Systematic Culturomics Shows that Half of Chicken Caecal Microbiota Members can be Grown in Vitro Except for Two Lineages of Clostridiales and a Single Lineage of Bacteroidetes
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples for Analysis
2.2. OTU Selection and Definition of In Vitro Growth
2.3. 16S rRNA Alignment
2.4. Ethics Statement
3. Results
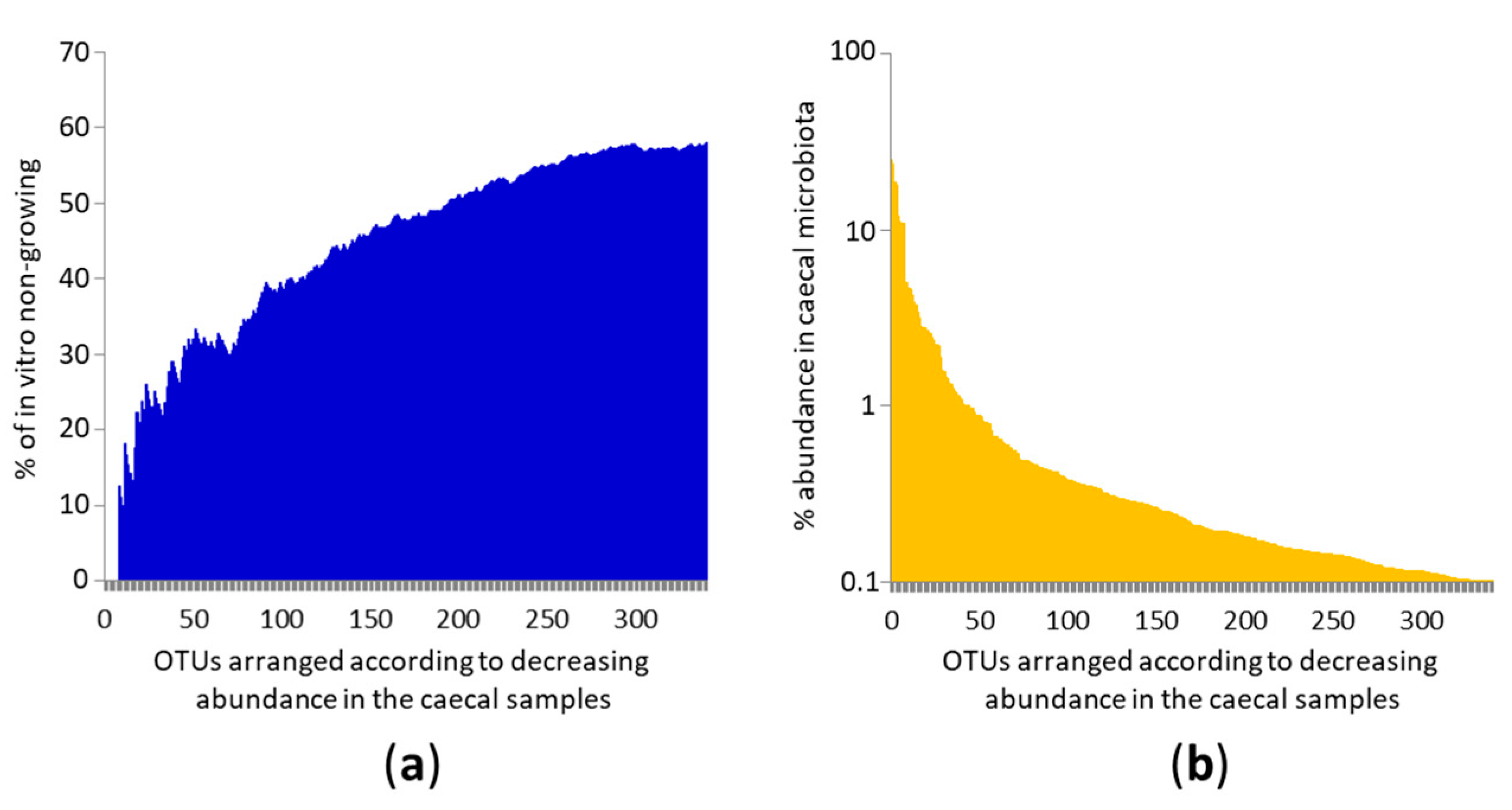
3.1. Identification of OTUs Common to the Chicken Caecum and Their Growth In Vitro
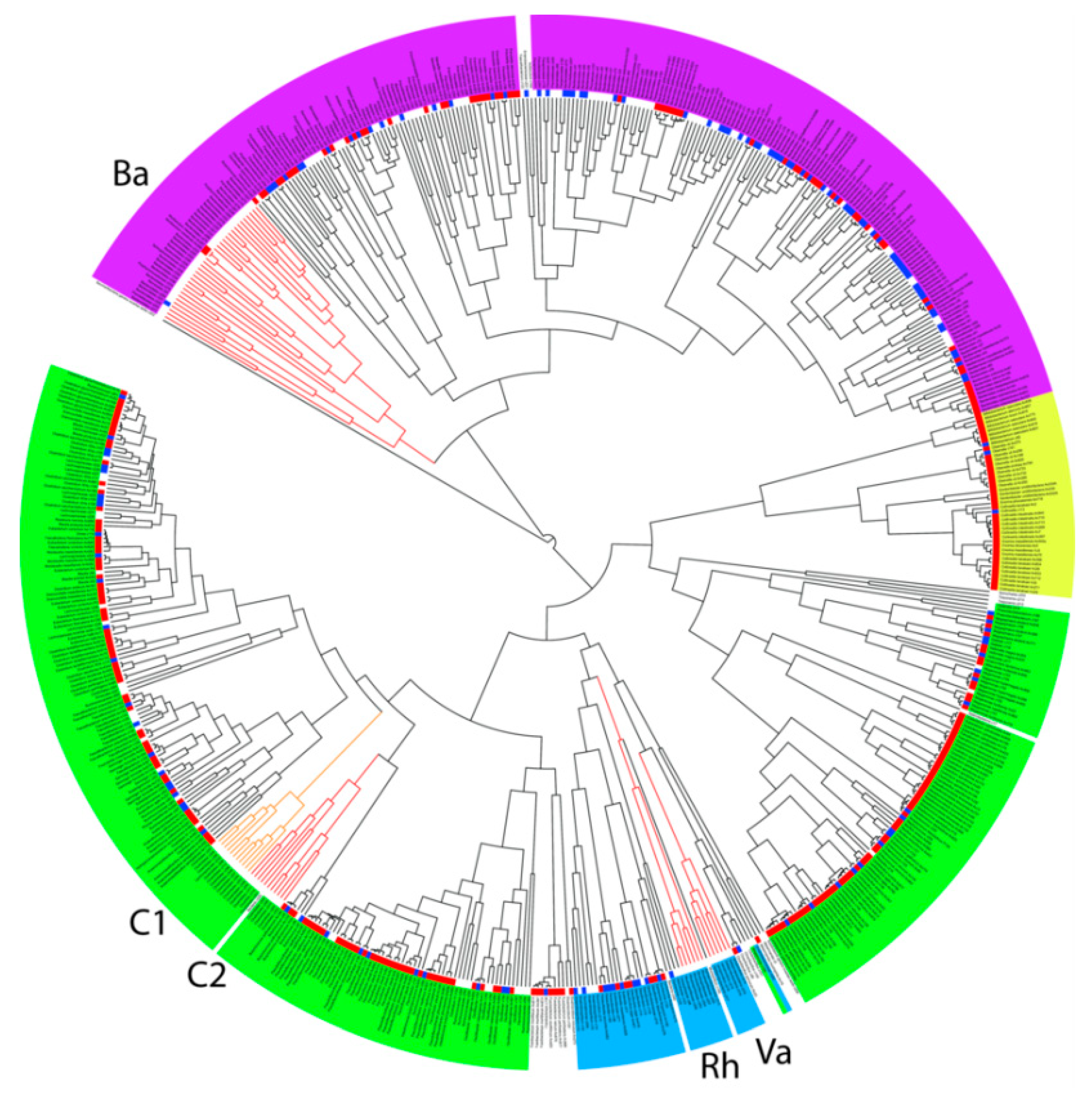
3.2. Specific Analysis of 16S rRNA Sequences
3.3. Culture Conditions Selective for Isolation of Particular Taxa
3.4. Firmicutes
3.5. Bacteroidetes
3.6. Proteobacteria, Actinobacteria and Fusobacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Videnska, P.; Sedlar, K.; Lukac, M.; Faldynova, M.; Gerzova, L.; Cejkova, D.; Sisak, F.; Rychlik, I. Succession and replacement of bacterial populations in the caecum of egg laying hens over their whole life. PLoS ONE 2014, 9, e115142. [Google Scholar] [CrossRef]
- Videnska, P.; Sisak, F.; Havlickova, H.; Faldynova, M.; Rychlik, I. Influence of Salmonella enterica serovar Enteritidis infection on the composition of chicken cecal microbiota. BMC Vet. Res. 2013, 9, 140. [Google Scholar] [CrossRef]
