The Metallochaperone Encoding Gene hypA Is Widely Distributed among Pathogenic Aeromonas spp. and Its Expression Is Increased under Acidic pH and within Macrophages
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. In Silico Search of hypA and ure Genes in the Aeromonas Genomes
2.3. Protein Sequence Analysis and 3D-Structure Prediction
2.4. Cell Line Culture, Infection and Induction Experiments
2.5. RNA Extraction and Quantitative RT-PCR
2.6. Statistical Analysis
3. Results
3.1. Identification of hypA Gene in Aeromonas Species
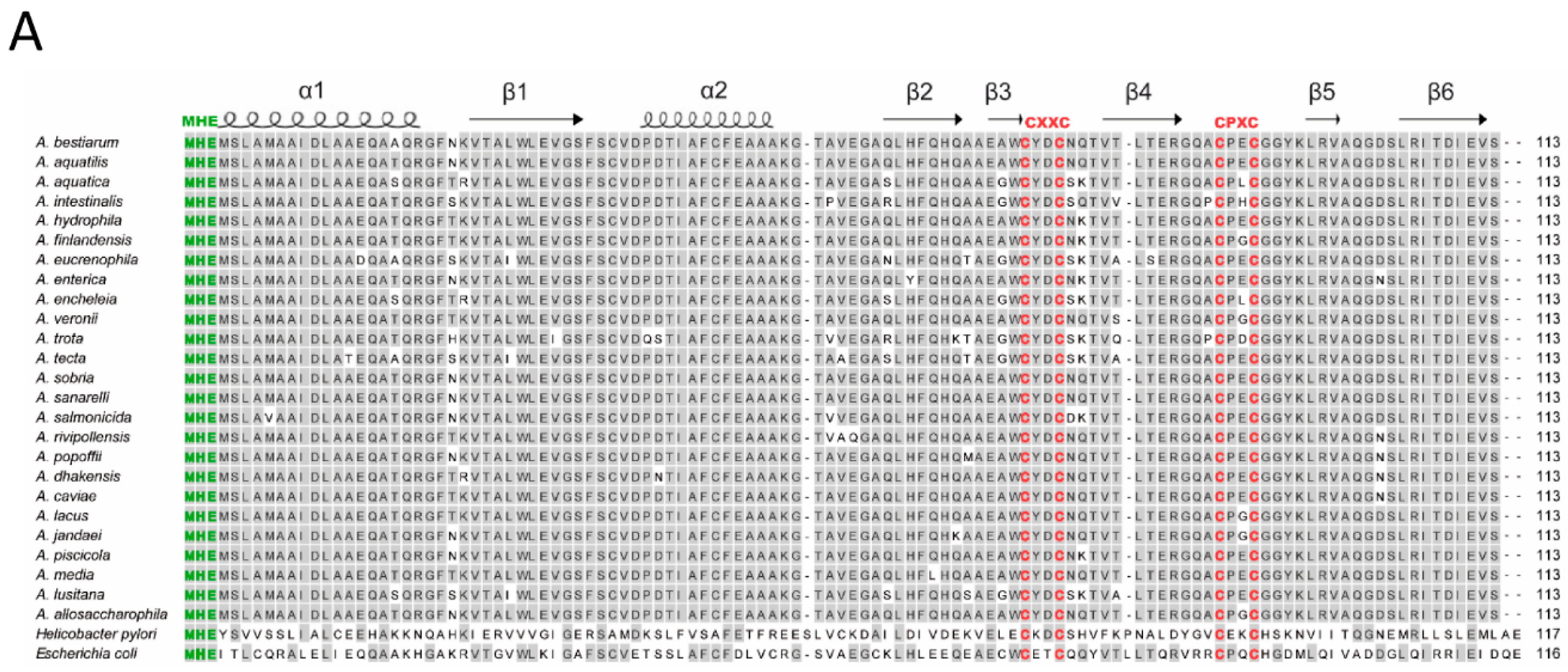
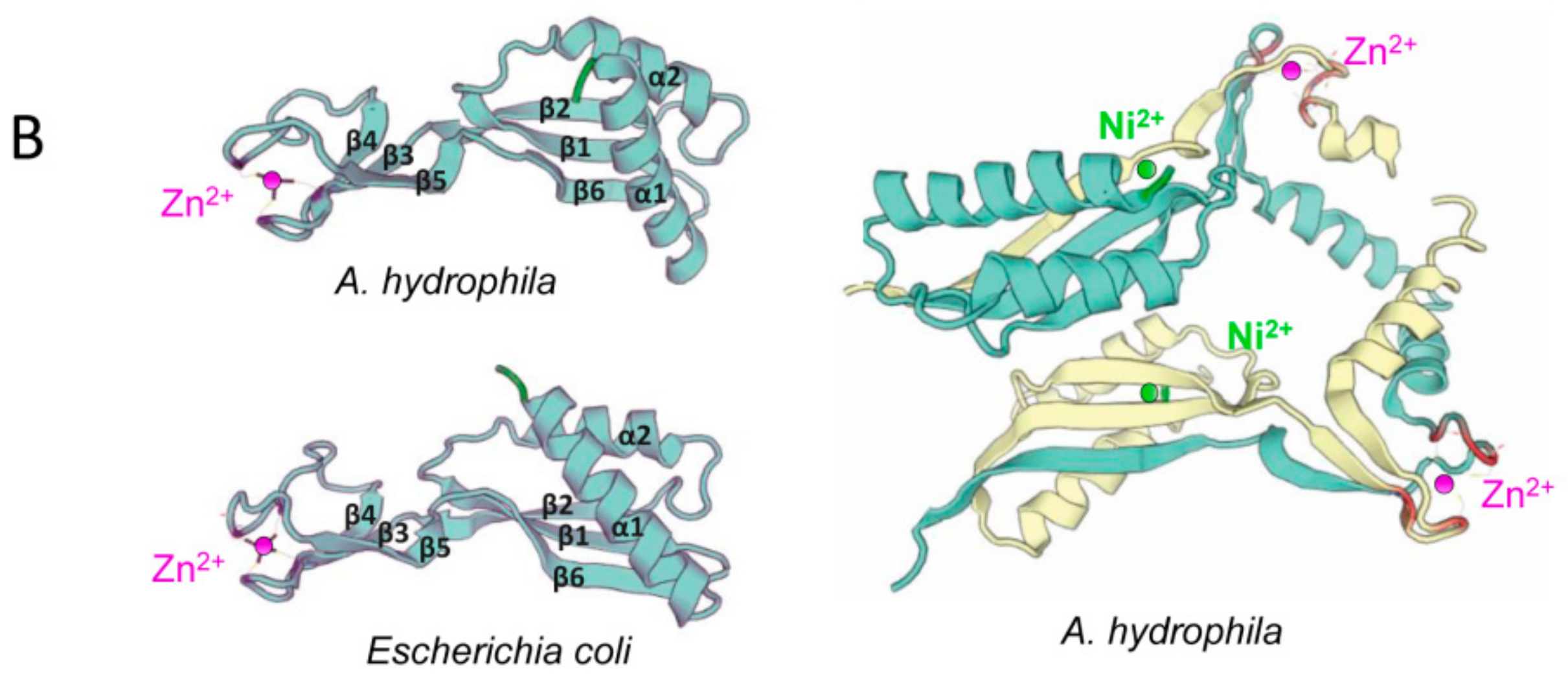
3.2. Sequence Analyses of hypA Proteins Shows Specific Motifs Associated to Metal Binding
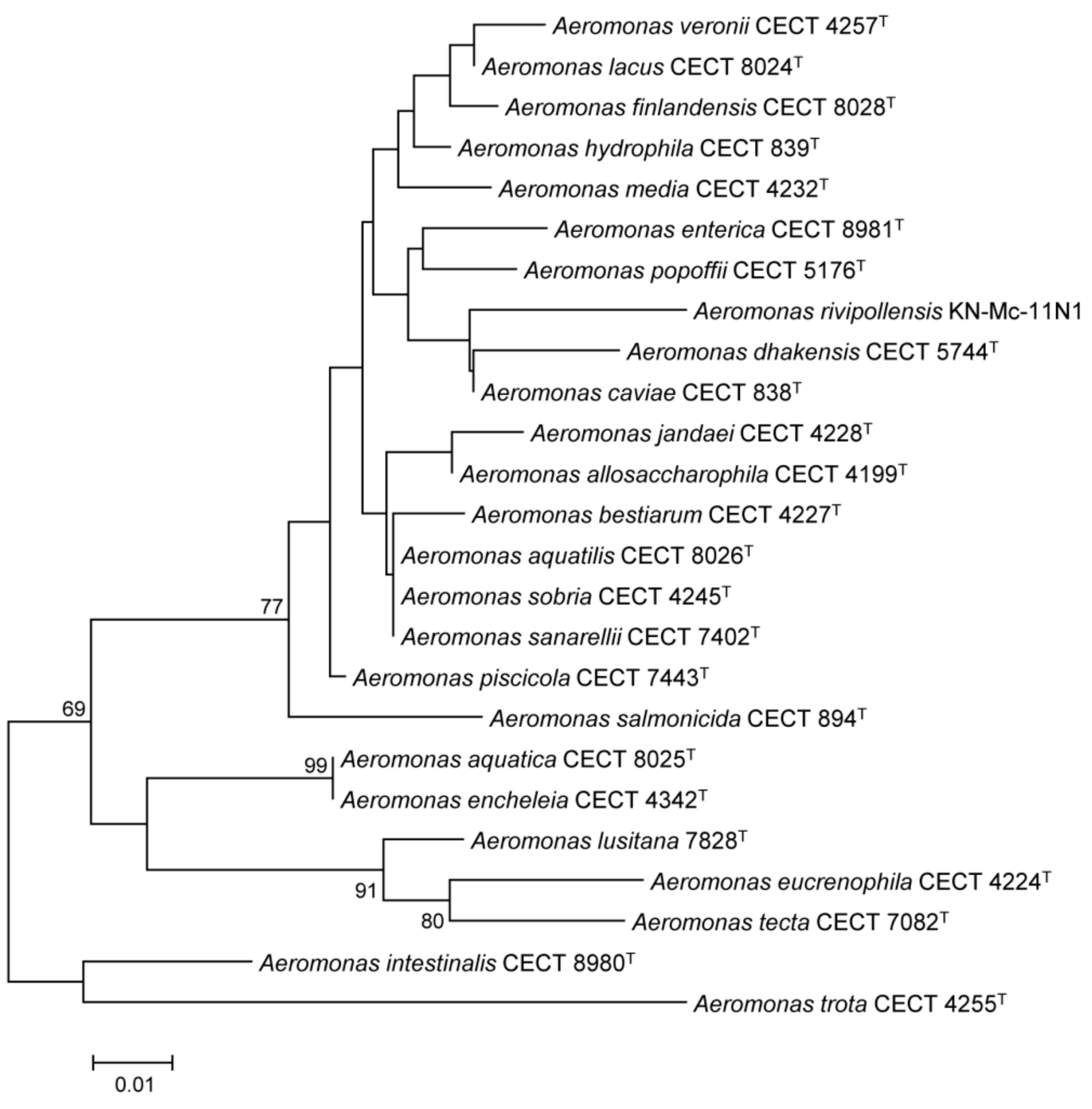
3.3. Conservation and Phylogenetic Relationships of hypA in Aeromonas
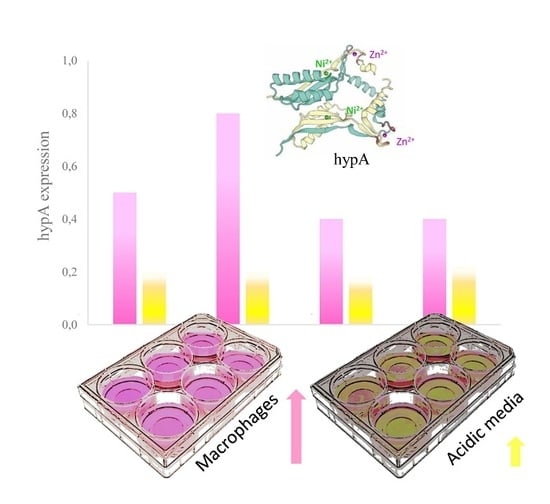
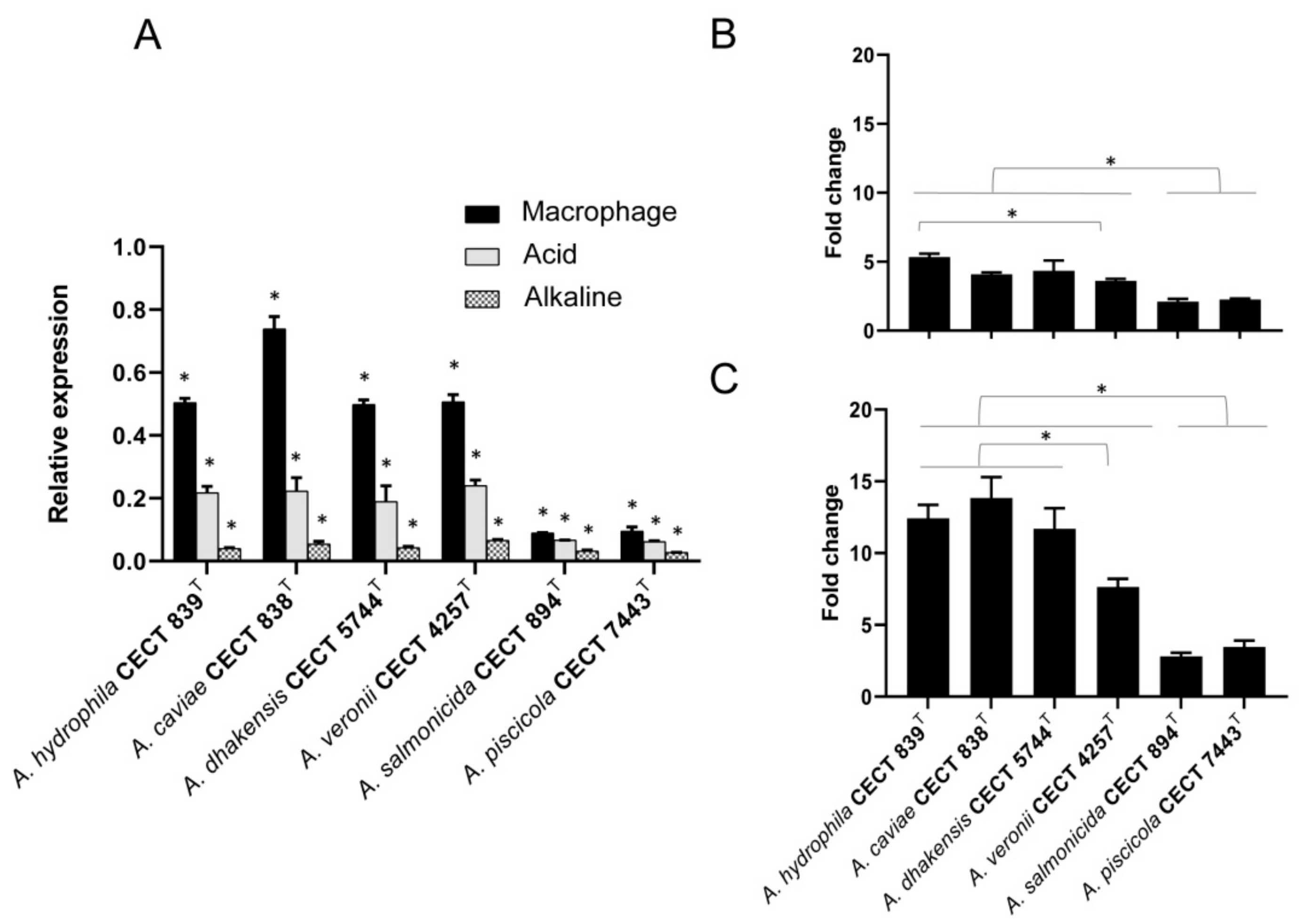
3.4. Transcriptional Regulation of hypA under Different pH-Condition and Macrophage
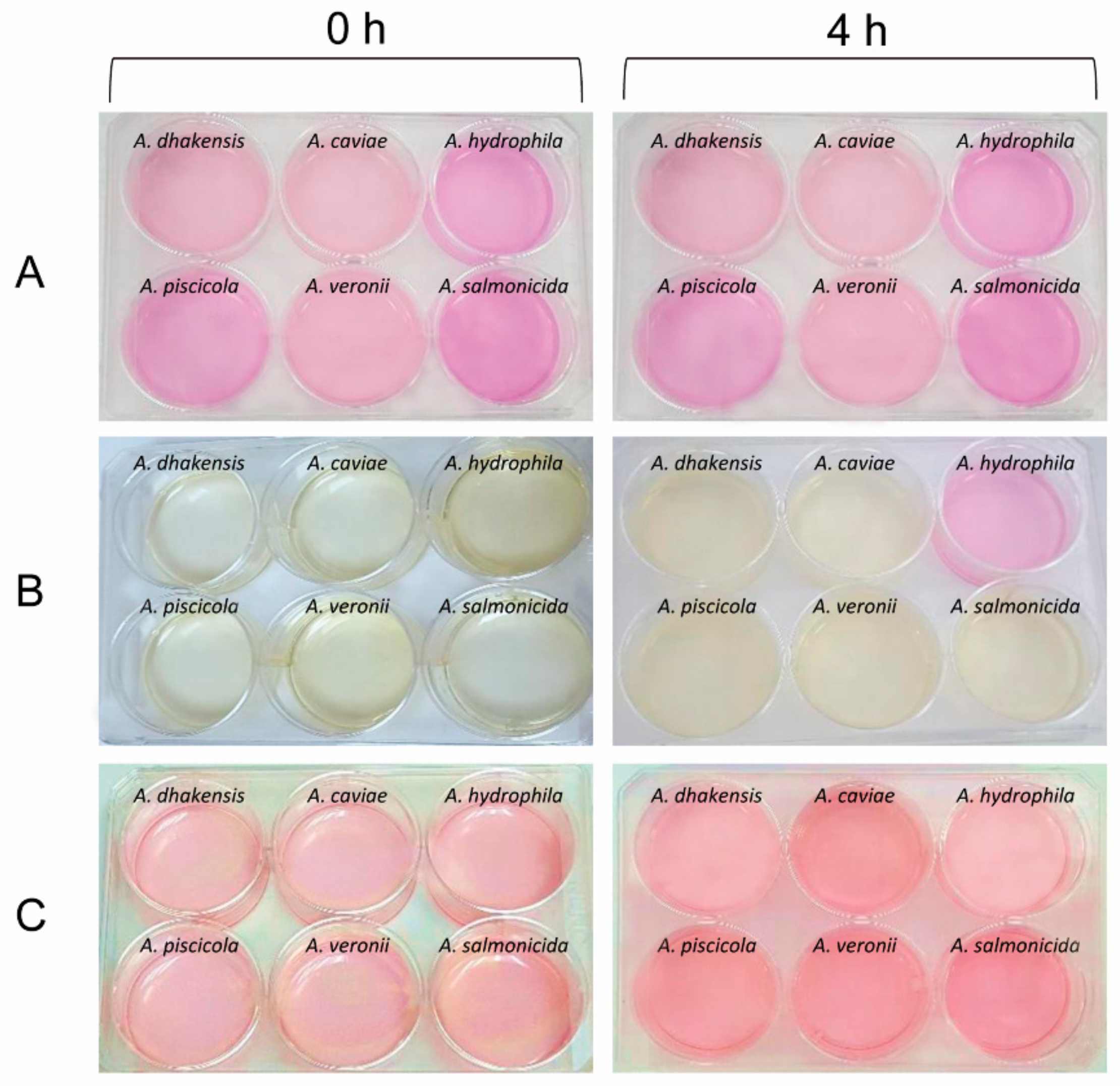
3.5. Urease Activity and Urease Genes in Aeromonas Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- O’Halloran, T.V.; Culotta, V.C. Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 2000, 275, 25057–25060. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yeung, N.; Sieracki, N.; Marshall, N.M. Design of functional metalloproteins. Nature 2009, 460, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Aschner, M.; Syversen, T.; Souza, D.O.; Rocha, J.B. Metallothioneins: Mercury species-specific induction and their potential role in attenuating neurotoxicity. Exp. Biol. Med. 2006, 231, 1468–1473. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, A.C. Metallochaperones: Bind and deliver. Chem. Biol. 2002, 9, 673–677. [Google Scholar] [CrossRef]
