1. Introduction
At present, widely recognized mechanisms of plant disease resistance are divided into two categories [
1,
2,
3,
4]. The first layer employs pattern recognition receptors (PRRs) to detect conserved pathogen-associated molecular patterns (PAMP) and to trigger PAMP-triggered immunity (PTI) [
5,
6,
7,
8,
9]. The second layer, effector-triggered immunity (ETI), involves a rapid and robust defense activation triggered by the direct or indirect recognition between an isolate-specific pathogen avirulence (Avr) effector and its cognate host resistance (R) protein, often accompanied by a hypersensitive reaction (HR) at the attempted pathogen infection sites, which activates a set of innate immunity signaling pathways [
10,
11,
12,
13,
14].
Recently, small RNAs (sRNAs) from pathogens have been found to secrete to host plants, causing cross-species of RNA interference reaction and leading to a loss of function of the host plant resistance [
15]. The biological function of these small RNA molecules from pathogens is similar to non-toxic effector proteins. For example, some
Botrytis cinerea sRNAs have been proved to hijack the host RNA interference (RNAi) machinery by loading into
Arabidopsis Argonaute 1 to selectively silence host immunity genes, demonstrating that a fungal pathogen transfers “virulent” sRNA effector into host cells to achieve infection, which reveals a naturally occurring cross-kingdom RNAi [
15]. Additionally,
Arabidopsis cells also have been proved to secrete exosome-like extracellular vesicles to deliver sRNAs into fungal pathogen
Botrytis cinerea. Transferred host sRNAs induce silencing of fungal genes critical for pathogenicity. Thus,
Arabidopsis has adapted exosome-mediated cross-kingdom RNA interference as part of its immune responses during the evolutionary arms race with the pathogen [
16]. In addition, wheat microRNA (miRNA) related research shows that, Tae-miR1023 can suppress the invasion of
Fusarium graminearum (
F. graminearum) by targeting and silencing
FGSG_03101 which codes an alpha/beta hydrolase gene in
F. graminearum [
17]. However, there are no reports on whether endogenous sRNAs from
F. graminearum can be transported into common wheat and play a biological role. In terms of the two main types of sRNAs (siRNAs and miRNAs), although they differ in their biosynthetic mechanisms [