- Colles, F.M.; Cain, R.J.; Nickson, T.; Smith, A.L.; Roberts, S.J.; Maiden, M.C.; Lunn, D.; Dawkins, M.S. Monitoring chicken flock behaviour provides early warning of infection by human pathogen Campylobacter. Proc. Biol. Sci. 2016, 283, 20152323. [Google Scholar]
- Kraimi, N.; Dawkins, M.; Gebhardt-Henrich, S.G.; Velge, P.; Rychlik, I.; Volf, J.; Creach, P.; Smith, A.; Colles, F.; Leterrier, C. Influence of the microbiota-gut-brain axis on behavior and welfare in farm animals: A review. Physiol. Behav. 2019, 112658. [Google Scholar] [CrossRef] [PubMed]
- Kubasova, T.; Kollarcikova, M.; Crhanova, M.; Karasova, D.; Cejkova, D.; Sebkova, A.; Matiasovicova, J.; Faldynova, M.; Pokorna, A.; Cizek, A.; et al. Contact with adult hen affects development of caecal microbiota in newly hatched chicks. PLoS ONE 2019, 14, e0212446. [Google Scholar] [CrossRef] [PubMed]
- Impey, C.S.; Mead, G.C.; George, S.M. Competitive exclusion of salmonellas from the chick caecum using a defined mixture of bacterial isolates from the caecal microflora of an adult bird. J. Hyg. (Lond) 1982, 89, 479–490. [Google Scholar] [CrossRef]
- Penha Filho, R.A.; Diaz, S.J.; Fernando, F.S.; Chang, Y.F.; Andreatti Filho, R.L.; Berchieri Junior, A. Immunomodulatory activity and control of Salmonella Enteritidis colonization in the intestinal tract of chickens by Lactobacillus based probiotic. Vet. Immunol. Immunopathol. 2015, 167, 64–69. [Google Scholar] [CrossRef]