- Waldron, K.J.; Rutherford, J.C.; Ford, D.; Robinson, N.J. Metalloproteins and metal sensing. Nature 2009, 460, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Capdevila, D.A.; Edmonds, K.A.; Giedroc, D.P. Metallochaperones and metalloregulation in bacteria. Essays Biochem. 2017, 61, 177–200. [Google Scholar] [CrossRef]
- Palmer, L.D.; Skaar, E.P. Transition metals and virulence in bacteria. Annu. Rev. Genet. 2016, 50, 67–91. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, L.A.; Parkin, A. Electrochemical insights into the mechanism of NiFe membrane-bound hydrogenases. Biochem. Soc. Trans. 2016, 44, 315–328. [Google Scholar] [CrossRef]
- Winter, G.; Buhrke, T.; Lenz, O.; Jones, A.K.; Forgber, M.; Friedrich, B. A model system for [NiFe] hydrogenase maturation studies: Purification of an active site-containing hydrogenase large subunit without small subunit. FEBS Lett. 2005, 579, 4292–4296. [Google Scholar] [CrossRef]
- Lubitz, W.; Ogata, H.; Rudiger, O.; Reijerse, E. Hydrogenases. Chem. Rev. 2014, 114, 4081–4148. [Google Scholar] [CrossRef]
- Chan, K.H.; Lee, K.M.; Wong, K.B. Interaction between hydrogenase maturation factors HypA and HypB is required for [NiFe]-hydrogenase maturation. PLoS ONE 2012, 7, e32592. [Google Scholar] [CrossRef] [PubMed]
- Liebgott, P.P.; Leroux, F.; Burlat, B.; Dementin, S.; Baffert, C.; Lautier, T.; Fourmond, V.; Ceccaldi, P.; Cavazza, C.; Meynial-Salles, I.; et al. Relating diffusion along the substrate tunnel and oxygen sensitivity in hydrogenase. Nat. Chem. Biol. 2010, 6, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Kawashima, T.; Nishitani, Y.; Kanai, T.; Wada, T.; Inaba, K.; Atomi, H.; Imanaka, T.; Miki, K. Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer. Proc. Natl. Acad. Sci. USA 2015, 112, 7701–7706. [Google Scholar] [CrossRef] [PubMed]
