Abstract
Aeromonas hydrophila is a well-known bacterial pathogen associated with mass mortalities in aquaculture. Yet, few reports are available on whiteleg shrimp-pathogenic A. hydrophila. In the present study, a virulent isolate WS05 was confirmed as a causative agent of diseased freshwater-cultured whiteleg shrimp and showed a mean lethal dose (LD50) value of 4.8 × 104 CFU mL−1. It was identified phenotypically and molecularly as an A. hydrophila strain, and exhibited susceptibility to several veterinary antibiotics extensively used in aquaculture, including cotrimoxazole, doxycycline, florfenicol, neomycin, and tetracycline. In view of the strongest inhibition zone of florfenicol against isolate WS05, the synergistic effect of the combinations of florfenicol and herb extracts was further evaluated, and the result indicated that Punica granatum extract was a potential synergist of florfenicol against isolate WS05 and the fractional inhibitory concentration index (FICI) for the florfenicol-P. granatum extract was calculated as 0.31. When combined with 7.81 mg mL−1 P. granatum extract, the minimum inhibitory concentration (MIC) of florfenicol against isolate WS05 was reduced from 0.50 to 0.03 mg L−1, and its activity against isolate WS05 was also enhanced with a significant reduction of ≥3.61 log in cell density after 24 h of treatment compared with that in the single drug treatment. In addition, the protective effect was potentiated by the combination of florfenicol and P. granatum extract, with a cumulative mortality of 36.66% (p < 0.05) and 33.33% (p < 0.05) lower than that in the single treatment with florfenicol and P. granatum extract after the challenge with isolate WS05 for seven days. As far as we know, this is the first study to describe whiteleg shrimp-pathogenic A. hydrophila and suggest P. granatum extract as a potential synergist of florfenicol against the A. hydrophila pathogen.
1. Introduction
The whiteleg shrimp Litopenaeus vannamei is one of the most important commercial shrimp species and is extensively cultivated in Central and South America, the United States of America (USA), East and South-East Asia, the Middle East, and Africa [1], which accounts for 75% of the global shrimp products [2]. Especially in China, with the rapid development of farming techniques, the whiteleg shrimp has been successfully cultured in freshwater since 2000 [3], with an annual freshwater aquaculture production of over 591,000 tons in 2017 [4]. However, this industry has been seriously affected by bacterial diseases [5], which are caused by several bacterial pathogens, such as Vibrio parahaemolyticus, Vibrio harveyi, Vibrio cholerae, Proteus penneri, Aeromonas schubertii, and Shewanella algae [6,7,8,9,10,11]. In order to prevent and control bacterial diseases, antibiotics are widely used in whiteleg shrimp aquaculture [12]. However, the widespread and frequent use of antibiotics in shrimp aquaculture has resulted in the development of antibiotic resistance among pathogens infecting cultured animals and humans [12]. Herbs with potential antibacterial activity against Aeromonas hydrophila have a crucial role in disease management in aquaculture [13]. Extracts from herbs, such as Acorus calamus, Indigofera aspalathoides, Coleus aromaticus, Thymus vulgaris, and Trifolium pannonicum, have been reported to effectively inhibit pathogenic A. hydrophila [14,15,16,17]. Many studies have also proved that oral administration of extracts from herbs, such as Allium sativum, Epilobium hirsutum, Panax quinquefolium, and Toona sinensis, can greatly enhance resistance against A. hydrophila infection in fish [18,19,20,21]. Therefore, in order to limit the use of antibiotics, the use of herb extracts in combination with conventional antibiotics is commonly suggested for the treatment of bacterial pathogens in aquaculture [22], which could significantly reduce the dosage of antibiotics against bacterial pathogens [23]. Jedlickova et al. (1992) found that the combinations of 1,8-cineol, linalool, and terpinen-4-ol with amikacin, gentamicin, and tobramycin could strongly inhibit the growth of Escherichia coli [24]; Jayaraman et al. (2010) showed that combinations of sulfamethoxazole and myricetin could significantly enhance antibacterial activities against Pseudomonas aeruginosa strains [25]. Yet, little has been documented on whiteleg shrimp-pathogenic Aeromonas hydrophila and its control with combinations of herb extracts and antibiotics.
In late May of 2019, a severe disease with the typical signs of hepatopancreatic shrinking occurred in nearly the whole shrimp farming regions of Fengxian, Shanghai, China, and resulted in a cumulative mortality of 50% to 100%. In the present study, we confirm that a virulent isolate of A. hydrophila was the causative agent of the disease in freshwater-cultured whiteleg shrimp, and its taxonomic position, virulence, and antibiotic susceptibility were examined. Furthermore, in view of the strongest inhibition zone of florfenicol against the isolate, the control of the isolate with combinations of florfenicol and herb extracts was further evaluated. To our knowledge, this is the first report of A. hydrophila as a bacterial pathogen of freshwater-cultured whiteleg shrimp, and the findings of this study can be used as a reference for disease control and health management in shrimp aquaculture.
2. Materials and Methods
2.1. Shrimp and Reagents
Twenty moribund freshwater-cultured whiteleg shrimp (6.80 ± 0.05 g in weight) suffering from hepatopancreatic shrinking were sampled from an infected freshwater pond of a shrimp farm in Fengxian, Shanghai, China during May 2019 and were placed into sterile bags and kept on ice during the 1-h transport to the laboratory as recommended by Jayasinghe et al. (2008) [26]. In total, 340 healthy shrimp (5.26 ± 0.36 g) for the infection assay were obtained from unaffected ponds of a shrimp farm in Lianyungang Aquaculture Co., Ltd., Jiangsu, China and maintained in 34 glass aquaria (76 cm × 50 cm × 48 cm) (10 shrimp per aquarium) with 100 L aerated filtered farm water as recommended by Moss et al. (1992, 2001) [27,28] with an initial pH of 7.50, 6.2 mg L−1 of dissolved oxygen, 0.11 mg L−1 of total ammonia, and 0.01 mg L−1 of nitrite at 28 °C for 14 days. Next, 240 healthy shrimp (0.62 ± 0.11 g) for the protective effect assay were obtained from unaffected ponds of a shrimp farm in Rudong Aquaculture Co., Ltd., Jiangsu, China, and maintained in 24 glass aquaria (76 cm × 50 cm × 48 cm) (10 shrimp per aquarium) supplied with 100 L aerated filtered farm water with an initial pH of 7.64, 6.6 mg L−1 of dissolved oxygen, 0.10 mg L−1 of total ammonia, and 0.01 mg L−1 of nitrite at 28 °C for 14 days. Their health status was assessed through a careful examination of the external appearance, gut condition, growth situation, physical behavior, and feeding trends as recommended by the Marine Products Export Development Authority and the Network of Aquaculture Centers in Asia-Pacific (2003) [29], as well as by culturing hemolymph and hepatopancreas smears from a few sampled animals on nutrient agar (NA) plates (Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) for the absence of any bacterial pathogens as recommended by Biswas et al. (2012) [30]. Filtered farm water used in our laboratory was prepared by passing 6 m3 of water, which was obtained from the natural Luchaoyin River, successively through 15-denier-size polyester fiber and a 60-pore per inch (ppi) polyurethane sponge in a water-filtration device (Shanghai Haisheng Biotech. Co., Ltd., Shanghai, China), with a flow rate of 0.24 m3 min−1 and a circulation rate of 8 times d−1 as recommended by Luo et al. (2008) [31], and was sterilized at 121 °C for 20 min to remove bacteria prior to its use. Florfenicol (98%) was purchased from Tianchen Biotech. Co. Ltd., Wuhan, China. Herbs were obtained from Marine Biology Institute of Shandong, Shandong, China. Reagents were of analytical grade from Sinopharm Chemical Reagent Co., Ltd., Shanghai, China.