- Netherwood, T.; Gilbert, H.J.; Parker, D.S.; O’Donnell, A.G. Probiotics shown to change bacterial community structure in the avian gastrointestinal tract. Appl. Environ. Microbiol. 1999, 65, 5134–5138. [Google Scholar]
- La Ragione, R.M.; Woodward, M.J. Competitive exclusion by Bacillus subtilis spores of Salmonella enterica serotype Enteritidis and Clostridium perfringens in young chickens. Vet. Microbiol. 2003, 94, 245–256. [Google Scholar] [CrossRef]
- Videnska, P.; Faldynova, M.; Juricova, H.; Babak, V.; Sisak, F.; Havlickova, H.; Rychlik, I. Chicken faecal microbiota and disturbances induced by single or repeated therapy with tetracycline and streptomycin. BMC Vet. Res. 2013, 9, 30. [Google Scholar] [CrossRef]
- Medvecky, M.; Cejkova, D.; Polansky, O.; Karasova, D.; Kubasova, T.; Cizek, A.; Rychlik, I. Whole genome sequencing and function prediction of 133 gut anaerobes isolated from chicken caecum in pure cultures. BMC Genomics 2018, 19, 561. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.C.; Khelaifia, S.; Alou, M.T.; Ndongo, S.; Dione, N.; Hugon, P.; Caputo, A.; Cadoret, F.; Traore, S.I.; Seck, E.H.; et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016, 1, 16203. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.T.; Whelan, F.J.; Herath, I.; Lee, C.H.; Collins, S.M.; Bercik, P.; Surette, M.G. Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. Genome Med. 2016, 8, 72. [Google Scholar] [CrossRef] [PubMed]
