Insights into Biodegradation Related Metabolism in an Abnormally Low Dissolved Inorganic Carbon (DIC) Petroleum-Contaminated Aquifer by Metagenomics Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Sample Collection and Chemical Analysis
2.3. DNA Isolation, Sequencing, and Library Construction
2.4. Bioinformatics Analysis
3. Results and Discussion
3.1. Hydrochemical Characteristics of Groundwater
3.2. Sequencing Statistics
3.3. Metabolism Analysis
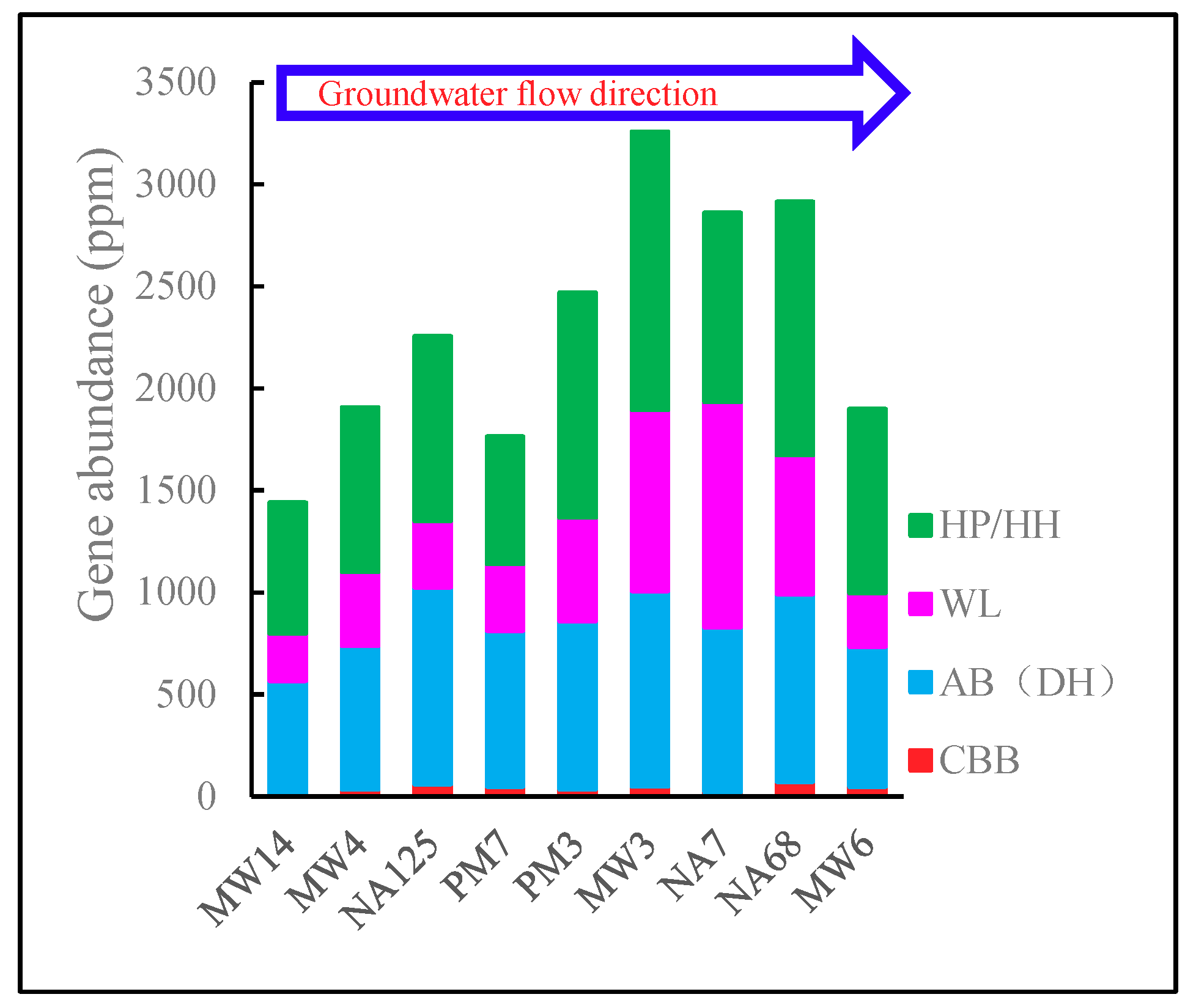
3.3.1. Carbon Fixation Metabolism
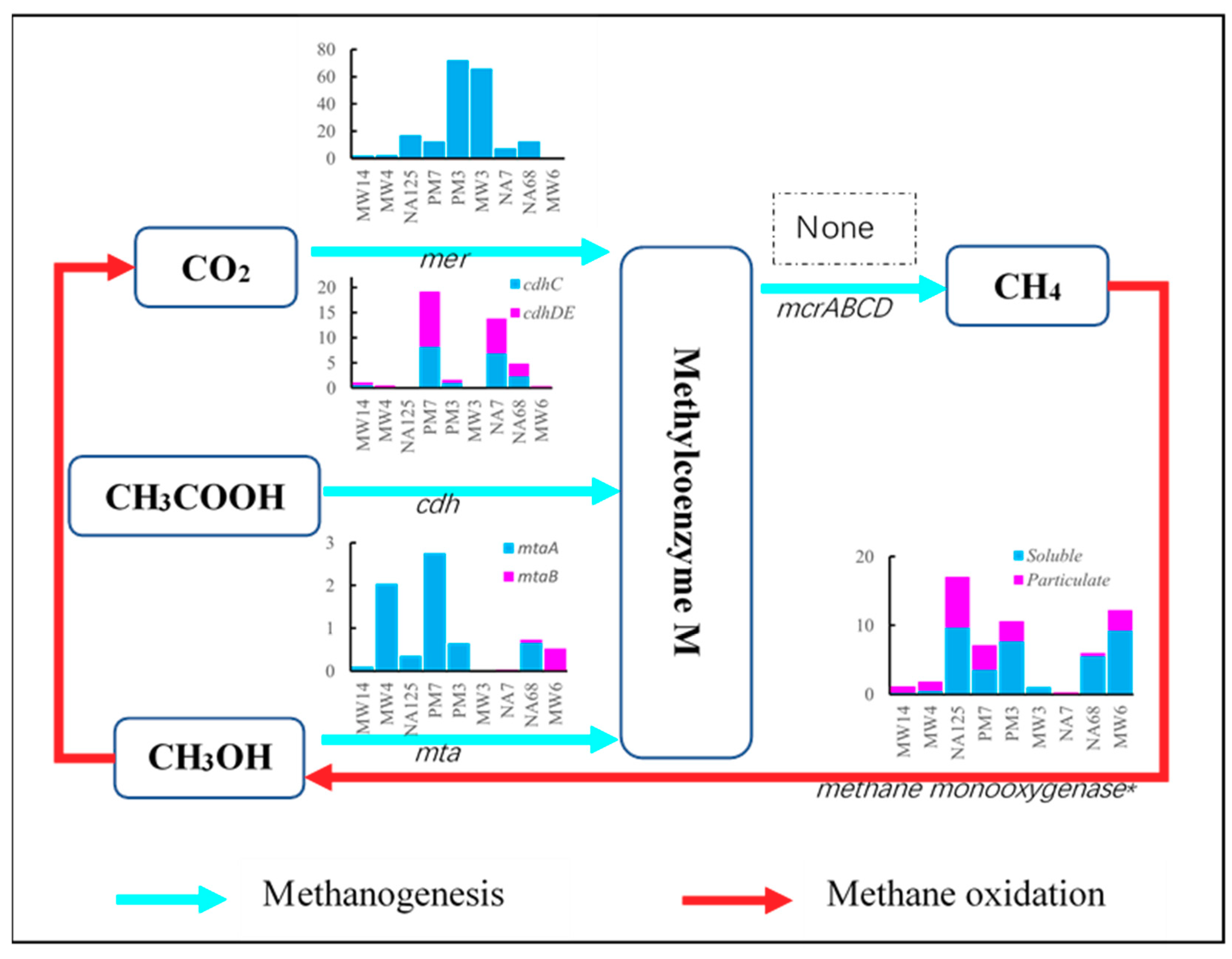
3.3.2. Methane Metabolism
3.3.3. Nitrogen Metabolism
3.3.4. Sulfur Metabolism
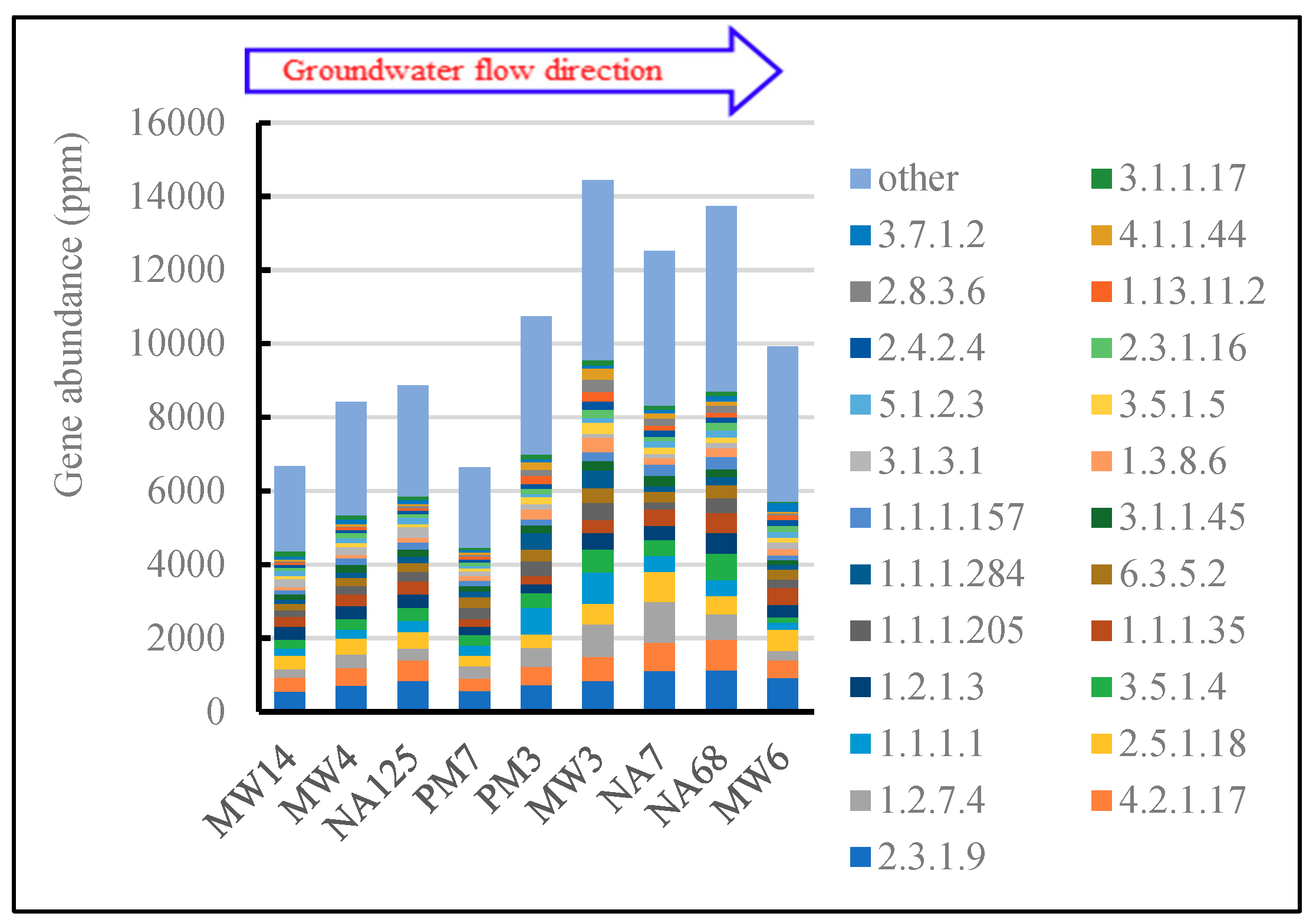
3.3.5. Other Biodegradation Related Metabolism
3.4. Possible Mechanism of Biodegradation Related Metabolism
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AB | Arnon–Buchanan |
| APS | adenylyl sulfate |
| BTEX | benzene, toluene, ethylbenzene, m-xylene, p-xylene, and o-xylene |
| CBB | Calvin–Benson–Bassham |
| COD | chemical oxygen demand |
| DH | dicarboxylate–hydroxybutyrate |
| DIC | dissolved inorganic carbon |
| DNRA | dissimilatory nitrate reduction to ammonia |
| DO | dissolved oxygen |
| EC | electrical conductivity |
| HH | hydroxypropionate-hydroxybutyrate |
| HP | 3-hydroxypropionate |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| NCBI | National Center for Biotechnology Information |
| ORP | oxidation–reduction potential |
| PAPS | 3’-phosphoadenylyl sulfate |
| PHC | petroleum hydrocarbon contaminated |
| ppm | Parts per million |
| VOCs | volatile organic compounds |
| WL | Wood–Ljungdahl |
References
- Verginelli, I.; Pecoraro, R.; Baciocchi, R. Using dynamic flux chambers to estimate the natural attenuation rates in the subsurface at petroleum contaminated sites. Sci. Total Environ. 2018, 619, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Safdari, M.-S.; Kariminia, H.