2.2. Confirmation of the Causative Pathogen
Each sampled diseased whiteleg shrimp was disinfected externally with 75% alcohol and dissected in the laboratory according to Cao et al. (2014) [9]. To verify the potential pathogens, a squash of organs (gill, hepatopancreas, muscle, intestine) were made and carefully examined for parasites under the microscope (YS100, Nikon, Tokyo, Japan) as described by Ekanem et al. (2011) [32]. Meanwhile, the virological examination was also conducted as described by Pan et al. (2009) [33]. Briefly, the homogenate of organs (gill, hepatopancreas, muscle, intestine) was made and filtered through 0.22-μm-pore-size membrane filter to remove bacteria. Two aquaria of 10 healthy shrimp were injected muscularly with 0.1 mL of each bacteria-free organ filtrate. Another two aquaria of 10 healthy shrimp, which were exposed to the same experimental conditions and injected muscularly with 0.1 mL of normal saline, served as the control. Experimental shrimp were kept at 28 °C without water change. The mortality and any visible changes of the experimental shrimp were recorded every day for 15 days. In addition, samples from organs (hepatopancreas, muscle) of each diseased shrimp were cut and directly streaked onto NA plates as recommended by Cao et al. (2018) [34]. After incubation for 24 h at 28 °C, the dominant uniform isolates were purified by streaking and re-streaking onto NA plates (Sinopharm Chemical Reagent Co., Ltd., Shanghai, China). Only the dominant isolates with dense virtually pure culture growth on NA plates were obtained as recommended by Orozova et al. (2012) [35] and Cao et al. (2014) [9] After being initially identified by 16S rRNA gene sequencing as recommended by Petti et al. (2005) [36], the pure isolates were further inoculated onto an NA plate, incubated at 28 °C for 24 h, and washed with sterile normal saline into a sterile tube. Their cell densities were determined by counting the colony forming units (CFU) on NA plates after a 10-fold serial dilution in sterile normal saline. Induced infection of the pure isolates was performed according to Cao et al. (2015) [10]. Briefly, two aquaria of 10 healthy shrimp were challenged by immersion in 100 L of water containing the isolates at a cell density of 5.0 × 106 CFU mL−1. Another two aquaria of 10 healthy shrimp, which were exposed to the same experimental conditions and remained unchallenged, served as the control. Experimental shrimp were kept at 28 °C without water change. The mortality and any visible changes of the experimental shrimp were recorded every day for 7 days. Dead shrimp were immediately removed and the hepatopancreas sampled on NA plates to confirm if the mortality was caused by the challenge isolate. Besides, ultrathin sections were prepared from hepatopancreas of the infected and healthy shrimp for light and electron microscopic examination as described by Zhou et al. (2012) [7]. Briefly, for light microscopy, the hepatopancreas was fixed in Boun’s solution, followed by dehydration in a graded series of alcohol (75%, 85%, 90%, 95%, 100%) (30 min in each dehydrating agent), cleared by xylol, embedded with paraffin, stained with hematoxylin and eosin, and then observed under a light microscope (Eclipse E100, Nikon, Tokyo, Japan). For transmission electron microscopy, the hepatopancreas was first fixed in 2.5% glutaraldehyde in 0.1 M PBS (pH 7.4) for 2 h at 4 °C, followed by post-fixation in 1% osmium tetroxide for 2 h, dehydrated in a graded series of alcohol (50%, 70%, 80%, 90%, 95%, 100%) and 100% acetone (15 min in each dehydrating agent), embedded in Spurr’s resin, stained with uranyl acetate and lead citrate, and then observed under a transmission electron microscope (HT770, Hitachi, Tokyo, Japan).
2.3. Identification of the Causative Pathogen
2.3.1. Molecular Identification
The genomic DNA was extracted from the pathogenic isolate using the TIANamp DNA Kit (Tiangen Biotech. Co., Ltd., Beijing, China). The 16S rRNA, gyrB, and rpoB genes from the pathogenic isolate were amplified by PCR according to Cao et al. (2010) [37], Yáňez et al. (2003) [38], and Küpfer et al. (2006) [39], and sequenced by an ABI 3730 XL DNA Sequencer (Applied Biosystems, Waltham, MA, USA). Homology searches for 16S rRNA, gyrB, and rpoB gene sequences were performed using the Basic Local Alignment Search Tool version 4 (BLAST) software (National Library of Medicine, Bethesda, MD, USA) available at the National Center for Biotechnology Information (NCBI). Sequence alignment was carried out with Molecular Evolutionary Genetics Analysis version 5 (MEGA5) software (Arizona State University, Tempe, AZ, USA), and the phylogenetic trees were constructed using the neighbor-joining method.
2.3.2. Phenotypic Identification
The phenotypic identification of the pathogenic isolate was performed by the API 20E system (Biomerieux, Lyon, France) as recommended by Abeyta et al. (2019) [40]. Briefly, the pathogenic isolate streaked on NA plates was incubated at 28 °C for 24 h, and then inoculated into the API 20E test strips (Biomerieux, Lyon, France) according to the instructions of the manufacturer. The test strips containing the isolate were incubated at 37 °C for 18 h and observed for biochemical reactions. The phenotypic characterization of A. hydrophila previously reported by Long et al. (2016) [41] and Ye et al. (2018) [42] served as a reference.
2.4. Bacterial Virulence Experiment
The bacterial virulence experiment consisted of one control and five treatment groups. Prior to this experiment, the pathogenic isolate was inoculated onto an NA plate, incubated at 28 °C for 24 h, and washed with sterile normal saline into a sterile tube. Its cell density was determined by counting the colony-forming units (CFU) on NA plates after a 10-fold serial dilution in sterile normal saline. Each group contained two glass aquaria (10 shrimp per aquarium) with 100 L aerated filtered farm water. In the treatment groups, the experimental shrimp in the glass aquaria were challenged by immersion with the pathogenic isolate at final cell densities of 2.0 × 103 to 2.0 × 107 CFU mL−1 in water as recommended by Teng et al. (2017) [43]. Another two aquaria of healthy shrimp, which were exposed to the same experimental conditions and remained unchallenged, served as the control. All of the test shrimp were kept at 28 °C without any water change. The mortality was recorded every day for 7 days. Dead shrimp were immediately removed and the hepatopancreas sampled on NA plates to confirm whether the mortality was caused by the challenge isolate. The mean lethal dose (LD50) value was calculated using the graphical probit method [44].
2.5. Antibiotics Susceptibility Assay
The susceptibility of the pathogenic isolate to veterinary antibiotics was examined using the Kirby–Bauer disk diffusion method described by Jones et al. (2001) [45] on NA plates. The susceptibility was determined by measuring inhibition zones after incubation at 28 °C for 24 h and was evaluated according to the guidelines of the manufacturer (Hangzhou Binhe Microorganism Reagent Co., Ltd., Hangzhou, China) as recommended by Cao et al. (2018) [34].
2.6. Synergistic Effect of Florfenicol–Herb Extracts Assay
Because of the strongest inhibition zone of florfenicol against the pathogenic isolate as shown in antibiotics susceptibility assay, florfenicol was chosen for further study. The synergistic effect of florfenicol in combination with herb extracts on the pathogenic isolate was evaluated in a 24-well microplate using the checkerboard method described by Chen et al. (2014) [46]. Prior to this assay, 1000 mg L−1 of florfenicol was prepared according to Liu et al. (2011) [47]. Extracts from peels of pomegranate Punica granatum, fruits of Prunus mume and Fructus toosendan, leaves of Artemisia argyi, Polygonum aviculare, Cephalanoplos segetum, and Artemisia capillaries and roots of the other herbs were prepared with distilled water as recommended by Rattanachaikunsopon and Phumkhachorn (2009) [48]. Briefly, each herb was oven-dried at 80 °C for 72 h, finely ground in a mortar, and extracted with the distilled water at a ratio of 1:10 (w/v) using a full-automatic herb-decocting kettle (Guangzhou Haizhu Electric Co., Ltd., Guangzhou, China). The mixture was then centrifuged at 13,000 rpm for 10 min at room temperature and the supernatant was collected as the herb extract. The concentration of each herb extract for each assay was adjusted to 1000 mg (dry weight) mL−1. Immediately after the pathogenic isolate was added to each plate well to a final cell density of 5 × 105 CFU mL−1 as recommended by Liu et al. (2000) [49], antimicrobial agents were loaded per well with serial concentrations ranging from 0.125 to 512 mg L−1 for florfenicol and from 1.95 to 500 mg mL−1 for the herb extracts. The plates were incubated at 28 °C for 24 h. The minimum inhibitory concentrations (MICs) of the antimicrobial agents alone or in combination were determined after 24 h of incubation, which were defined as the lowest concentration showing no color change that exhibited complete inhibition of growth according to Haroun and Al-Kayali (2016) [50]. The fractional inhibitory concentration index (FICI) was calculated according to the formula as recommended by Liu et al. (2017) [51]: FICI = MIC (florfenicol in combination)/MIC (florfenicol alone) + MIC (herb extract in combination)/MIC (herb extract alone). The joint effect was evaluated according to the following criteria as suggested by Gong et al. (2005) [52]: Synergistic effect (FICI ≤ 0.5); additive effect (0.5 < FICI ≤ 1), no interaction (1 < FICI < 4) and antagonistic effect (FICI ≥ 4).