- Lacasse, M.J.; Douglas, C.D.; Zamble, D.B. Mechanism of selective nickel transfer from HypB to HypA, Escherichia coli [NiFe]-hydrogenase accessory proteins. Biochemistry 2016, 55, 6821–6831. [Google Scholar] [CrossRef] [PubMed]
- Atanassova, A.; Zamble, D.B. Escherichia coli HypA is a zinc metalloprotein with a weak affinity for nickel. J. Bacteriol. 2005, 187, 4689–4697. [Google Scholar] [CrossRef]
- Xia, W.; Li, H.; Sze, K.H.; Sun, H. Structure of a nickel chaperone, HypA, from Helicobacter pylori reveals two distinct metal binding sites. J. Am. Chem. Soc. 2009, 131, 10031–10040. [Google Scholar] [CrossRef] [PubMed]
- Blum, F.C.; Hu, H.Q.; Servetas, S.L.; Benoit, S.L.; Maier, R.J.; Maroney, M.J.; Merrell, D.S. Structure-function analyses of metal-binding sites of HypA reveal residues important for hydrogenase maturation in Helicobacter pylori. PLoS ONE 2017, 12, e0183260. [Google Scholar] [CrossRef]
- Hu, H.Q.; Johnson, R.C.; Merrell, D.S.; Maroney, M.J. Nickel ligation of the N-terminal amine of HypA is required for urease maturation in Helicobacter pylori. Biochemistry 2017, 56, 1105–1116. [Google Scholar] [CrossRef]
- Johnson, R.C.; Hu, H.Q.; Merrell, D.S.; Maroney, M.J. Dynamic HypA zinc site is essential for acid viability and proper urease maturation in Helicobacter pylori. Metallomics 2015, 7, 674–682. [Google Scholar] [CrossRef]
- Vignais, P.M.; Billoud, B. Occurrence, classification, and biological function of hydrogenases: An overview. Chem. Rev. 2007, 107, 4206–4272. [Google Scholar] [CrossRef]