18], they are extremely similar in terms of product size, sequence characteristics, and specific silencing patterns, which implies that there are inevitable similarities between the biological functions and mechanisms of siRNAs and miRNAs.
The traditional methods of plant disease research techniques commonly used are host-induced gene silencing (HIGS), which is a method of reverse genetics technique widely used. It can be artificially induced by pathogens associated with double stranded RNA fragments, so that plants get new disease-resistant function via HIGS [
19,
20]. For example, using a gene gun bombardment, transient expression barley powdery mildew toxic effector gene AVRa10 of RNA fragments in barley leaves, can effectively inhibit the barley powdery mildew infection of barley [
20].
Agrobacterium tumefaciens expression of mitogen activated protein kinase RNA fragments can effectively enhance the wheat leaf rust resistance [
21]. By stable transgenic methods, the
F. graminearum cytochrome P450 lanosterol C-14α-demethylase (CYP51) genes fragment was stably transformed into
Arabidopsis and barley plants, and found that stable transgenic plants obtained for resistance to
F. graminearum by means of HIGS [
22]. These HIGS technology applications are based on artificially induced plant pathogens which produce exogenous siRNAs, however, a direct over-expression or silencing of
F. graminearum small RNA molecules in common wheat has not been found [
23].
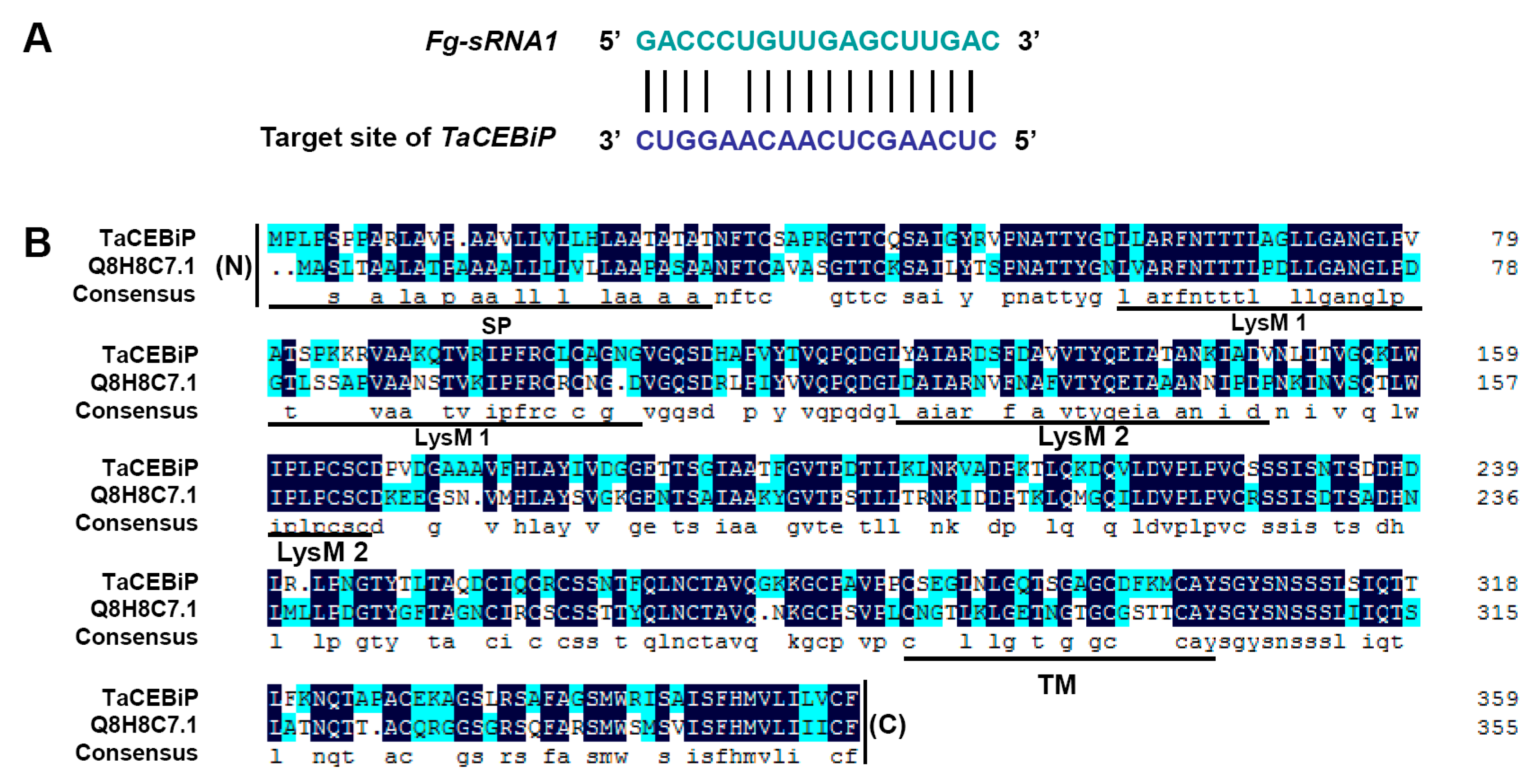
In order to detect whether F. graminearum endogenous sRNAs can be transferred into wheat to exert a biological function, we decided to screen F. graminearum sRNAs, which could target the wheat genome, and investigated the effect of silencing of target candidate genes. Fortunately, we found one F. graminearum endogenous sRNA could target the wheat CEBiP gene, and negatively regulate wheat resistance.
2. Materials and Methods
2.1. Plant Materials
Nicotiana benthamiana (N. benthamiana) plants are grown in a controlled environment at 25 °C with a 14-h-light/10-h-darkness photoperiod. Wheat plants (Chinese spring) used for the BSMV-based sRNA silencing experiment and F. graminearum (strain PH-1) punch inoculation experiment are grown in pots in a greenhouse with 16-h-light/8-h-darkness cycle until the two-leaf stage. After inoculated with BSMV, wheat plants are transferred to a climate chamber at 23–25 °C for the evaluation. For each