2.7. The in Vitro Activity of Florfenicol-Herb Extracts Assay
The in vitro activity of florfenicol in combination with the herb extract with a synergistic effect was examined by the time-kill curve method as recommended by Dong et al. (2016) [23]. The assay was carried out in 50-mL glass flasks and consisted of one control and three treatment groups (three flasks per group). Immediately after the pathogenic isolate was inoculated in 20 mL of autoclaved nutrient broth to a final cell density of 5.0 × 105 CFU mL−1 as recommended by Liu et al. (2000) [49], antimicrobials were independently inoculated into autoclaved nutrient broth in the treatment groups to final concentrations of 0.03 mg L−1 florfenicol, 7.81 mg mL−1 P. granatum extract, and 0.03 mg L−1 florfenicol plus 7.81 mg mL−1 P. granatum extract, which were determined by the synergistic effect assay above. All of the mixtures were then incubated at 28 °C for 48 h as recommended by Dong et al. (2016) [23]. In the control group, only the pathogenic isolate was inoculated in autoclaved nutrient broth to a final cell density of 5.0 × 105 CFU mL−1 and incubated as mentioned above. The cell density of the pathogenic isolate was measured every 12 h by spread plate counts on the NA plate [23,37].
2.8. Protective Effect of Florfenicol-Herb Extracts Assay
The protective effect assay consisted of one blank control, one negative control, three positive control, and three treatment groups. Prior to this assay, the pathogenic isolate was inoculated onto an NA plate, incubated at 28 °C for 24 h, and washed with sterile normal saline into a sterile tube. Its cell density was determined by counting the CFU on NA plates after a 10-fold serial dilution in sterile normal saline. Each group contained three glass aquaria (10 shrimp per aquarium) with 100 L aerated filtered farm water. In the treatment groups, antimicrobials were independently inoculated into the aerated filtered farm water in the aquaria to final concentrations of 0.03 mg L−1 florfenicol, 7.81 mg mL−1 P. granatum extract, and 0.03 mg L−1 florfenicol plus 7.81 mg mL−1 P. granatum extract, which were determined by the synergistic effect assay above. Thereafter, all of the shrimp were challenged by immersion through continuous exposure to the pathogenic isolate at a final cell density of 2.0 × 106 CFU mL−1 in water as determined above. In the positive control groups, the shrimp were only treated with the antimicrobials. In the negative control group, the shrimp were only challenged by immersion with the pathogenic isolate at a final cell density of 2.0 × 106 CFU mL−1 in water as described above. Another three aquaria of healthy shrimp, which were exposed to the same experimental conditions and remained unchallenged, served as the blank control. All of the test shrimp were kept at 28 °C without water change. The mortality was recorded every day for 7 days. Dead shrimp were immediately removed and the hepatopancreas sampled on NA plates to confirm if the mortality was caused by the challenge isolate.
2.9. Statistical Analysis
Statistical analysis was carried out using the statistical software SPSS 15.0 (SPSS, Inc.) to observe the difference in each assay. All of the data are presented as the mean ± standard deviation (SD) for the indicated number of each assay. Differences were considered statistically significant at p < 0.05 using analysis of variance according to Duncan’s test.
2.10. Ethics Statements
The experimental protocol strictly followed the guidelines for the ethical review of animal welfare and the general requirements for animal experiments, China. The present experiment was approved by the Institutional Animal Ethics Committee of Shanghai Ocean University (establishment date: 18 February 2016) with the permission No. 20171025 valid until 31 December 2020.
3. Results
3.1. Confirmation of the Causative Pathogen
No parasites were found in the diseased whiteleg shrimp, and all of the experimental shrimp challenged with the bacteria-free organ filtrate survived with no visible changes (data not shown), indicating that the disease was not caused by parasites or viruses. Five dominant isolates (numbered from WS01 to WS05) were recovered from the hepatopancreas of the diseased shrimp, which were identified as the members of the genus Bacillus and Aeromonas based on the analysis of their 16S rRNA genes (GenBank accession nos. MN367958, MN371228, MN367959, MN367960, MN148709) (Figure S1 and Figure 1), and none were isolated from the muscles. The result of the bacterial challenge in shrimp indicated that only the isolate (WS05) was pathogenic to whiteleg shrimp, showing an LD50 value of 4.8 × 104 CFU mL−1 (Figure 2). In contrast, no visible changes or death were recorded in the control shrimp. The artificially infected shrimp exhibited the sign of hepatopancreas shrinkage similar to the naturally infected shrimp (Figure 3), and the same strain (WS05) was re-isolated from the experimentally diseased shrimp and confirmed by phenotypical and molecular identification. Furthermore, the histopathological changes in the hepatopancreas from naturally infected and artificially infected shrimp showed a disordered arrangement of hepatopancreatic tubules and inflammatory cell infiltration (Figure 4). Moreover, an electron microscopic analysis further showed the hepatopancreatic cell was damaged in both naturally infected and artificially infected shrimp, including abnormal formation of the mitochondrial myelin body and dilatational endoplasmic reticulum (Figure 5). These findings demonstrated that isolate WS05 was the causative agent of this disease.
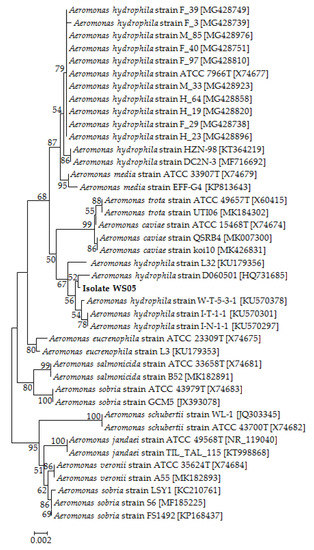
Figure 1.
The 16S rRNA phylogenetic tree of 40 known bacteria and isolate WS05 constructed using the neighbor-joining method. The length of aligned sequences is 1430 bp. The bootstrap values (%) are shown besides the clades, accession numbers are indicated beside the name of strains, and scale bars represent distance values.
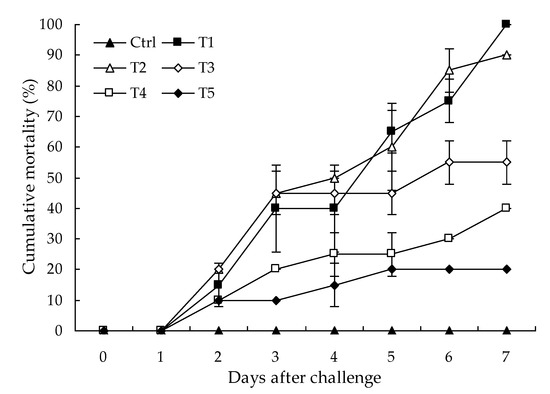
Figure 2.
Virulence of isolate WS05 at final cell densities of 2.0 × 103 to 2.0 × 107 CFU mL−1 for experimental whiteleg shrimp. Ctrl, Control group; T1, Treatment with isolate WS05 at 2.0 × 107 CFU mL−1; T2, Treatment with isolate WS05 at 2.0 × 106 CFU mL−1; T3, Treatment with isolate WS05 at 2.0 × 105 CFU mL−1; T4, Treatment with isolate WS05 at 2.0 × 104 CFU mL−1; T5, Treatment with isolate WS05 at 2.0 × 103 CFU mL−1. Data are presented as the mean ± SD.