- Browne, H.P.; Forster, S.C.; Anonye, B.O.; Kumar, N.; Neville, B.A.; Stares, M.D.; Goulding, D.; Lawley, T.D. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 2016, 533, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic. Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. Trimal: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, S.; Martinez, O.A.; Fernandez, H.; Collado, L. Comparison of media and growth conditions for culturing enterohepatic Helicobacter species. Lett. Appl. Microbiol. 2019, 69, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.C.; Poppleton, D.; Klingl, A.; Criscuolo, A.; Dupuy, B.; Brochier-Armanet, C.; Beloin, C.; Gribaldo, S. Phylogenomic analysis supports the ancestral presence of LPS-outer membranes in the Firmicutes. Elife 2016, 5, e14589. [Google Scholar] [CrossRef] [PubMed]
- Rutgeerts, P.; Ghoos, Y.; Vantrappen, G. The enterohepatic circulation of bile acids during continuous liquid formula perfusion of the duodenum. J. Lipid. Res. 1983, 24, 614–619. [Google Scholar] [PubMed]
- Volf, J.; Polansky, O.; Varmuzova, K.; Gerzova, L.; Sekelova, Z.; Faldynova, M.; Babak, V.; Medvecky, M.; Smith, A.L.; Kaspers, B.; et al. Transient and Prolonged Response of Chicken Cecum Mucosa to Colonization with Different Gut Microbiota. PLoS ONE 2016, 11, e0163932. [Google Scholar] [CrossRef] [PubMed]