- Hayes, E.T.; Wilks, J.C.; Sanfilippo, P.; Yohannes, E.; Tate, D.P.; Jones, B.D.; Radmacher, M.D.; BonDurant, S.S.; Slonczewski, J.L. Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol. 2006, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, K.; Riggins, D.P.; Eldahan, K.C.; Kitko, R.D.; Slonczewski, J.L. Hydrogenase-3 contributes to anaerobic acid resistance of Escherichia coli. PLoS ONE 2010, 5, e10132. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, O.; Wust, P.K.; Hellmuth, S.; Borst, K.; Horn, M.A.; Drake, H.L. Novel [NiFe]- and [FeFe]-hydrogenase gene transcripts indicative of active facultative aerobes and obligate anaerobes in earthworm gut contents. Appl. Env. Microbiol. 2011, 77, 5842–5850. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef]
- Figueras, M.J.; Beaz-Hidalgo, R. Aeromonas infections in humans. In Aeromonas; Graf, J., Ed.; Caister Academic Press: Norfolk, UK, 2015; pp. 65–108. [Google Scholar]
- Teunis, P.; Figueras, M.J. Reassessment of the enteropathogenicity of mesophilic Aeromonas species. Front. Microbiol. 2016, 7, 1395. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Soler, L.; Yanez, M.A.; Chacon, M.R.; Aguilera-Arreola, M.G.; Catalan, V.; Figueras, M.J.; Martinez-Murcia, A.J. Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes. Int. J. Syst. Evol. Microbiol. 2004, 54, 1511–1519. [Google Scholar] [CrossRef]
- Canals, R.; Altarriba, M.; Vilches, S.; Horsburgh, G.; Shaw, J.G.; Tomas, J.M.; Merino, S. Analysis of the lateral flagellar gene system of Aeromonas hydrophila AH-3. J. Bacteriol. 2006, 188, 852–862. [Google Scholar] [CrossRef]
- Menanteau-Ledouble, S.; Kattlun, J.; Nobauer, K.; El-Matbouli, M. Protein expression and transcription profiles of three strains of Aeromonas salmonicida ssp. salmonicida under normal and iron-limited culture conditions. Proteome Sci. 2014, 12, 29. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Friedrich, A.W.; Kock, R.; Bielaszewska, M.; Zhang, W.; Karch, H.; Mathys, W. Distribution of the urease gene cluster among and urease activities of enterohemorrhagic Escherichia coli O157 isolates from humans. J. Clin. Microbiol. 