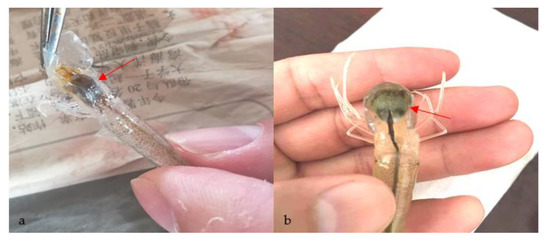
Figure 3.
Gross signs of affected shrimp in the disease outbreak region. (a) Diseased shrimp. Arrow shows the atrophic hepatopancreas; (b) healthy shrimp. Arrow shows the normal hepatopancreas.
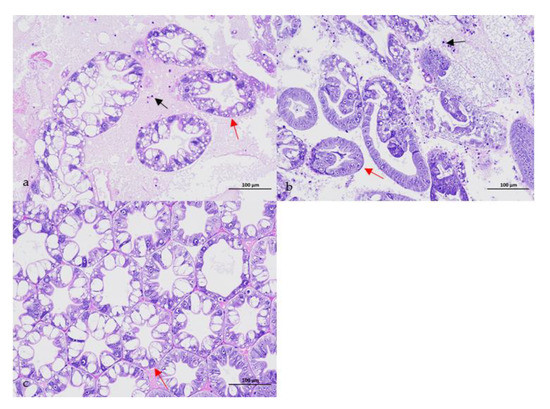
Figure 4.
Histopathological changes in the atrophic hepatopancreas of affected shrimp. (a) Disordered arrangement of hepatopancreatic tubules (red arrow) and inflammatory cell infiltration (black arrow) in the naturally infected hepatopancreas; (b) disordered arrangement of hepatopancreatic tubules (red arrow) and inflammatory cell infiltration (black arrow) in the artificially infected hepatopancreas; (c) normal arrangement of hepatopancreas tubules (red arrow) in healthy hepatopancreas.
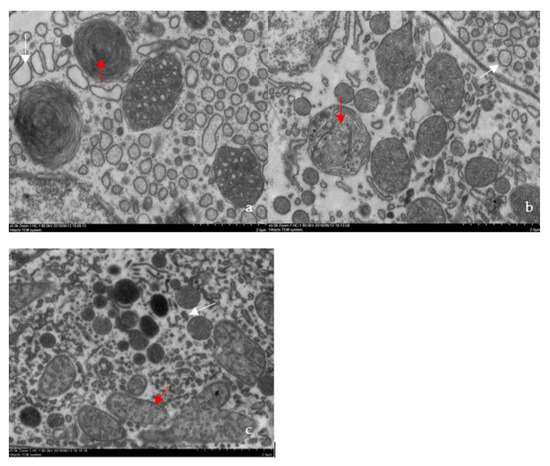
Figure 5.
Ultrastructural changes in the atrophic hepatopancreas of affected shrimp. (a) Mitochondrial myelin body formation (red arrow) and dilatational endoplasmic reticulum (white arrow) in the naturally infected cell cytoplasm (5000 ×); (b) mitochondrial myelin body (red arrow), which is forming, and dilatational endoplasmic reticulum (white arrow) in the artificially infected cell cytoplasm (5000 ×); (c) normal mitochondria (red arrow) and endoplasmic reticulum (white arrow) in healthy cell cytoplasm (5000 ×).
3.2. Identification of the Pathogenic Isolate
Isolate WS05 was identified phenotypically and molecularly as A. hydrophila. It exhibited the same phenotypic traits with the reference strains of A. hydrophila [41,42]. Isolate WS05 was positive for utilization of arginine, lysine, citrate, and tryptophan, and was negative for growth on amygdalin, inositol, melibiose, rhamnose, and sorbitol (Table 1). Furthermore, the 16S rRNA, gyrB, rpoB gene sequences of isolate WS05 (GenBank accession nos. MN148709, MN394631, MN394672) respectively showed similarities of 99% with the known strains of A. hydrophila in the GenBank database. Phylogenetic trees constructed by the neighbor-joining method demonstrated isolate WS05 to be an A. hydrophila strain (Figure 1, Figure 6 and Figure 7), which is in agreement with the phenotypic identification of isolate WS05 (Table 1).

Table 1.
Phenotypic characterization of isolate WS05.
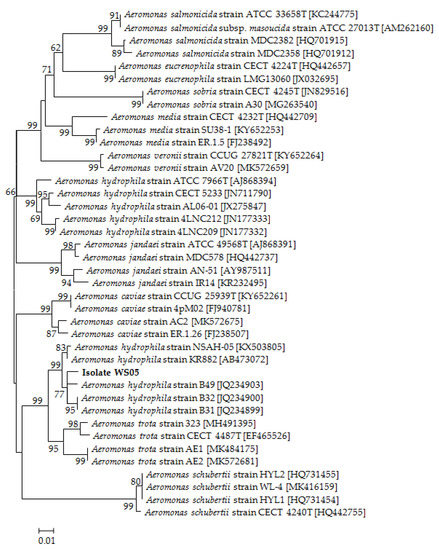
Figure 6.
The phylogenetic tree based on gyrB gene sequences of 39 Aeromonas strains and isolate WS05 constructed using the neighbor-joining method. The length of aligned sequences is 765 bp. The bootstrap values (%) are shown besides the clades, accession numbers are indicated beside the name of strains, and scale bars represent distance values.
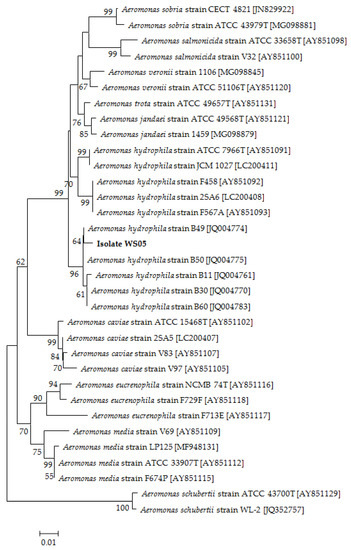
Figure 7.
The phylogenetic tree based on rpoB gene sequences of 32 Aeromonas strains and isolate WS05 constructed using the neighbor-joining method. The length of aligned sequences is 423 bp. The bootstrap values (%) are shown besides the clades, accession numbers are indicated beside the name of strains, and scale bars represent distance values.
3.3. Antibiotic Susceptibility of the Pathogenic Isolate
The data (Table S1) indicated that isolate WS05 was susceptible to a range of veterinary antibiotics except novobiocin and rifampicin, including amoxicillin, cotrimoxazole, cefotaxime, doxycycline, enrofloxacin, florfenicol, gentamicin, kanamycin, nalidixic acid, neomycin, netilmicin, oxacillin, polymyxin B, streptomycin, tetracycline, and tobramycin. In particular, isolate WS05 apparently exhibited susceptibility to cotrimoxazole, doxycycline, florfenicol, neomycin, and tetracycline, which are currently approved for use in aquaculture [53].
3.4. Synergistic Effect of Florfenicol–Herb Extracts
The MICs of florfenicol and herb extracts alone or in combination against isolate WS05 are presented in Table 2. The result indicated that only the combination of florfenicol and P. granatum extract had a synergistic antibacterial effect on isolate WS05. When combined with 7.81 mg mL−1 P. granatum extract, the MIC of florfenicol against isolate WS05 was reduced from 0.50 to 0.03 mg L−1. The FICI for the combination of florfenicol and P. granatum extract was calculated as 0.31. However, florfenicol exhibited additive effects on isolate WS05 when combined with extracts from A. argyi, A. capillaries, Galla chinensis, P. mume, Radix aucklandiae, and Radix sophorae flavescentis. The joint effects of florfenicol with extracts from other herbs (C. segetum, F. toosendan, P. aviculare, Polygonum cuspidatum, Radix et rhizoma rhei, Radix scutellariae, Radix sanguisorbae, Radix bupleuri, Rhizoma acori tatarinowii) showed no interactions. This suggests that P. granatum extract can be considered as a potential synergist of florfenicol against isolate WS05.

Table 2.