- Schnorr, S.L.; Candela, M.; Rampelli, S.; Centanni, M.; Consolandi, C.; Basaglia, G.; Turroni, S.; Biagi, E.; Peano, C.; Severgnini, M.; et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014, 5, 3654. [Google Scholar] [CrossRef]
- Gorvitovskaia, A.; Holmes, S.P.; Huse, S.M. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome 2016, 4, 15. [Google Scholar] [CrossRef]
- Moran, E.T., Jr. Nutrition of the developing embryo and hatchling. Poult. Sci. 2007, 86, 1043–1049. [Google Scholar] [CrossRef]
- Noy, Y.; Sklan, D. Yolk utilisation in the newly hatched poult. Br. Poult. Sci. 1998, 39, 446–451. [Google Scholar] [CrossRef]
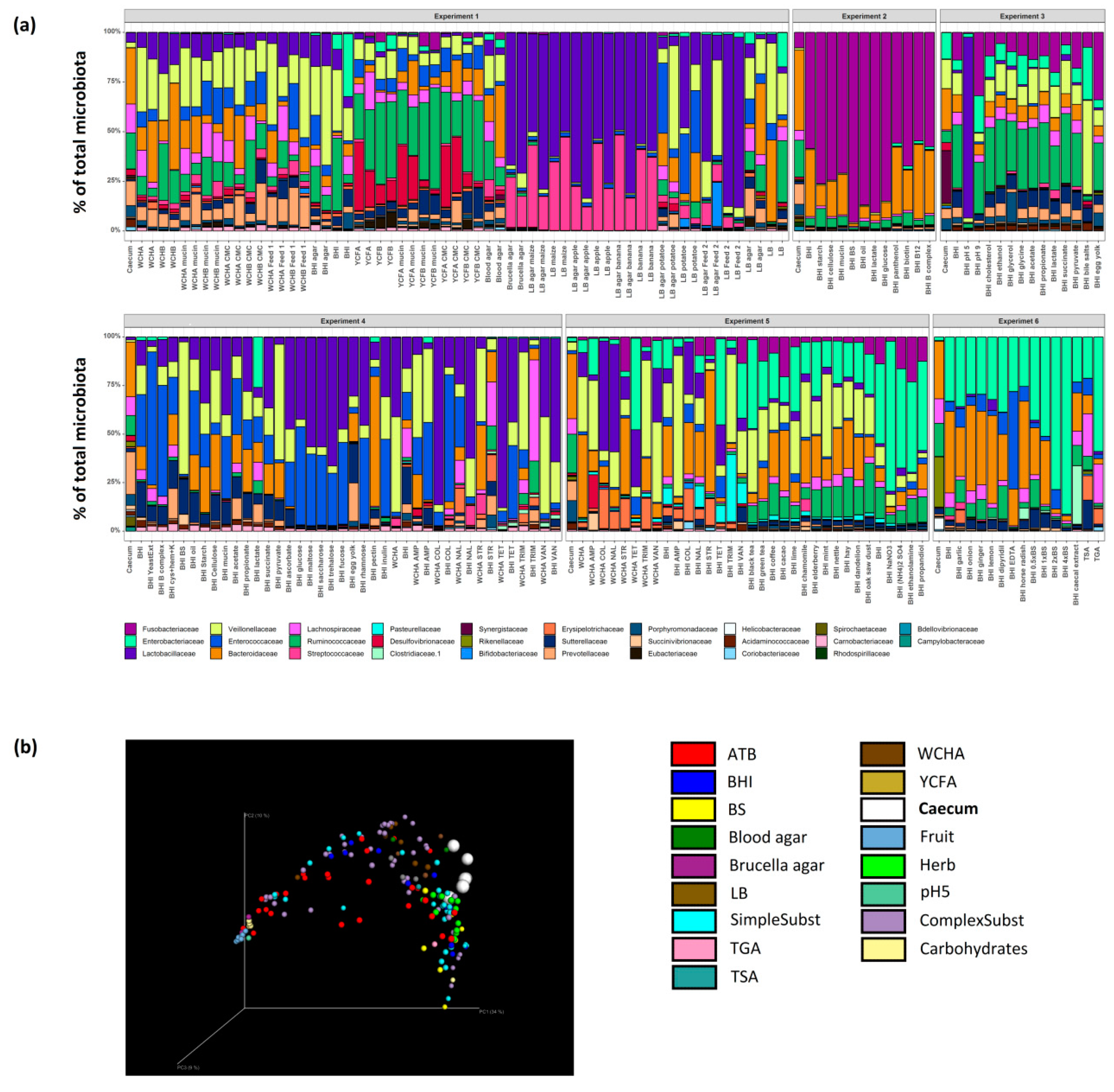
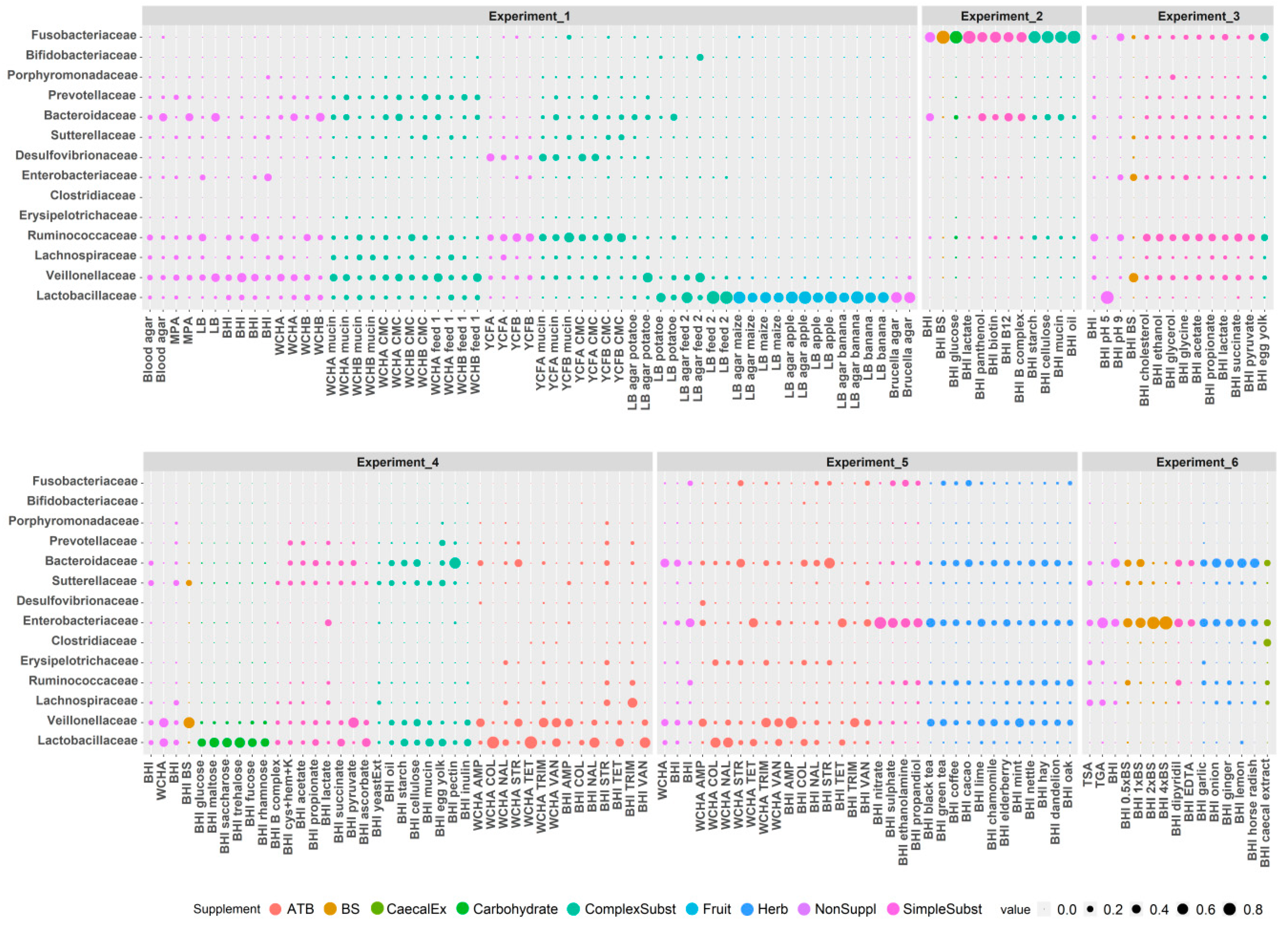
| Category | Used Nutrient Broths and Tested Supplements |
|---|---|
| Non supplemented broths | Blood agar, Brucella agar, TGA, BHI, TSA, WCHA, WCHB, YCFB, YCFA, LB agar, LB broth |
| Complex substrates | caecal extract, carboxymethyl cellulose, hay, mucin, oil, starch, cellulose, egg yolk, feed, inulin, oak saw dust, pectin, potato, yeast extract |
| Simple substrates | (NH4)2 SO4, acetate, ascorbate, B complex, vitamine B12, biotin, cystein+hemin+vitamine K, dipyridil, EDTA, ethanol, ethanolamine, glycerol, glycine, cholesterol, lactate, NaNO3, panthenol, propandiol, propionate, pyruvate, succinate |