-R.; Rahmati, M.; Fazlollahi, F.; Polasko, A.; Mahendra, S.; Wilding, W.V.; Fletcher, T.H. Development of bioreactors for comparative study of natural attenuation, biostimulation, and bioaugmentation of petroleum-hydrocarbon contaminated soil. J. Hazard. Mater. 2018, 342, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.-H.; Dahlgren, R.A.; Gao, S.; Tanji, K.K. Characterization of redox processes in shallow groundwater of owens dry lake, california. Environ. Sci. Technol. 2004, 38, 5950–5957. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, G.; Qian, Y.; Yang, Y.; Zhang, F. Microbial functional gene patterns related to soil greenhouse gas emissions in oil contaminated areas. Sci. Total Environ. 2018, 628, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Alfreider, A.; Schirmer, M.; Vogt, C. Diversity and expression of different forms of rubisco genes in polluted groundwater under different redox conditions. Fems. Microbiol. Ecol. 2012, 79, 649–660. [Google Scholar] [CrossRef]
- Tischer, K.; Kleinsteuber, S.; Schleinitz, K.M.; Fetzer, I.; Spott, O.; Stange, F.; Lohse, U.; Franz, J.; Neumann, F.; Gerling, S.; et al. Microbial communities along biogeochemical gradients in a hydrocarbon-contaminated aquifer. Environ. Microbiol. 2013, 15, 2603–2615. [Google Scholar] [CrossRef]
- Yergeau, E.; Sanschagrin, S.; Maynard, C.; St-Arnaud, M.; Greer, C.W. Microbial expression profiles in the rhizosphere of willows depend on soil contamination. Isme J. 2013, 8, 344. [Google Scholar] [CrossRef]
- Main, C.E.; Ruhl, H.A.; Jones, D.O.B.; Yool, A.; Thornton, B.; Mayor, D.J. Hydrocarbon contamination affects deep-sea benthic oxygen uptake and microbial community composition. Deep Sea Res. Part I Oceanogr. Res. Pap. 2015, 100, 79–87. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R. Predictive functional profiling of microbial communities using 16s rrna marker gene sequences. Nat. Biotechnol. 2013, 31, 814. [Google Scholar] [CrossRef]