MICs of florfenicol and herb extracts alone or in combination against isolate WS05.
3.5. The in Vitro Activity of Florfenicol-P. granatum Extract
The time–kill curve with florfenicol and P. granatum extract alone or in combination is presented in Figure 8. No reduction in the cell density of isolate WS05 was observed in the control group, and the single treatment with florfenicol and P. granatum extract only showed a slight antibacterial activity in comparison with the control. In contrast, the combination of florfenicol and P. granatum extract exhibited a significant antibacterial activity against isolate WS05, with a significant reduction of ≥3.61 log in cell density after 24 h of treatment compared with that in the single antimicrobial treatment (p < 0.05). According to the criteria suggested by Pillai et al. (2005) [54] that synergism is defined as a respective decrease of ≥2 log CFU mL−1 in antimicrobial activity produced by the combination compared with that by the more active agent alone after 24 h, the in vitro antibacterial activity of the combination of florfenicol and P. granatum extract can be categorized as synergism, which is in line with the findings in the synergistic effect assay above.
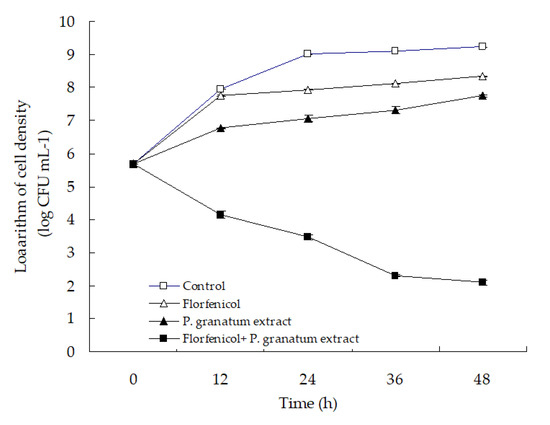
Figure 8.
The time-kill curve with 0.03 mg L−1 florfenicol and 7.81 mg mL−1 P. granatum extract alone or in combination against isolate WS05.
3.6. Protective Effect of Florfenicol-P. granatum Extract
The protective effect of florfenicol and P. granatum extract alone or in combination is shown in Figure 9. No mortality was observed in the test whiteleg shrimp in the blank and positive control groups (data not shown), and the single treatment with florfenicol and P. granatum extract only showed a slightly protective effect in comparison with the negative control, with only a 13.34% and 16.67% decrease in the cumulative mortality of shrimp as compared with the negative control. However, a significant protection was produced by the combination of florfenicol and P. granatum extract, with a cumulative mortality of 36.66% (p < 0.05) and 33.33% (p < 0.05) lower than that in the single treatment with florfenicol and P. granatum extract after the challenge with isolate WS05 for seven days. The death of all of the test shrimp was caused by the challenge strains, as determined by bacterial isolation and identification (data not shown). These findings indicated that the combination of florfenicol and P. granatum extract could potentiate the protective effect against A. hydrophila infection in freshwater-farmed whiteleg shrimp.
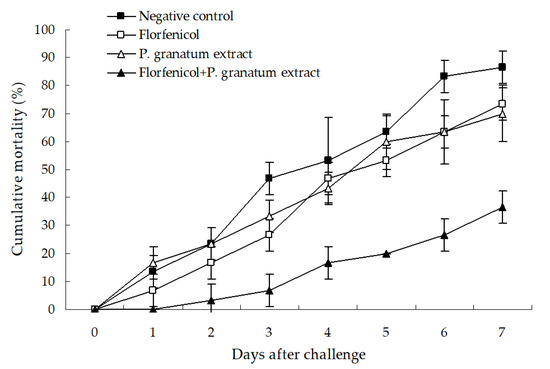
Figure 9.
Protection of 0.03 mg L−1 florfenicol and 7.81 mg mL−1 P. granatum extract alone or in combination against the challenge of isolate WS05 in whiteleg shrimp. Data are presented as the mean ± SD.
4. Discussion
The impact of A. hydrophila in aquaculture has been well documented, with massive mortality reported in fairy shrimp Branchipus schaefferi (Fisher), crayfish Pacifastacus leniusculus, climbing perch Anabas testudineus, grass carp Ctenopharynngodon idellus, silver carp Hypophthalmichthys molitrix, southern catfish Silurus meridionalis, white bream Parabramis pekinensis, and Murray cod Maccullochella peelii [55,56,57,58,59,60,61,62]. However, there is limited information on A. hydrophila infection in freshwater-cultured whiteleg shrimp. Therefore, more attention should be given for whiteleg shrimp–pathogenic A. hydrophila.
Multiple virulence factors have been demonstrated to play an important role in the pathogenicity of A. hydrophila, including flagella, elastase, haemolysins, enterotoxins, and aerolysin [63,64,65,66,67]. Diseases caused by A. hydrophila in aquaculture are usually associated with the production of these virulent factors [68]. In general, the isolated bacteria in aquaculture were classified as virulent, weakly virulent, or avirulent based on the dose and survival time after challenge [56]. A virulent Vibrio parahaemolyticus strain VP1 isolated from vibriosis-infected penaeid shrimp in India was reported to kill 35% of shrimp after 7 days of bath challenge at final concentrations of 2.1 × 104 to 1.3 × 105 CFU mL−1 [69]. Our data indicated that isolate WS05 of A. hydrophila could cause an average mortality of 40% to 55% mortality in healthy shrimp after 7 days of challenge at 2.0 × 104 to 2.0 × 105 CFU mL−1, with an LD50 value of 4.8 × 104 CFU mL−1. This reveals that A. hydrophila isolate WS05 is more virulent than V. parahaemolyticus VP1 and can pose a more serious threat to penaeid shrimp farming. In addition to the virulence of A. hydrophila isolate WS05, there might be other secondary factors that induce this disease, which should also be raised as concerns, such as ammonia and sulfide, which can increase the susceptibility of whiteleg shrimp to bacterial infections by depression of immune capability [70,71].
In many cases, A. hydrophila is abundant in water fauna and is commonly associated with copepods, which can further transport this bacterium to other farming regions to result in aeromonasis [56]. Therefore, A. hydrophila isolate WS05 may constitute a danger for many Aeromonas-susceptible aquatic animals and the control of this isolate should not be underestimated. Several studies have reported that over 300 strains of A. hydrophila from aquatic animals have exhibited a relatively high resistance to rifampicin and novobiocin [72,73]. This is also observed in A. hydrophila isolate WS05. In addition, our data indicated that A. hydrophila isolate WS05 was susceptible to cotrimoxazole, doxycycline, florfenicol, neomycin, and tetracycline, which can be used in the treatment of this disease. However, one of the most important problems involving treatments with antibiotics against A. hydrophila in aquaculture is that antibiotic resistance develops readily [74]. Thus, new techniques to prevent antibiotic resistance development in pathogenic A. hydrophila are needed to make the aquaculture industry more sustainable [75].
Currently, an effective approach to overcome the resistance of bacterial pathogens is a combination of herb extracts with antibiotics. Lee et al. (1998) found that combinations of chloramphenicol and extracts from several herbs, such as Acanthopanax koreanum, Artemisia iwayomogi, Clerodendron trichotomum, Juniperus rigida, Magnolia sieboldii, Pinus densiflora, Pinus koraiensis, and Zanthoxylum piperitum, had resistance inhibitory activities against multidrug-resistant Staphylococcus aureus [76]; Liu et al. (2010) demonstrated that baicalin isolated from Scutellaria amoena had the potential to restore the effectiveness of β-lactam antibiotics against β-lactam-resistant Staphylococcus aureus and could potentiate the killing of β-lactam-resistant S. aureus cells [49]; and Haroun and Al-Kayali (2006) confirmed that Thymbra spicata extracts could significantly increase the antibacterial action of cefotaxime against multidrug-resistant S. aureus and Klebsiella pneumoniae strains [50]. In our study, P. granatum extract potentiated the antimicrobial action of florfenicol, suggesting a possible utilization of this herb extract in combination with florfenicol against A. hydrophila pathogens because a large majority of A. hydrophila strains are susceptible to phenicol antibiotics, like florfenicol [72,73]. Similar findings were also made by Endo et al. (2010) [77], in that a combination of P. granatum extract and fluconazole has a synergistic action against fungal pathogens. Besides its antibacterial activity [78], the synergistic activity of P. granatum extract may be attributed to the ability of its active component, like punicalagin, gallic acid, and chlorogenic acid, to disturb the cell wall and depolarize the cytoplasmic membrane and then facilitate the influx of florfenicol inside bacterial cells to strongly inhibit bacterial protein synthesis [79,80,81].