| Carbohydrates | glucose, maltose, saccharose, trehalose, fucose, rhamnose |
| Fruit | apple, banana, maize |
| Herb | black tea, green tea, coffee, cacao, lime, chamomile, elderberry, mint, nettle, dandelion, garlic, onion, ginger, lemon, horse radish |
| Antibiotics | ampicillin, colistin, nalidixic acid, streptomycin, tetracycline, trimethoprim, vancomycin |
| Miscellaneous | bile salts, pH 5, pH 9 |
| Phylum | All OTUs | Non-Growing | Growing | % of Growing |
|---|---|---|---|---|
| Archaea | 1 | 1 | 0 | 0 |
| Unclassified Bacteria | 2 | 2 | 0 | 0 |
| Candidatus Saccharibacteria | 1 | 1 | 0 | 0 |
| Deferribacteres | 1 | 1 | 0 | 0 |
| Spirochaetes | 3 | 3 | 0 | 0 |
| Elusimicrobia | 1 | 1 | 0 | 0 |
| Tenericutes | 1 | 1 | 0 | 0 |
| Verrucomicrobia | 2 | 2 | 0 | 0 |
| Fusobacteria | 1 | 0 | 1 | 100 |
| Synergistetes | 3 | 2 | 1 | 33.33 |
| Actinobacteria | 3 | 0 | 3 | 100 |
| Bacteroidetes | 175 | 101 | 74 | 42.29 |
| Firmicutes | 115 | 62 | 53 | 46.09 |
| Proteobacteria | 32 | 20 | 12 | 37.5 |
| Total | 341 | 197 | 144 | 42.23 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crhanova, M.; Karasova, D.; Juricova, H.; Matiasovicova, J.; Jahodarova, E.; Kubasova, T.; Seidlerova, Z.; Cizek, A.; Rychlik, I. Systematic Culturomics Shows that Half of Chicken Caecal Microbiota Members can be Grown in Vitro Except for Two Lineages of Clostridiales and a Single Lineage of Bacteroidetes. Microorganisms 2019, 7, 496. https://doi.org/10.3390/microorganisms7110496