biological replicate, six wheat seeds are sown in one pot of 12 cm diameter, and two pots per BSMV construct. Totally, 10–12 wheat plants of two-leaf stage are prepared for BSMV inoculation. Twenty segments of 4th wheat leaves displaying the BSMV infected symptom, are collected from three biological replicates for the F. graminearum punch inoculation experiment.
2.2. Small RNA Isolation and Deep Sequencing
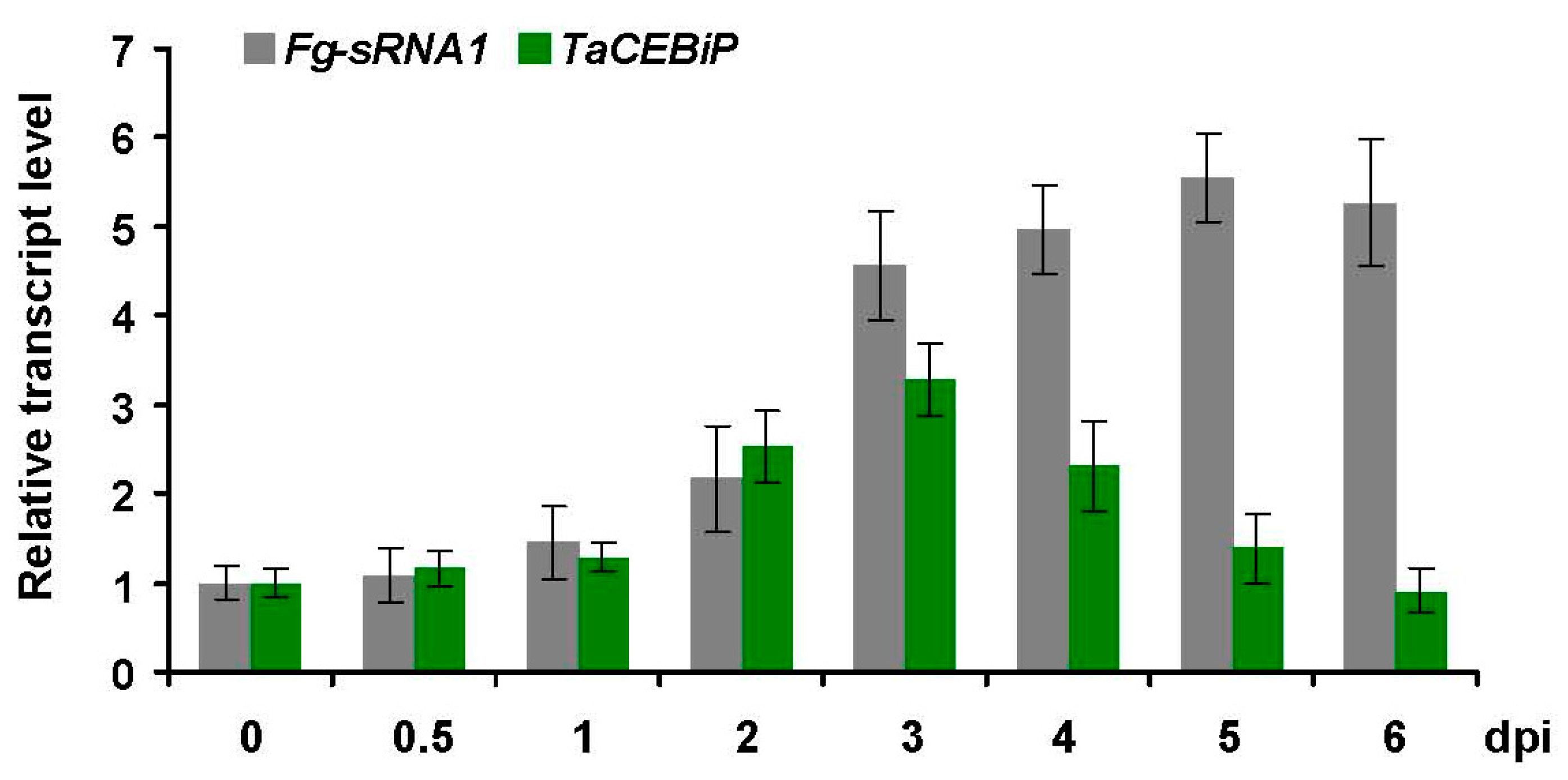
Fifteen-day-old leaves of wheat were inoculated with F. graminearum strain PH-1 for 0, 24 and 72 h, and total RNAs were isolated using TRIzol solution according to the manufacturer’s instructions. Small RNAs of 18–30 nt were excised and isolated from 5 to 10 mg total RNAs electrophoresed on 15% polyacrylamide denaturing gel, and then were ligated with 59 nt and 39 nt adapters (BGI, Beijing). The ligated small RNAs were used as templates for cDNA synthesis followed by PCR amplification, and synthetic cDNA were prepared for sequencing. The obtained libraries were sequenced using the Solexa sequencing platform (BGI, Beijing).
2.3. Fungal Strains, Culture Conditions and Punch Inoculation Experiment
F. graminearum strain PH-1 is used as the wild-type (WT) strain in this study. The WT strains are routinely cultured on potato dextrose agar (PDA) (200 g potato, 20 g dextrose, 20 g agar and 1 L water) at 25 °C with a 12-h-light/12-h-darkness cycle. The WT strains are grown on carrot agar for induction of sexual development near-UV light (wavelength, 365 nm; HKiv Co., Ltd., Xiamen, China), and in mung bean broth (MBB) for conidiation assays under continuous light. Assays for
F. graminearum punch inoculation are performed as described previously [
24]. For all leaf inoculation assays, the
F. graminearum conidia concentration is adjusted to 5 × 10
4 conidia per mL
−1. Inoculations of 4th wheat leaves displaying Barley stripe mosaic virus (BSMV) symptom, or non-BSMV infected wheat leaves, are done by wound inoculation of detached leaves segments with
F. graminearum strains. Fifteen leaves are detached for each biological replicate and transferred in 1% agar plates supplemented with 85 μM Benzimidazole. For assessing the progression of
F. graminearum disease symptoms, the lesion size is measured from the digital images using the free software ImageJ program (
http://rsb.info.nih.gov/ij/index.html). Each experiment is repeated three times.