In conclusion, for the first time, the present study identified A. hydrophila as a causative agent of whiteleg shrimp, and indicated that P. granatum extract potentiated the antimicrobial action of florfenicol, suggesting a possible utilization of this herb in combination with florfenicol against shrimp-pathogenic A. hydrophila.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/7/10/450/s1.
Author Contributions
H.Z. and C.G. conducted the experimental studies; G.Y. cultured the experimental healthy shrimp; J.A. provided the experimental healthy shrimp; K.L. and L.X. performed the data analysis; H.C. designed the experimental studies and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work has been financially supported by the Fishery Sci-Tech Innovation and Popularization Project of Jiangsu Province (No. Y2017-6 and Y2018-8), the Earmarked Fund for China Modern Shrimp Industry Technology Research (No. CARS-48) and Qingpu District Sci-Tech Development Program, Shanghai China (No. QSD2018-11).
Acknowledgments
The authors thank J.S. Li for taking photo of diseased shrimp. Also, we thank the anonymous academic editor and reviewers for their insightful comments on the manuscript.
Conflicts of Interest
The authors have no conflicts of interest.
References
- Benzie, J.A.H. Use and exchange of genetic resources of penaeid shrimps for food and aquaculture. Rev. Aquacult. 2009, 1, 232–250. [Google Scholar] [CrossRef]
- Zhou, J. Development path and technological change of shrimp aquaculture in China. Chin. Agric. Sci. Bull. 2016, 32, 22–29. [Google Scholar]
- Ding, S.; Wang, F.; Dong, S.; Li, Y. Comparison of the respiratory metabolism of juvenile Litopenaeus vannamei cultured in seawater and freshwater. J. Ocean. Univ. China (Ocean. Coast. Sea Res.) 2014, 13, 331–337. [Google Scholar] [CrossRef]
- Ministry of Agriculture and Rural Affairs of China. China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2018; p. 24.
- Aguirre-Guzmán, G.; Ruíz, H.M.; Ascencio, R.F. A review of extracellular virulence product of Vibrio species important in diseases of cultivated shrimp. Aquac. Res. 2004, 35, 1395–1404. [Google Scholar] [CrossRef]
- Sudheesh, P.S.; Xu, H. Pathogenicity of Vibrio parahaemolyticus in tiger prawn Penaeus monodon Fabricius: Possible role of extracellular proteases. Aquaculture 2001, 196, 37–46. [Google Scholar] [CrossRef]
- Zhou, J.; Fang, W.; Yang, X.; Zhou, S.; Hu, L.; Li, X.; Qi, X.; Su, H.; Xie, L. A nonluminescent and highly virulent Vibrio harveyi strain is associated with “bacterial white tail disease” of Litopenaeus vannamei shrimp. PLoS ONE 2012, 7, e29961. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; An, J.; Zheng, W.; He, S. Vibrio cholerae pathogen from the freshwater-cultured whiteleg shrimp Penaeus vannamei and control with Bdellovibrio bacteriovorus. J. Invertebr. Pathol. 2015, 130, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; He, S.; Lu, L.; Yang, X.; Chen, B. Identification of a Proteus penneri isolate as the causal agent for the red body disease of the cultured white shrimp Penaeus vannamei and its control with Bdellovibrio bacteriovorus. Anton. Leeuwen. 2014, 105, 423–430. [Google Scholar] [CrossRef]
- Cao, H.; An, J.; He, S.; Lu, L.; Yang, X.; Zheng, W. Aeromonas schubertii: A potential pathogen for the freshwater cultured whiteleg shrimp Penaeus vannamei. Isr. J. Aquacult. Bamid. 2015, 67, 1–6. [Google Scholar]
- Cao, H.; Chen, S.; Lu, L.; An, J. Shewanella algae: An emerging pathogen of black spot disease in freshwater-cultured whiteleg shrimp (Penaeus vannamei). Isr. J. Aquacult. Bamid. 2018, 70, 1–7. [Google Scholar]
- Holmstrom, K.; Graslund, S.; Wahlstrom, A.; Poungshompoo, S.; Bengtsson, B.E.; Kautsky, N. Antibiotics use in shrimp farming and implications for environmental impacts and human health. Int. J. Food Sci. Tech. 2003, 38, 255–266. [Google Scholar] [CrossRef]
- Harikrishnan, R.; Balasundaram, C. Modern trends in Aeromonas hydrophila disease management with fish. Rev. Fish. Sci. 2005, 13, 281–320. [Google Scholar] [CrossRef]
- Bhuvaneswari, R.; Balasundaram, C. Traditional Indian herbal extracts used in vitro against growth of the pathogenic bacteria—Aeromonas hydrophila. Isr. J. Aquacult. Bamid. 2006, 58, 89–96. [Google Scholar]
- Haniffa, M.A.; Kavitha, K. Antibacterial activity of medicinal herbs against the fish pathogen Aeromonas hydrophila. J. Agric. Tech. 2012, 8, 205–211. [Google Scholar]
- Al Laham, S.A.; Al Fadel, F.M. Antibacterial activity of various plants extracts against antibiotic-resistant Aeromonas hydrophila. Jundishapur. J. Microbiol. 2014, 7, e11370. [Google Scholar] [CrossRef]
- Türker, H.; Yildirim, A.B.; Karakaş, F.P.; Köylüoğlu, H. Antibacterial activities of extracts from some Turkish endemic plants on common fish pathogens. Turk. J. Biol. 2009, 33, 73–78. [Google Scholar]
- Nya, E.J.; Austin, B. Use of garlic, Allium sativum, to control Aeromonas hydrophila infection in rainbow trout, Oncorhynchus mykiss (Walbaum). J. Fish. Dis. 2009, 32, 963–970. [Google Scholar] [CrossRef]
- Pakravan, S.; Hajimoradloo, A.; Ghorbani, R. Effect of dietary willow herb, Epilobium hirsutum extract on growth performance, body composition, haematological parameters and Aeromonas hydrophila challenge on common carp, Cyprinus carpio. Aquac. Res. 2012, 43, 861–869. [Google Scholar] [CrossRef]
- Abdel-Tawwab, M. The use of American ginseng (Panax quinquefolium) in practical diets for Nile tilapia (Oreochromis niloticus): Growth performance and challenge with Aeromonas hydrophila. J. Appl. Aquacult. 2012, 24, 366–376. [Google Scholar] [CrossRef]
- Wu, C.C.; Liu, C.H.; Chang, Y.P.; Hsieh, S.L. Effects of hot-water extract of Toona sinensis on immune response and resistance to Aeromonas hydrophila in Oreochromis mossambicus. Fish Shellfish. Immun. 2010, 29, 258–263. [Google Scholar] [CrossRef]
- Cheesman, M.J.; Ilanko, A.; Blonk, B.; Cock, I.E. Developing new antimicrobial therapies: Are synergistic combinations of plant extracts/compounds with conventional antibiotics the solution? Pharmacogn. Rev. 2017, 11, 57–72. [Google Scholar] [PubMed]
- Dong, J.; Ruan, J.; Xu, N.; Yang, Y.; Ai, X. In vitro synergistic effects of fisetin and norfloxacin against aquatic isolates of Serratia marcescens. FEMS Microbiol. Lett. 2016, 363, 1–5. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jedlicková, Z.; Mottl, O.; Sery, V. Antibacterial properties of the Vietnamese cajeput oil and ocimum oil in combination with antibacterial agents. J. Hyg. Epidemiol. Microbiol. Immunol. 1992, 36, 303–309. [Google Scholar] [PubMed]