2005, 43, 546–550. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Weiss, G.; Carver, P.L. Role of divalent metals in infectious disease susceptibility and outcome. Clin. Microbiol. Infect. 2018, 24, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Falush, D. Toward the use of genomics to study microevolutionary change in bacteria. PLoS Genet. 2009, 5, e1000627. [Google Scholar] [CrossRef] [PubMed]
- Mehta, N.; Olson, J.W.; Maier, R.J. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J. Bacteriol. 2003, 185, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Abbott, S.L.; Cheung, W.K.; Janda, J.M. The genus Aeromonas: Biochemical characteristics, atypical reactions, and phenotypic identification schemes. J. Clin. Microbiol. 2003, 41, 2348–2357. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.L.; Shaw, J.G. Aeromonas spp. clinical microbiology and disease. J. Infect. 2011, 62, 109–118. [Google Scholar] [CrossRef]
- James, S.J.; Cutler, P.; Melnyk, S.; Jernigan, S.; Janak, L.; Gaylor, D.W.; Neubrander, J.A. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am. J. Clin. Nutr. 2004, 80, 1611–1617. [Google Scholar] [CrossRef]
- Hwang, O. Role of oxidative stress in Parkinson’s disease. Exp. Neurobiol. 2013, 22, 11–17. [Google Scholar] [CrossRef]
- Handa, O.; Naito, Y.; Yoshikawa, T. Redox biology and gastric carcinogenesis: The role of Helicobacter pylori. Redox. Rep. 2011, 16, 1–7. [Google Scholar] [CrossRef]
- Segal, A.W. How neutrophils kill microbes. Annu. Rev. Immunol. 2005, 23, 197–223. [Google Scholar] [CrossRef]
- Shiloh, M.U.; Nathan, C.F. Reactive nitrogen intermediates and the pathogenesis of Salmonella and mycobacteria. Curr. Opin. Microbiol. 2000, 3, 35–42. [Google Scholar] [CrossRef]