- Stapleton, R.D.; Sayler, G.S.; Boggs, J.M.; Libelo, E.L.; Stauffer, T.; MacIntyre, W.G. Changes in subsurface catabolic gene frequencies during natural attenuation of petroleum hydrocarbons. Environ. Sci. Technol. 2000, 34, 1991–1999. [Google Scholar] [CrossRef]
- Stapleton, R.; Sayler, G. Assessment of the microbiological potential for the natural attenuation of petroleum hydrocarbons in a shallow aquifer system. Microb. Ecol. 1998, 36, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Kellermann, C.; Selesi, D.; Lee, N.; Hügler, M.; Esperschütz, J.; Hartmann, A.; Griebler, C. Microbial CO2 fixation potential in a tar-oil-contaminated porous aquifer. Fems Microbiol. Ecol. 2012, 81, 172–187. [Google Scholar] [CrossRef] [PubMed]
- Beller, H.R.; Kane, S.R.; Legler, T.C.; Alvarez, P.J.J. A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ. Sci. Technol. 2002, 36, 3977–3984. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Bhadury, P. Effect of pollution on aquatic microbial diversity. In Environmental Microbial Biotechnology; Springer: Berlin, Germany, 2015; pp. 53–75. [Google Scholar]
- Pérez-Jiménez, J.R.; Young, L.Y.; Kerkhof, L.J. Molecular characterization of sulfate-reducing bacteria in anaerobic hydrocarbon-degrading consortia and pure cultures using the dissimilatory sulfite reductase (dsrab) genes. Fems Microbiol. Ecol. 2001, 35, 145–150. [Google Scholar] [CrossRef]
- Reid, T.; Chaganti, S.R.; Droppo, I.G.; Weisener, C.G. Novel insights into freshwater hydrocarbon-rich sediments using metatranscriptomics: Opening the black box. Water Res. 2018, 136, 1–11. [Google Scholar] [CrossRef] [PubMed]
- United States Environmental Protection Agency. Method 8260b Volatile Organic Compounds by Gas Chromatography/Mass Spectrometry (gc/ms); United States Environmental Protection Agency: Washington, DC, USA, 1996; p. 86.