- Jayaraman, P.; Sakharkar, M.K.; Lim, C.S.; Tang, T.H.; Sakharkar, K.R. Activity and interactions of antibiotic and phytochemical combinations against Pseudomonas aeruginosa in vitro. Int. J. Biol. Sci. 2010, 6, 556–558. [Google Scholar] [CrossRef] [PubMed]
- Jayasinghe, C.V.L.; Ahmed, S.B.N.; Kariyawasam, M.G.I.U. The isolation and identification of Vibrio species in marine shrimps of Sri Lanka. J. Food Agric. 2008, 1, 36–44. [Google Scholar] [CrossRef]
- Moss, S.M.; Pruder, G.D.; Leber, K.M.; Wyban, J.A. The relative enhancement of Penaeus vannamei growth by selected fractions of shrimp pond water. Aquaculture 1992, 101, 229–239. [Google Scholar] [CrossRef]
- Moss, S.M.; Divakaran, S.; Kim, B.G. Stimulating effects of pond water on digestive enzyme activity in the Pacific white shrimp, Litopenaeus vannamei (Boone). Aquac. Res. 2001, 32, 125–131. [Google Scholar] [CrossRef]
- Network of Aquaculture Centers in Asia-Pacific. Shrimp Health Management Extension Manual; MPEDA House: Cochin, India, 2003; p. 23. [Google Scholar]
- Biswas, G.; Korenaga, H.; Nagamine, R.; Kono, T.; Shimokawa, H.; Itami, T.; Sakai, M. Immune stimulant effects of a nucleotide-rich baker’s yeast extract in the Kuruma shrimp, Marsupenaeus japonicus. Aquaculture 2012, 366, 40–45. [Google Scholar] [CrossRef]
- Luo, G.; Tan, H.; Zhu, X. The effects of several water-treatment units in a recirculating aquaculture system. J. Dalian Fish. Univ. 2008, 23, 68–72. [Google Scholar]
- Ekanem, A.P.; Eyo, V.O.; Sampson, A.F. Parasites of landed fish from Great Kwa River, Calabar, Cross River State, Nigeria. Int. J. Fish. Aquacult. 2011, 3, 225–230. [Google Scholar]
- Pan, X.; Shen, J.; Li, J.; He, B.; Yin, W.; Hao, G.; Xu, Y.; Yao, J. Identification and biological characteristics of the pathogen causing Macrobrachium nipponense soft-shell syndrome. Microbiol. China 2009, 36, 1571–1576. [Google Scholar]
- Cao, H.; Yang, Y.; Chen, S.; Ai, X. Providencia alcalifaciens: A causal agent of red leg disease in freshwater-cultured whiteleg shrimp Penaeus vannamei. Isr. J. Aquacult. Bamid. 2018, 70, 1–7. [Google Scholar]
- Orozova, P.; Sirakov, I.; Petkov, I.; Crumlish, M.; Austin, B. Recovery of Aeromonas hydrophila associated with bacteraemia in captive snakes. FEMS Microbiol. Lett. 2012, 334, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Petti, C.A.; Polage, C.R.; Schreckenberger, P. The role of 16S rRNA gene sequencing in identification of microorganisms misidentified by conventional methods. J. Clin. Microbiol. 2005, 43, 6123–6125. [Google Scholar] [CrossRef]
- Cao, H.; He, S.; Lu, L.; Hou, L. Characterization and phylogenetic analysis of the bitrichous pathogenic Aeromonas hydrophila isolated from diseased Siberian sturgeon. Isr. J. Aquacult. Bamid. 2010, 62, 181–189. [Google Scholar]
- Yáňez, M.A.; Catalán, V.; Apráiz, D.; Figueras, M.J.; Martínez-Murcia, A.J. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int. J. Syst. Evol. Microbiol. 2003, 53, 875–883. [Google Scholar] [CrossRef]
- Küpfer, M.; Kuhnert, P.; Korczak, B.M.; Peduzzi, R.; Demarta, A. Genetic relationships of Aeromonas strains inferred from 16S rRNA, gyrB and rpoB gene sequences. Int. J. Syst. Evol. Microbiol. 2006, 56, 2743–2751. [Google Scholar] [CrossRef]
- Abeyta, C.; Kaysner, C.A.; Wekell, M.M.; Sullivan, J.J.; Stelma, G.N. Recovery of Aeromonas hydrophila from oysters implicated in an outbreak of foodborne illness. J. Food Protect. 2019, 49, 643–646. [Google Scholar] [CrossRef]
- Long, S.; Han, S.; Niu, Z.; Liang, J.; Hu, D.; Huang, J.; Li, H.; Liu, Q.; Su, J. Isolation and identification of pathogenic Aeromonas in Clarias fuscus and analysis of the correlation between its pathogenicity and virulence genorypes. J. Fish. China 2016, 40, 308–317. [Google Scholar]
- Ye, W.; Guo, C.; Cao, H.; Yang, X. Isolation, identification, pathogenicity and in vitro antibacterial drugs of pathogenic Aeromonas hydrophila from Micropterus salmoides with hemorrhagic disease. Freshw. Fish. 2018, 5, 54–60. [Google Scholar]
- Teng, T.; Liang, L.; Chen, K.; Xi, B.; Xie, J.; Xu, P. Isolation, identification and phenotypic and molecular characterization of pathogenic Vibrio vulnificus isolated from Litopenaeus vannamei. PLoS ONE 2017, 12, e0186135. [Google Scholar] [CrossRef] [PubMed]
- Ogbuagu, D.H.; Iwuchukwu, E.I. Evaluation of the toxicity of three hair shampoos on the catfish (Clarias gariepinus) fingerlings. Appl. Ecol. Environ. Sci. 2014, 2, 86–89. [Google Scholar]
- Jones, R.N.; Ballow, C.H.; Biedenbach, D.J. Multi-laboratory assessment of the linezolid spectrum of activity using the Kirby-Bauer disk diffusion method: Report of the Zyvox (R) antimicrobial potency study (ZAPS) in the United States. Diagn. Microbiol. Infect. Dis. 2001, 40, 59–66. [Google Scholar] [CrossRef]
- Chen, Z.; Li, X.; Wu, X.; Wang, W.; Wang, W.D. Synergistic activity of econazole-nitrate and chelerythrine against clinical isolates of Candida albicans. Iran. J. Pharm. Res. 2014, 13, 567–573. [Google Scholar] [PubMed]
- Liu, Q.; Li, J.; Wang, Q.; Dai, F. Pharmacodynamic Test Technical Specification for Aquaculture Antimicrobial Agent—Part1: Antibiotics Susceptibility Test of Macro-Broth Dilution Method; China Standards Press: Beijing, China, 2011; pp. 1–6. [Google Scholar]
- Rattanachaikunsopon, P.; Phumkhachorn, P. Prophylactic effect of Andrographis paniculata extracts against Streptococcus agalactiae infection in Nile tilapia (Oreochromis niloticus). J. Biosci. Bioeng. 2009, 107, 579–582. [Google Scholar] [CrossRef]
- Liu, I.X.; Durham, D.G.; Richards, R.M.E. Baicalin synergy with β-lactam antibiotics against methicillin–resistant Staphylococcus aureus and other β-lactam-resistant strains of S. aureus. J. Pharm. Pharmacol. 2000, 52, 361–366. [Google Scholar] [CrossRef]
- Haroun, M.; Al-Kayali, R.S. Synergistic effect of Thymbra spicata L. extracts with antibiotics against multidrug-resistant Staphylococcus aureus and Klebsiella pneumoniae strains. Iran. J. Basic Med. Sci. 2016, 19, 1193–1200. [Google Scholar]
- Liu, H.; Zhang, W.; Wu, Y.; Sun, L.; Wang, Y.; Liu, Y.; Zhang, X.; Hong, P.; Ji, H. Synergistic antimicrobial effect and mechanism of lipopeptides and tea polyphenols against Vibrio parahaemolyticus. Food Sci. 2017, 38, 14–19. [Google Scholar]
- Gong, X.; Le, G.; Li, Y. Antibacterial spectrum of antibacterial peptides from Musca dommestica larvae and synergic interaction between the peptides and antibiotics. Acta Microbiol. Sin. 2005, 45, 516–520. [Google Scholar]