2.4. Vector Constructions
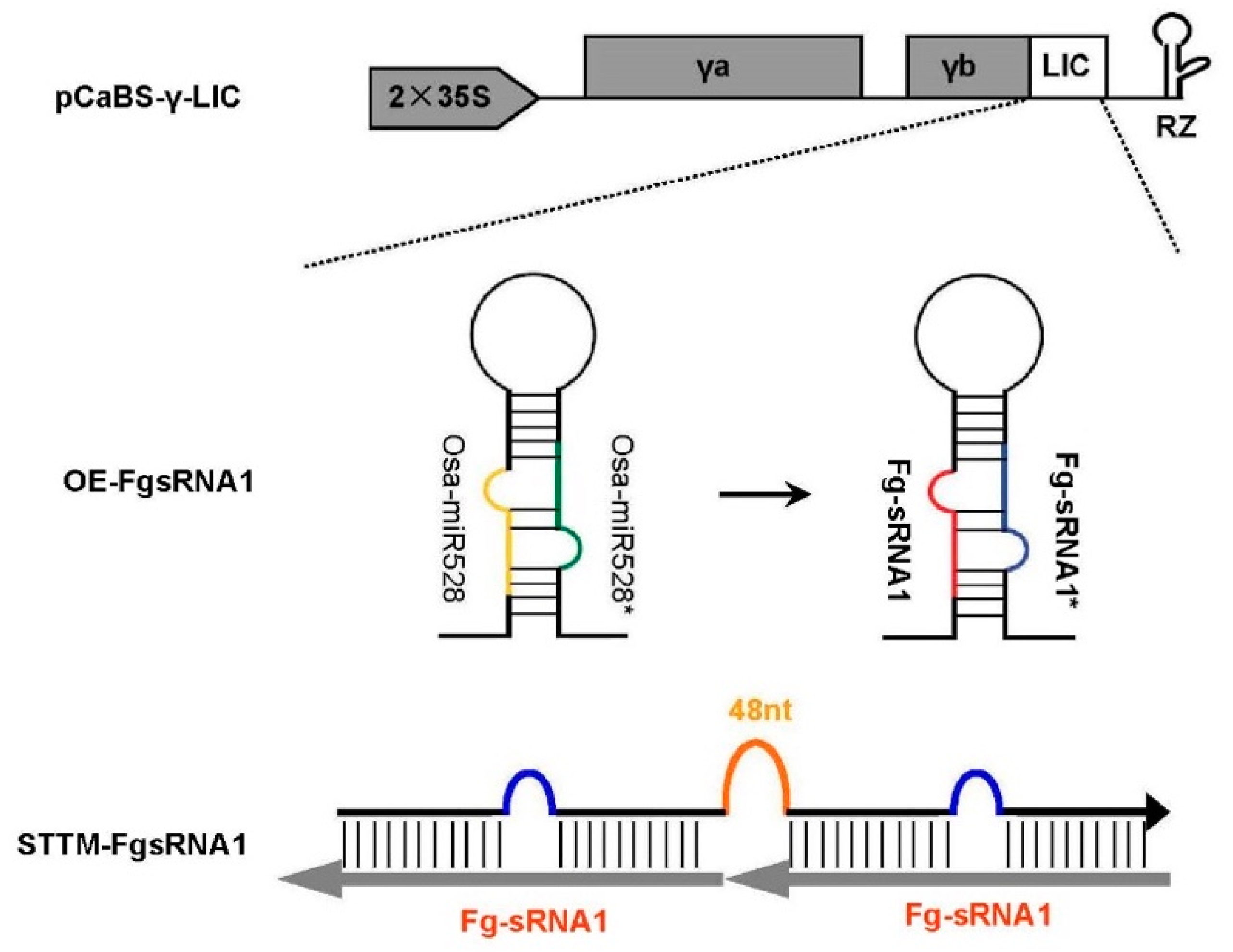
For construct BSMV: OE-FgsRNA1,
Fg-sRNA1 is engineered into
Osa-miR528 precursor backbone using overlap PCR to replace endogenous miRNA sequence. Both miRNA and the partially complementary miRNA
* sequences in
Osa-miR528 precursor, are substituted by
Fg-sRNA1 and
Fg-sRNA1*, respectively. The reconstructed precursor is added ligation-independent-cloning (LIC) adaptors for linking with BSMV vector. For construct BSMV: STTM-FgsRNA1 (for silencing of
Fg-sRNA1 using short tandem target mimic (STTM) strategy) is constructed as follows. Primers with LIC adaptor, corresponding target mimic of
Fg-sRNA1, and STTM 48 nt spacer (5′-GTTGTTGTTGTTATGGTCTAATTTAAATATGGTCTAAAGAAGAAGAAT-3′) are employed to PCR amplify STTM-FgsRNA1 molecules. STTM-FgsRNA1 is added LIC adaptors for linking with BSMV vector. The
Osa-miR528 precursor harboring
Fg-sRNA1 fragment or STTM fragment with LIC adaptors are cloned into BSMV-γb using the LIC protocol as described [
25]. The plasmids in Western blotting experiment and YFP observation assay are constructed by Gateway technology (Invitrogen Thermo Fisher Scientific-CN) following the instructions of the manufacturer (
http://www.invitrogen.com/content/sfs/manuals/gatewayman.pdf). The two 35S: pKANNIBAL-FgsRNA1 or -FgsRNA2 expressing vectors are constructed by PCR amplification of
Osa-miR528 precursor harboring
Fg-sRNA1, or
Fg-sRNA2, followed by sequential digestion with
HindIII and
KpnI and subsequent cloning into the pKANNIBAL destination vector. All constructs are confirmed by DNA sequencing.
2.5. BSMV-Based Experiments
BSMV-based sRNA over-expression and silencing experiments are performed as described [
17,
25]. Constructs of pCaBS-α, pCaBS-β, and pCaBS-γ-LIC derivatives (OE-FgsRNA1 and STTM-FgsRNA1) are transformed into
Agrobacterium (
A. tumefaciens strain EHA105). The
Agrobacterium suspensions of OD
600 = 0.8 are mixed at 1:1:1 ratio (pCaBS-α: pCaBS-β: each pCaBS-γ-LIC derivative) and infiltrated in
N. benthamiana leaves. Agroinfiltrated
N. benthamiana leaves can provide excellent sources of virus for secondary BSMV infections in wheat plants. The
N. benthamiana sap is extracted from leaves with BSMV symptom at about 12 days post infiltration, ground in 20 mM Na-phosphate buffer (pH 7.2) containing 1% celite, and the sap is mechanically inoculated onto the first two emerging leaves of wheat. Infected wheat plants are further grown for 14–21 d to allow emergence of new leaves displaying viral symptoms. Segments of the 4th leaves of BSMV-infected wheat plants are collected for further experiments from three biological replicates per construct.