- United States Environmental Protection Agency. Non-Halogenated Organics Using gc/fid; United States Environmental Protection Agency: Washington, DC, USA, 2003.
- Standard, A. Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 1998. [Google Scholar]
- Zhao, Y.; Zhang, X.-X.; Zhao, Z.; Duan, C.; Chen, H.; Wang, M.; Ren, H.; Yin, Y.; Ye, L. Metagenomic analysis revealed the prevalence of antibiotic resistance genes in the gut and living environment of freshwater shrimp. J. Hazard. Mater. 2018, 350, 10–18. [Google Scholar] [CrossRef]
- Newell, C.J.; McLeod, R.K.; Gonzales, J.R. Bioscreen: Natural Attenuation Decision Support System. User’s Manual Version 1.3; National Risk Management Research Laboratory, Office of Research and Development, United States Environmental Protection Agency: Washington, DC, USA, 1996.
- Ning, Z.; Guo, C.; Cai, P.; Zhang, M.; Chen, Z.; He, Z. Geochemical evaluation of biodegradation capacity in a petroleum contaminated aquifer. China Environ. Sci. 2018, 38, 4068–4074. [Google Scholar]
- American Society for Testing and Materials. Standard guide for remediation of ground water by natural attenuation at petroleum release sites. ASTM. Int. 2015, E1943–E1998. [Google Scholar]
- Suarez, M.P.; Rifai, H.S. Evaluation of btex remediation by natural attenuation at a coastal facility. Ground Water Monit. Remediat. 2010, 22, 62–77. [Google Scholar] [CrossRef]
- Marić, N.; Matić, I.; Papić, P.; Beškoski, V.P.; Ilić, M.; Gojgić-Cvijović, G.; Miletić, S.; Nikić, Z.; Vrvić, M.M. Natural attenuation of petroleum hydrocarbons—A study of biodegradation effects in groundwater (vitanovac, serbia). Environ. Monit. Assess. 2018, 190, 89. [Google Scholar] [CrossRef]