- Zou, W.; Yang, C.; Jiang, I.; Wu, S.; Yi, Q.; Wu, J. Guideline of fishery drug application (NY5071-2002). Ocean. Fish. 2007, 12, 39–41. [Google Scholar]
- Pillai, S.K.; Moellering, R.C.; Eliopoulos, G.M. Antimicrobial combinations. In Antibiotics in Laboratory Medicine, 5th ed.; Lorian, V., Ed.; Williams & Wilkins: Baltimore, MD, USA, 2005; pp. 365–440. [Google Scholar]
- Dierckens, K.R.; Vandenberghe, J.; Beladjal, L.; Huys, G.; Mertens, J.; Swings, J. Aeromonas hydrophila causes ‘black disease’ in fairy shrimps (Anostraca; Crustacea). J. Fish. Dis. 1998, 21, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Jiravanichpaisal, P.; Roos, S.; Edsman, L.; Liu, H.; Soderhall, K. A highly virulent pathogen, Aeromonas hydrophila, from the freshwater crayfish Pacifastacus leniusculus. J. Invertebr. Pathol. 2009, 101, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.F.; Rashid, M.M.; Sayed, M.A. Experimental infection of indigenous climbing perch Anabas testudineus with Aeromonas hydrophila bacteria. Prog. Agric. 2011, 22, 105–114. [Google Scholar] [CrossRef]
- Zheng, W.; Cao, H.; Yang, X. Grass carp (Ctenopharynngodon idellus) infected with multiple strains of Aeromonas hydrophila. Afr. J. Microbiol. Res. 2012, 21, 4512–4520. [Google Scholar]
- Rashid, M.M.; Hossain, M.S.; Ali, M.F. Isolation and identification of Aeromonas hydrophila from silver carp and its culture environment from Mymensingh region. J. Bangladesh Agric. Univ. 2013, 2, 373–376. [Google Scholar] [CrossRef]
- Zhu, C.; Zhou, X.; Zhang, Q. Pathogenic bacterium identification and histopathology of septicemia of juvenile southern catfish, Silurus meridionalis Chen. J. Fish. Sci. China 2011, 2, 360–370. [Google Scholar] [CrossRef]
- Ye, Y.W.; Fan, T.F.; Li, H.; Lu, J.F.; Jiang, H.; Hu, W.; Jiang, Q.H. Characterization of Aeromonas hydrophila from hemorrhagic diseased freshwater fishes in Anhui Province, China. Int. Food Res. J. 2013, 3, 1449–1452. [Google Scholar]
- Gai, C.; Ye, W.; Lu, L.; Li, Y.; Yang, X.; Cao, H. Aeromonas hydrophila: A causative agent for tail rot disease in freshwater cultured Murray cod Maccullochella peelii. Isr. J. Aquacult. Bamid. 2016, 68, 1–8. [Google Scholar]
- Altarriba, M.; Merino, S.; Gavin, R.; Canals, R.; Rabaan, A.; Shaw, J.G.; Tomas, J.M. A polar flagella operon (flg) of Aeromonas hydrophila contains genes required for lateral flagella expression. Microb. Pathol. 2003, 34, 249–259. [Google Scholar] [CrossRef]
- Cascón, A.; Yugueros, J.; Temprano, A.; Sánchez, M.; Hernanz, C.; Luengo, J.M.; Naharro, G. A major secreted elastase is essential for pathogenicity of Aeromonas hydrophila. Infect. Immun. 2000, 68, 3233–3241. [Google Scholar] [CrossRef]
- Gonzalez-Serrano, C.J.; Santos, J.A.; García-López, M.L.; Otero, A. Virulence markers in Aeromonas hydrophila and Aeromonas veronii biovar sobria isolates from freshwater fish and from a diarrhea case. J. Appl. Microbiol. 2002, 93, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Erova, T.E.; Pillai, L.; Fadl, A.A.; Sha, J.; Wang, S.; Galindo, C.L.; Chopra, A.K. DNA adenine methyltransferase influences the virulence of Aeromonas hydrophila. Infect. Immun. 2006, 74, 410–424. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dong, J.; Liu, Y.; Xu, N.; Yang, Q.; Yang, Y.; Ai, X. Expression, purification and characterization of aerolysin from Aeromonas hydrophila. Chin. Fish. Qual. Stand. 2019, 9, 27–33. [Google Scholar]
- Jiang, Q.; Ye, Y.; Hu, W.; Jiang, H.; Lu, J. Research progresses of virulence factors and control technologies in Aeromonas hydrophila. Mod. Agric. Sci. Technol. 2012, 6, 324–327. [Google Scholar]
- Haldar, S.; Chatterjee, S.; Asakura, M.; Vijayakumaran, M.; Yamasaki, S. Isolation of Vibrio parahaemolyticus and Vibrio cholerae (Non-O1 and O139) from moribund shrimp (Penaeus monodon) and experimental challenge study against post larvae and juveniles. Ann. Microbiol. 2007, 57, 55–60. [Google Scholar] [CrossRef]
- Liu, C.H.; Chen, J.C. Effect of ammonia on the immune response of white shrimp Litopenaeus vannamei and its susceptibility to Vibrio alginolyticus. Fish. Shellfish. Immunol. 2004, 16, 321–334. [Google Scholar] [CrossRef]
- Hsu, S.W.; Chen, J.C. The immune response of white shrimp Penaeus vannamei and its susceptibility to Vibrio alginolyticus under sulfide stress. Aquaculture 2007, 271, 61–69. [Google Scholar] [CrossRef]
- Vivekanandhan, G.; Savithamani, K.; Hatha, A.A.M.; Lakshmanaperumalsamy, P. Antibiotic resistance of Aeromonas hydrophila isolated from marketed fish and prawn of South India. Int. J. Food Microbiol. 2002, 76, 165–168. [Google Scholar] [CrossRef]
- Hatha, M.; Vivekanandhan, A.A.; Joice, G.J.; Christol. Antibiotic resistance pattern of motile aeromonads from farm raised fresh water fish. Int. J. Food Microbiol. 2005, 98, 131–134. [Google Scholar] [CrossRef]
- Belém-Costa, A.; Cyrino, J.E.P. Antibiotic resistance of Aeromonas hydrophila isolated from Piaractus mesopotamicus (Holmberg, 1887) and Oreochromis niloticus (Linnaeus, 1758). Sci. Agric. 2006, 63, 281–284. [Google Scholar] [CrossRef]
- Defoirdt, T.; Sorgeloos, P.; Bossier, P. Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr. Opin. Microbiol. 2011, 14, 251–258. [Google Scholar] [CrossRef]
- Lee, C.K.; Kim, H.; Moon, K.H.; Shin, K.H. Screening and isolation of antibiotic resistance inhibitors from herb materials-resistance inhibition of volatile components of Korean aromatic herbs. Arch. Pharm. Res. 1998, 21, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Endo, E.H.; Cortez, D.A.G.; Ueda-Nakamura, T.; Nakamura, C.V.; Filho, B.P.D. Potent antifungal activity of extracts and pure compound isolated from pomegranate peels and synergism with fluconazole against Candida albicans. Res. Microbiol. 2010, 161, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, D.; Asha, M.K.; Amit, A. Antibacterial activity of Punica granatum. Fitoterapia 2001, 72, 171–173. [Google Scholar] [CrossRef]
- Li, G.; Guo, W.; Gao, X.; Wang, Y.; Sun, S.; Xia, X. Synergistic antimicrobial effect and mechanism of punicalagin and chlorogenic acid against Staphyloccocus aureus. Sci. Tech. Food Ind. 2018, 39, 17–21. [Google Scholar]
- Lu, X.; Zhou, L.; Xie, K.; Jing, Y.; Yun, B.; Wang, Y.; Xie, M. Antibacterial activity and mechanism of gallic acid on Staphylococcus aureus. Edib. Fung. China 2012, 31, 54–56. [Google Scholar]
- Choi, M.J.; Lee, E.; Lee, S.; Reza, M.A.; Le, J.; Gebru, E. The in vitro antibacterial activity of florfenicol in combination with amoxicillin or cefuroxime against pathogenic bacteria of animal origin. Pak. Vet. J. 2011, 31, 141–144. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).