2.6. Protein Analyses
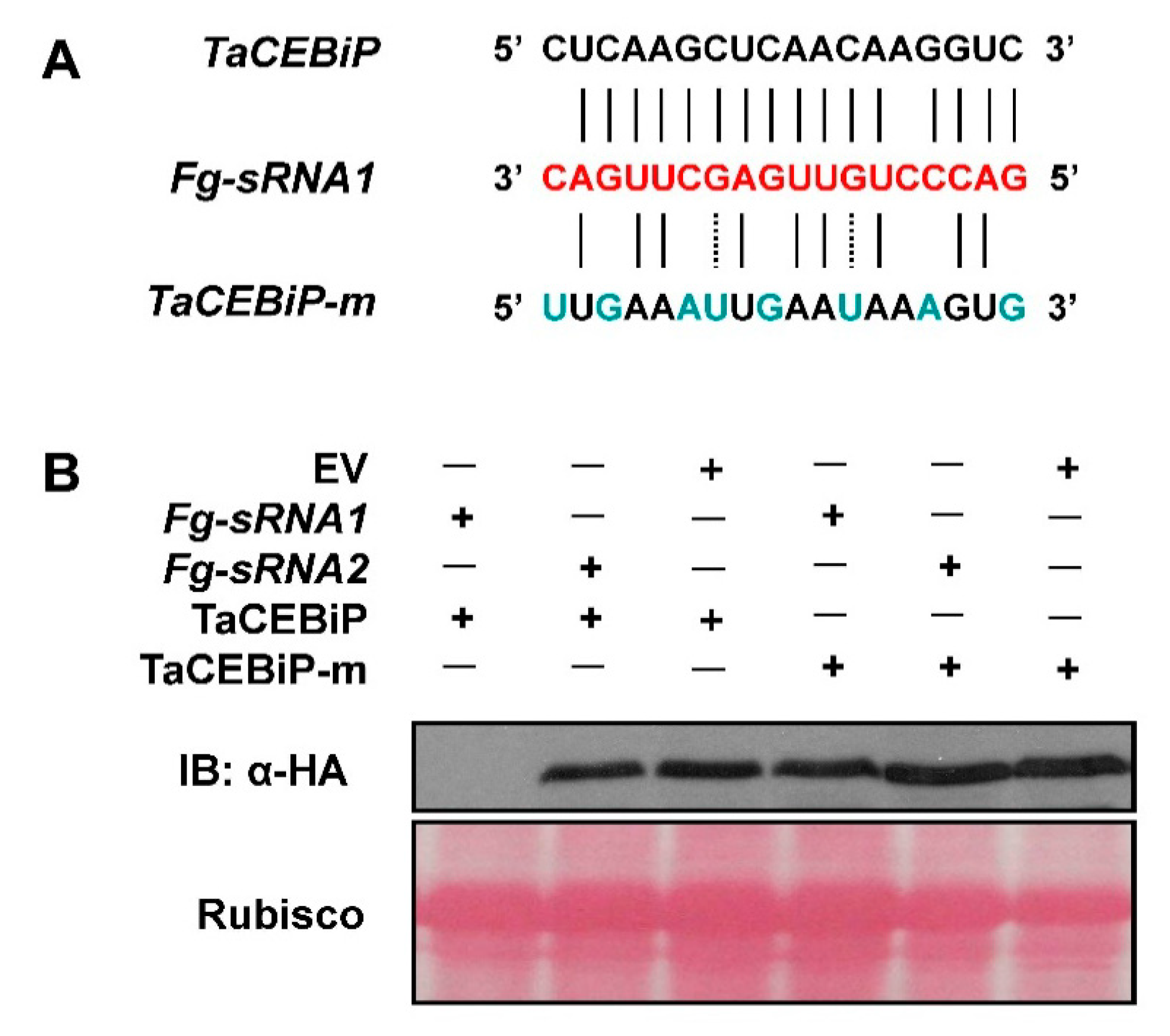
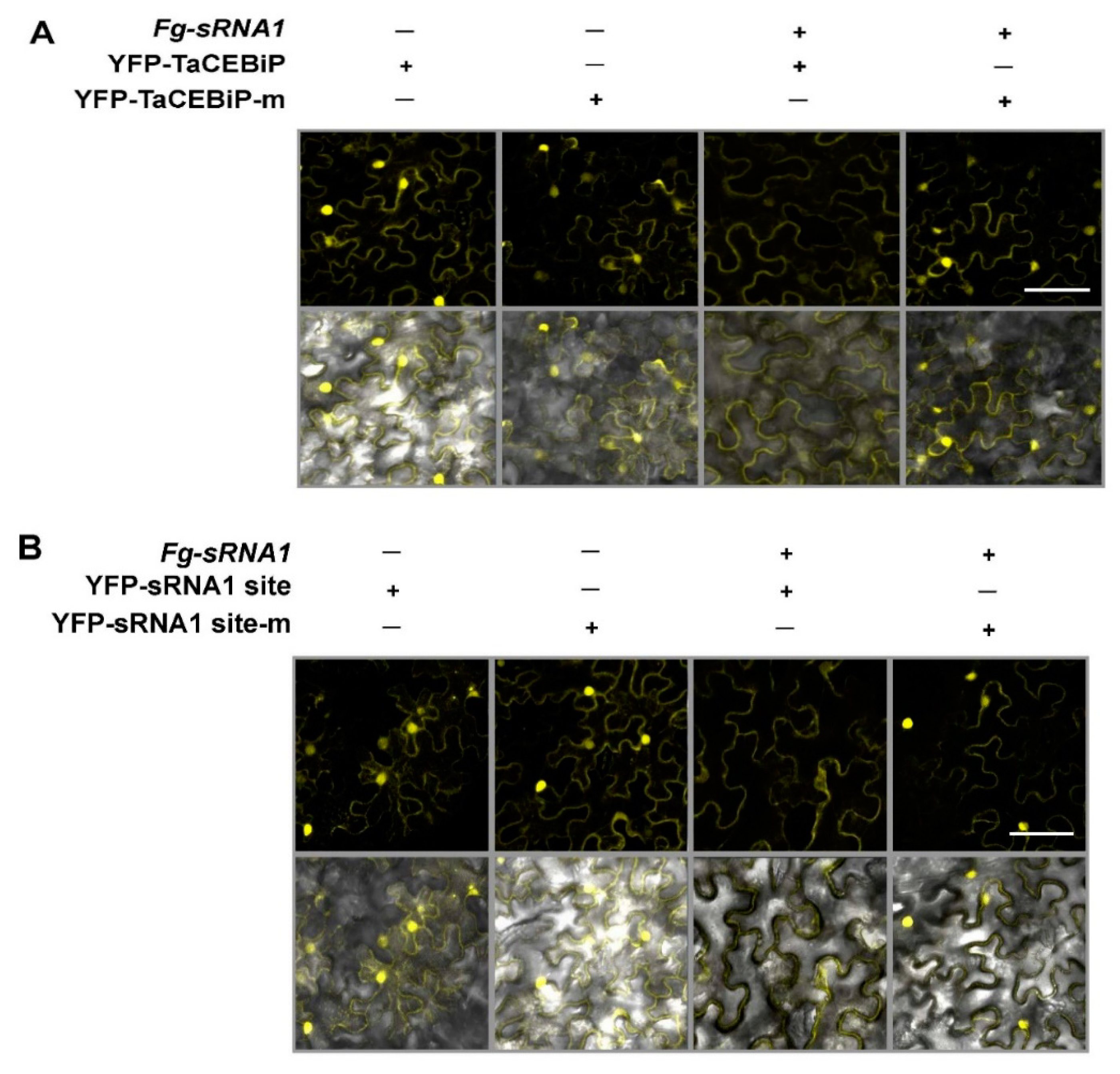
Western blotting experiment is performed in order to analyze whether Fg-sRNA1 or Fg-sRNA2 can cleavage TaCEBiP, respectively. First, 35S: pKANNIBAL-FgsRNA1 or -FgsRNA2 expressing vectors are transiently transformed into N. benthamiana leaves by Agrobacterium-mediated transformation. After 24 hpi, 35S: cTAPi-TaCEBiP or -TaCEBiP mutant (TaCEBiP-m) proteins expressing vector are agroinfiltrated into the same N. benthamiana leaves, respectively. For TaCEBiP & TaCEBiP-m proteins accumulation analysis, leaf samples are collected at 24–36 h post the 2nd agroinoculation. HA-tagged protein extraction, separation and fraction are detected by immunoblotting using rat anti-HA antibody (Roche, Indianapolis, IN, USA) and anti-rat IgG conjugated with horseradish peroxidase (HRP) (Sigma, St Louis, MO, USA). For YFP observation assay, cell suspensions of A. tumefaciens strain GV3101 containing the indicated constructs were infiltrated into N. benthamiana leaves. Confocal images were taken using a confocal laser-scanning microscope Zeiss LSM 710 (Carl Zeiss, Oberkochen, Germany).