- Bolliger, C.; Hohener, P.; Hunkeler, D.; Haberli, K.; Zeyer, J. Intrinsic bioremediation of a petroleum hydrocarbon-contaminated aquifer and assessment of mineralization based on stable carbon isotopes. Biodegradation 1999, 10, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Su, X.-S.; Lue, H.; Zhang, W.-J.; Zhang, Y.-L.; Jiao, X. Super (13) c and super (34) s isotope evidence for biodegradation of a petroleum hydrocarbon-contaminated aquifer in the northeast of china. J. Jilin Univ. (Earth Sci. Ed.) 2011, 41, 847–854. [Google Scholar]
- Ning, Z.; Cai, P.; Zhang, M.; Guo, C.; Shi, C.; He, Z. Abnormally Low dissolved inorganic carbon anomaly in petroleum contaminated groundwater caused by microbiological geochemistry. Acta Sci. Circumstantiae 2019, 39, 1140–1147. [Google Scholar]
- Slater, G.F.; Nelson, R.K.; Kile, B.M.; Reddy, C.M. Intrinsic bacterial biodegradation of petroleum contamination demonstrated in situ using natural abundance, molecular-level 14c analysis. Org. Geochem. 2006, 37, 981–989. [Google Scholar] [CrossRef]
- Alfreider, A.; Baumer, A.; Bogensperger, T.; Posch, T.; Salcher, M.M.; Summerer, M. CO2 assimilation strategies in stratified lakes: Diversity and distribution patterns of chemolithoautotrophs. Environ. Microbiol. 2017, 19, 2754–2768. [Google Scholar] [CrossRef] [PubMed]
- Hügler, M.; Sievert, S.M. Beyond the calvin cycle: Autotrophic carbon fixation in the ocean. Annu. Rev. Mar. Sci. 2011, 3, 261–289. [Google Scholar] [CrossRef] [PubMed]
- Malkawi, H.I.; Jahmani, M.; Hussein, E.; Al-Horani, F.; Al-Deeb, T. Investigation on the ability of soil bacterial isolates to degrade petroleum hydrocarbons. Int. J. Integr. Biol. 2009, 7, 92–99. [Google Scholar]
- Sohn, J.H.; Kwon, K.K.; Kang, J.-H.; Jung, H.-B.; Kim, S.-J. Novosphingobium pentaromativorans sp. Nov., a high-molecular-mass polycyclic aromatic hydrocarbon-degrading bacterium isolated from estuarine sediment. Int. J. Syst. Evol. Microbiol. 2004, 54, 1483–1487. [Google Scholar] [CrossRef]
- Lyu, Y.; Zheng, W.; Zheng, T.; Tian, Y. Biodegradation of polycyclic aromatic hydrocarbons by novosphingobium pentaromativorans us6-1. PLoS ONE 2014, 9, e101438. [Google Scholar] [CrossRef] [PubMed]