- Deffert, C.; Schappi, M.G.; Pache, J.C.; Cachat, J.; Vesin, D.; Bisig, R.; Ma Mulone, X.; Kelkka, T.; Holmdahl, R.; Garcia, I.; et al. Bacillus calmette-guerin infection in NADPH oxidase deficiency: Defective mycobacterial sequestration and granuloma formation. PLoS Pathog. 2014, 10, e1004325. [Google Scholar] [CrossRef] [PubMed]
- Graumann, J.; Lilie, H.; Tang, X.; Tucker, K.A.; Hoffmann, J.H.; Vijayalakshmi, J.; Saper, M.; Bardwell, J.C.; Jakob, U. Activation of the redox-regulated molecular chaperone Hsp33--a two-step mechanism. Structure 2001, 9, 377–387. [Google Scholar] [CrossRef]
- Wholey, W.Y.; Jakob, U. Hsp33 confers bleach resistance by protecting elongation factor Tu against oxidative degradation in Vibrio cholerae. Mol. Microbiol. 2012, 83, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Heo, D.H.; Choi, S.W.; Kim, K.N.; Shim, J.; Kim, C.W.; Sung, H.C.; Yun, C.W. Functional characterization of Hsp33 protein from Bacillus psychrosaccharolyticus; additional function of HSP33 on resistance to solvent stress. Biochem. Biophys. Res. Commun. 2007, 358, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Kumsta, C.; Jakob, U. Redox-regulated chaperones. Biochemistry 2009, 48, 4666–4676. [Google Scholar] [CrossRef] [PubMed]
- Seshadri, R.; Joseph, S.W.; Chopra, A.K.; Sha, J.; Shaw, J.; Graf, J.; Haft, D.; Wu, M.; Ren, Q.; Rosovitz, M.J.; et al. Genome sequence of Aeromonas hydrophila ATCC 7966T: Jack of all trades. J. Bacteriol. 2006, 188, 8272–8282. [Google Scholar] [CrossRef]
| Primers | Sequence 5′−3′ | |
|---|---|---|
| hypA | Forward | ATGCACGAAATGTCTCTGGC |
| Reverse | TCGTAATTTGTACCCGCCAC | |
| 16S rRNA | Forward | TGTGTCCTTGAGACGTGGC |
| Reverse | ACAAAGGACAGGGGTTGCG | |