2.7. RNA, DNA and PCR Analysis
Plant total RNAs are extracted from three independent biological replicates, BSMV-infected leaves and
F. graminearum-infected lesion area of wheat leaves with TRIzol reagent, as described by the manufacturer (Invitrogen Thermo Fisher Scientific-CN, Shanghai, China), and treated with DnaseI. DNA and Total RNA are extracted from cultured
F. graminearum strains using fungal DNA or Total RNA extraction kits. About 2 mg of total RNA and M-MLV Reverse Transcriptase (Promega) are further used for reverse transcription. For coding gene (
TaCEBiP) reverse transcription, first-strand cDNA is synthesized using Oligo (dT)
18. For sRNA reverse transcription, specifically designed end-point stem-loop reverse transcription primers are used, and follow the procedures described by Liu [
26]. Real-time RT-PCR assays with three technical replicates are performed using StepOne real-time system (Applied Biosystems, Foster City, CA, USA) and GoTaq qPCR Master Mix (Promega, A6001, Madison, WI, USA). sRNA forward primers are respectively used with universal reverse primer to quantify the relative transcript levels of mature
Fg-sRNA1 or
Fg-sRNA2. Real-time RT-PCR components for sRNA are as follows: 2× GoTaq qPCR Master Mix 5 μL, diluted cDNA 1 μL, sRNA forward primer 0.2 μL, sRNA universal reverse primer 0.2 μL, and ddH
2O up to 10 μL. Real-time RT-PCR conditions are as follows: 95 °C for 5 min, followed by 35–40 cycles of 95 °C for 5 s, 60 °C for 10 s, and 72 °C for 1 s. For melting curve analysis, denature samples at 95 °C, then cool to 65 °C at 20 °C per second [
17,
26]. For the determination of target gene
TaCEBiP, gene-specific primer pairs spanning the miRNA-guided cleavage site are used.
Tae-U6,
Tae-Actin and
Fg-Actin which served as internal reference genes for sRNAs and protein-coding genes are detected, respectively. GenBank accession numbers of
Tae-U6,
Tae-Actin,
Fg-Actin are X63066, KC775781, XM_011328784.1, respectively. Error bars representing standard error (SE) are calculated from three biological replicates per construct.
4. Discussion
Our research showed that a small RNA
Fg-sRNA1 of
F. graminearum can cross-species transport and cause silence of wheat resistance-related gene
TaCEBiP. BSMV-mediated
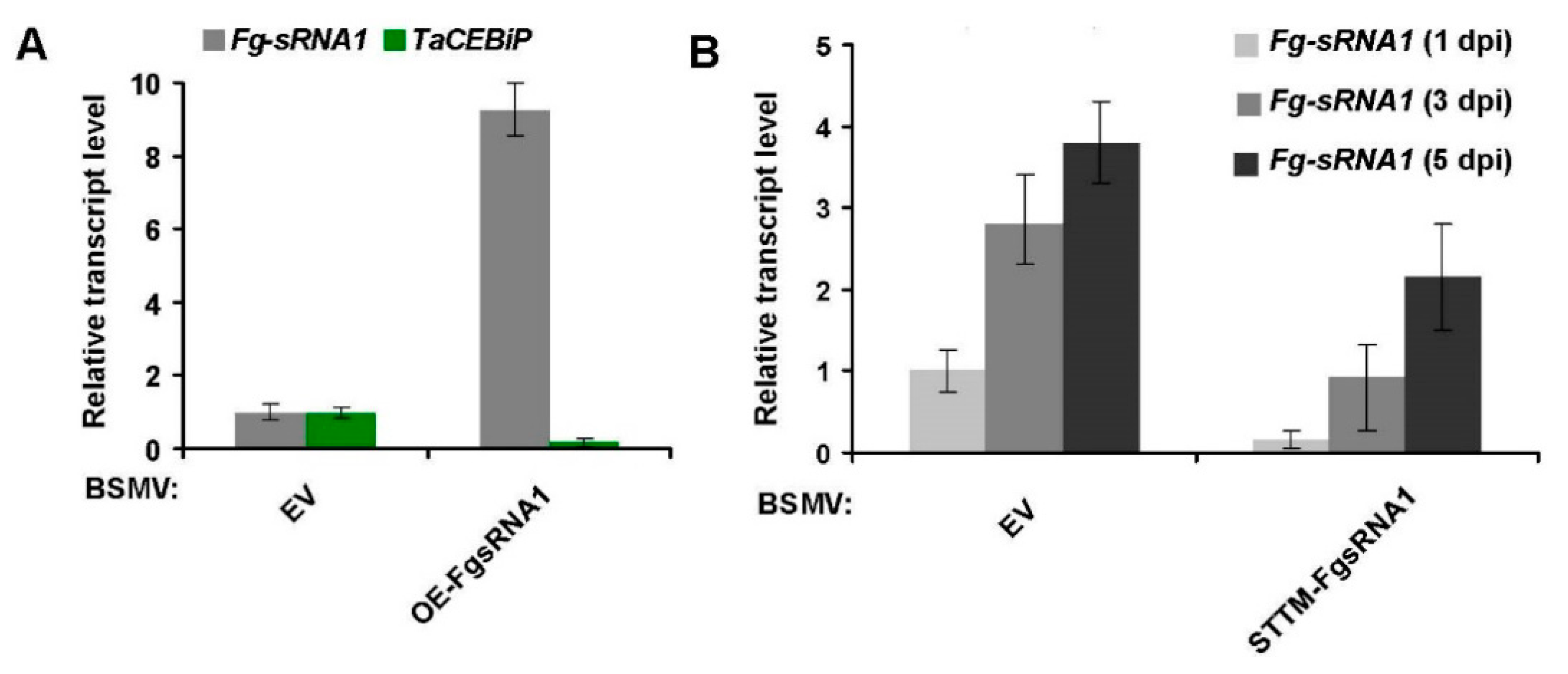
Fg-sRNA1 over-expression and silencing systems have been employed in this study. For example, enhanced expression of
Fg-sRNA1 results in a larger area of wheat leaf necrosis phenotype, and pre-expression of STTM molecules that silence
Fg-sRNA1 can effectively reduce the transcriptional expression of
Fg-sRNA1 after fungal infection, and effectively prevent the growth of necrotic spots in wheat leaves. Relative transcriptional level detection experiments indicate that
Fg-sRNA1 can target
TaCEBiP mRNA and trigger silencing of
TaCEBiP in vivo, and Western blotting experiments and YFP fluorescence observation proofs show that
Fg-sRNA1 can suppress the accumulation of TaCEBiP. Overall, our results suggest that
Fg-sRNA1 of
F. graminearum can target and silence wheat resistance-related gene
TaCEBiP to enhance invasion of
F. graminearum accompanying with the weakening of wheat resistance. Combined with previous reports that wheat miRNA1023 can be transported to
F. graminearum and fungal infection by silencing
F. graminearum endogenous gene [
12], our research indicates that some sRNAs are present in the trans-species transport and regulation mechanisms between common wheat and
