 2.87 ± 0.05 a,D | 0.28 ± 0.23 b,D | 0.10 ± 0.00 b,C | |
| Batch 1 | Batch 2 | ||||
|---|---|---|---|---|---|
| Asp 2 | Glu 2 | Asp 2 | Glu 2 | ||
| Day 0 | 4.22 ± 1.00 | 17.76 ± 3.10 | 3.27 ± 0.62 | 18.64 ± 3.49 | |
| Day 7 | Control | 4.77 ± 0.57 | 20.72 ± 0.99 | ||
| Lactococcus lactis | 13.06 ± 0.68 | 0.81 ± 0.07 | |||
| Lactobacillus curvatus | 3.22 ± 0.06 | 26.52 ± 0.20 | |||
| Lactobacillus farciminis | 10.43 ± 0.85 | 35.56 ± 1.91 | |||
| Lactobacillus plantarum | 7.85 ± 0.42 | 19.07 ± 0.13 | |||
| Lactobacillus sakei | 0.73 ± 0.20 | 24.92 ± 5.25 | 1.76 ± 0.01 | 33.08 ± 0.22 | |
| Pediococcus acidilactici | 1.31 ± 0.02 | 37.09 ± 1.14 | 2.37 ± 0.11 | 34.95 ± 0.35 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, B.; Sam, C.; Dries, V.; Ruben, S.; Christel, V.; Mik, V.D.B.; Bart, L.; Leen, V.C. Comparison of Six Commercial Meat Starter Cultures for the Fermentation of Yellow Mealworm (Tenebrio molitor) Paste. Microorganisms 2019, 7, 540. https://doi.org/10.3390/microorganisms7110540
An B, Sam C, Dries V, Ruben S, Christel V, Mik VDB, Bart L, Leen VC. Comparison of Six Commercial Meat Starter Cultures for the Fermentation of Yellow Mealworm (Tenebrio molitor) Paste. Microorganisms. 2019; 7(11):540. https://doi.org/10.3390/microorganisms7110540
Chicago/Turabian StyleAn, Borremans, Crauwels Sam, Vandeweyer Dries, Smets Ruben, Verreth Christel, Van Der Borght Mik, Lievens Bart, and Van Campenhout Leen. 2019. "Comparison of Six Commercial Meat Starter Cultures for the Fermentation of Yellow Mealworm (Tenebrio molitor) Paste" Microorganisms 7, no. 11: 540. https://doi.org/10.3390/microorganisms7110540
APA StyleAn, B., Sam, C., Dries, V., Ruben, S., Christel, V., Mik, V. D. B., Bart, L., & Leen, V. C. (2019). Comparison of Six Commercial Meat Starter Cultures for the Fermentation of Yellow Mealworm (Tenebrio molitor) Paste. Microorganisms, 7(11), 540. https://doi.org/10.3390/microorganisms7110540