| Species | Strain | Source | Presence of hypA Gene | Protein Accession Number (NCBI) |
|---|---|---|---|---|
| A. aquatica | CECT 8025T | Cyanobacterial bloom | Yes | WP_033130233.1 ¥ |
| A. aquatilisπ | CECT 8026T | Lake water | Yes | - |
| A. allosaccharophila | CECT 4199T | Eel | Yes | WP_042658210.1 |
| A. bestiarum | CECT 4227T | Sick fish | Yes | WP_043555398.1 |
| A. caviae | CECT 838T | Guinea pig | Yes | WP_017786826.1 |
| A. dhakensis | CECT 5744T | Children feces | Yes | WP_042008198.1 |
| A. eucrenophila | CECT 4224T | Fresh water fish | Yes | WP_042642402.1 |
| A. encheleia | CECT 4342T | Eel | Yes | WP_033130233.1 ¥ |
| A. entericaπ | CECT 8981T | Human feces | Yes | - |
| A. finlandensis | CECT 8028T | Cyanobacterial bloom | Yes | WP_033136932.1 |
| A. intestinalisπ | CECT 8980T | Human feces | Yes | - |
| A. jandaei | CECT 4228T | Human feces | Yes | WP_042029441.1 |
| A. hydrophila | CECT 839T | Milk | Yes | WP_005333346.1 |
| A. lacus | CECT 8024T | Cyanobacterial bloom | Yes | - |
| A. lusitana | CECT 7828T | Untreated water | Yes | WP_100861683.1 |
| A. media | CECT 4232T | Fisheries water | Yes | WP_042061779.1 |
| A. popoffi | CECT 5176T | Drinking water | Yes | WP_042034298.1 |
| A. piscicola | CECT 7443T | Sick fish | Yes | WP_021140355.1 |
| A. rivipollensis * | KN-Mc-11N1 | Wild nutria | Yes | WP_017778896.1 |
| A. salmonicida | CECT 894T | Salmon | Yes | WP_005315136.1 |
| A. sanarelli | CECT 7402T | Wound infection | Yes | WP_005301911.1 # |
| A. sobria | CECT 4245T | Fish | Yes | WP_005301911.1 # |
| A. tecta | CECT 7082T | Children feces | Yes | WP_050720085.1 |
| A. trota | CECT 4255T | Human feces | Yes | WP_026458218.1 |
| A. veronii | CECT 4257T | Sputum | Yes | WP_005351492.1 |
| A. australiensis | CECT 8023T | Irrigation water | No | - |
| A. bivalvium | CECT 7113T | Shellfish | No | - |