F. graminearum.
The main mechanism of plant disease resistance PTI and ETI can be grouped into four stages of immune response [
1,
2]. The first stage, the plant transmembrane pattern recognition receptors (Pattern recognition receptors, PRRs) recognize pathogen associated molecular model (Pathogen-associated molecular patterns, PAMPs), such as bacterial flagellin, and lead to pathogen associated molecular models of induced immune response to inhibit further colonization and spread of pathogens [
2,
4]. In stage two, successful pathogens deploy effectors that contribute to pathogen virulence. Effectors can interfere with PTI. This results in effector-triggered susceptibility (ETS) [
3,
5]. In the third stage, the plant will not sit still, evolved more antiviral protein, directly or indirectly identify the pathogen effector proteins, and trigger effector proteins induced immune response (Effector-triggered immunity, ETI). ETI is an accelerated and amplified PTI response, resulting in disease resistance and, usually, a hypersensitive cell death response (HR) at the infection site [
7,
10,
11]. The fourth stage, co-evolution of plants and pathogens, namely natural selection process, are other types of pathogenic bacteria by secreting effector protein or modifying the original effector proteins to break through the plant’s defense system, and the plant also evolved new antiviral proteins responding to new pathogen effector proteins, forward with this process of repeated co-evolution [
12].
Animal and plant pathogens have evolved virulence or effector proteins to counteract host immune responses. Various protein effectors have been predicted or discovered in fungal or oomycete pathogens from whole-genome sequencing and secretome analysis, although delivery mechanisms are still under active investigation [
12,
13,
14]. Jin’s lab shows that sRNAs as well can act as effectors through a mechanism that silences host genes in order to debilitate plant immunity and achieve infection. They find that sRNAs from
B. cinerea hijack the plant RNAi machinery by binding to AGO proteins, which in turn direct host gene silencing. The implications of these findings may extend beyond plant gray mold disease caused by
B. cinerea, and suggest an extra mechanism underlying pathogenesis promoted by sophisticated pathogens with the capability to generate and deliver small regulatory RNAs into hosts to suppress host immunity [
15].
From plant to pathogen small molecule RNA interference effect exercised across species has been reported [
21]. For example, using a gene gun bombardment, transient expression barley powdery mildew toxic effector gene AVRa10 of RNA fragments in barley leaves, can effectively inhibit the barley powdery mildew infection of barley [
20].
Agrobacterium tumefaciens expression of mitogen activated protein kinase RNA fragments, can effectively enhance the wheat leaf rust resistance [
21]. By stable transgenic methods, the
F. graminearum cytochrome P450 lanosterol C-14α-demethylase (
CYP51) genes fragment was stably transformed into
Arabidopsis and barley plants, and found that stable transgenic plants obtained for resistance to
F. graminearum by means of HIGS [
22]. These HIGS technology applications were based on artificially induced plant pathogens which produce exogenous siRNAs. Compared with these studies, a direct cross-kingdom expression of fungal endogenous small RNA molecule and acquired enhanced infectivity were reported in this study.
BSMV-based sRNA silencing and over-expression technology is the most widely used reverse-genetic strategy to study sRNA function [
31]. Transient virus-induced gene silencing displays several advantages when constitutive loss of gene function through stable transformation brings about sporophytic or gametophytic lethality [
31]. Moreover, the currently described BSMV-mediated sRNA silencing and over-expression system is efficient and quick, and it can be carried out for sRNAs silencing through intermediary of argoinocubated
N. benthamiana, a more simple but effective method without complicated experiment operations or expensive instruments [
25]. Here, we presented that BSMV-based sRNA silencing and over-expression system could be used to evaluate the function of
F. graminearum sRNA by simple agroinfiltration. The modified BSMV vector may facilitate to high-throughput screen the targets of fungal sRNAs, and to characterize their function in wheat crops [
25].
This paper verifies the F. graminearum small RNA molecule Fg-sRNA1 capable of transporting into common wheat to exercise RNA interference biological function. A variety of evidences show that Fg-sRNA1 can target and silence TaCEBiP gene, thereby negatively regulating wheat resistance and enhancing F. graminearum infection. In summary, this study demonstrates another evidence of co-evolution between F. graminearum and main food crops.