Crhanova M, Karasova D, Juricova H, Matiasovicova J, Jahodarova E, Kubasova T, Seidlerova Z, Cizek A, Rychlik I. Systematic Culturomics Shows that Half of Chicken Caecal Microbiota Members can be Grown in Vitro Except for Two Lineages of Clostridiales and a Single Lineage of Bacteroidetes. Microorganisms. 2019; 7(11):496. https://doi.org/10.3390/microorganisms7110496
Chicago/Turabian StyleCrhanova, Magdalena, Daniela Karasova, Helena Juricova, Jitka Matiasovicova, Eva Jahodarova, Tereza Kubasova, Zuzana Seidlerova, Alois Cizek, and Ivan Rychlik. 2019. "Systematic Culturomics Shows that Half of Chicken Caecal Microbiota Members can be Grown in Vitro Except for Two Lineages of Clostridiales and a Single Lineage of Bacteroidetes" Microorganisms 7, no. 11: 496. https://doi.org/10.3390/microorganisms7110496
APA StyleCrhanova, M., Karasova, D., Juricova, H., Matiasovicova, J., Jahodarova, E., Kubasova, T., Seidlerova, Z., Cizek, A., & Rychlik, I. (2019). Systematic Culturomics Shows that Half of Chicken Caecal Microbiota Members can be Grown in Vitro Except for Two Lineages of Clostridiales and a Single Lineage of Bacteroidetes. Microorganisms, 7(11), 496. https://doi.org/10.3390/microorganisms7110496