| A. cavernicola | CECT 7862T | Cavern creek water | No | - |
| A. crassostreaeπ | CECT 8982T | Shellfish | No | - |
| A. diversa | CECT 4254T | Wound infection | No | - |
| A. fluvialis | CECT 7401T | River water | No | - |
| A. molluscorum | CECT 5864T | Shellfish | No | - |
| A. rivuli | CECT 7518T | River water | No | - |
| A. schubertii | CECT 4240T | Skin abscess | No | - |
| A. simiae | IBS S-6874T | Monkey feces | No | - |
| A. taiwanensis | CECT 7403T | Wound infection | No | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Bravo, A.; López-Fernández, L.; Figueras, M.J. The Metallochaperone Encoding Gene hypA Is Widely Distributed among Pathogenic Aeromonas spp. and Its Expression Is Increased under Acidic pH and within Macrophages. Microorganisms 2019, 7, 415. https://doi.org/10.3390/microorganisms7100415
Fernández-Bravo A, López-Fernández L, Figueras MJ. The Metallochaperone Encoding Gene hypA Is Widely Distributed among Pathogenic Aeromonas spp. and Its Expression Is Increased under Acidic pH and within Macrophages. Microorganisms. 2019; 7(10):415. https://doi.org/10.3390/microorganisms7100415
Chicago/Turabian StyleFernández-Bravo, Ana, Loida López-Fernández, and Maria José Figueras. 2019. "The Metallochaperone Encoding Gene hypA Is Widely Distributed among Pathogenic Aeromonas spp. and Its Expression Is Increased under Acidic pH and within Macrophages" Microorganisms 7, no. 10: 415. https://doi.org/10.3390/microorganisms7100415
APA StyleFernández-Bravo, A., López-Fernández, L., & Figueras, M. J. (2019). The Metallochaperone Encoding Gene hypA Is Widely Distributed among Pathogenic Aeromonas spp. and Its Expression Is Increased under Acidic pH and within Macrophages. Microorganisms, 7(10), 415. https://doi.org/10.3390/microorganisms7100415