- Kertesz*, M.; Kawasaki, A. Hydrocarbon-degrading sphingomonads: Sphingomonas, sphingobium, novosphingobium, and sphingopyxis. Handb. Hydrocarb. Lipid Microbiol. 2010, 1693–1705. [Google Scholar] [CrossRef]
- Sheng, X.; He, L.; Zhou, L.; Shen, Y. Characterization of microbacterium sp. F10a and its role in polycyclic aromatic hydrocarbon removal in low-temperature soil. Can. J. Microbiol. 2009, 55, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Salam, L.B.; Obayori, O.S.; Olatoye, N.O. Biodegradation of anthracene by a novel actinomycete, microbacterium sp. Isolated from tropical hydrocarbon-contaminated soil. World J. Microbiol. Biotechnol. 2014, 30, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Siddiqi, M.A.; Maccubbin, A.E.; Kumar, S.; Sikka, H.C. Degradation of polynuclear aromatic hydrocarbons by sphingomonas paucimobilis. Environ. Sci. Technol. 1995, 30, 136–142. [Google Scholar] [CrossRef]
- Singleton, D.R.; Ramirez, L.G.; Aitken, M.D. Characterization of a polycyclic aromatic hydrocarbon degradation gene cluster in a phenanthrene-degrading acidovorax strain. Appl. Environ. Microbiol. 2009, 75, 2613–2620. [Google Scholar] [CrossRef] [PubMed]
- Song, W.-F.; Wang, J.-W.; Yan, Y.-C.; An, L.-Y.; Zhang, F.; Wang, L.; Xu, Y.; Tian, M.-Z.; Nie, Y.; Wu, X.-L. Shifts of the indigenous microbial communities from reservoir production water in crude oil- and asphaltene-degrading microcosms. Int. Biodeterior. Biodegrad. 2018, 132, 18–29. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Z.; Dong, X.; Jia, Z.; Sun, Q. Variance in bacterial communities, potential bacterial carbon sequestration and nitrogen fixation between light and dark conditions under elevated co2 in mine tailings. Sci. Total Environ. 2019, 652, 234–242. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, H.; Yang, K.; Sun, Y.; Tian, J.; Lv, B. Nitrate removal by a novel autotrophic denitrifier (microbacterium sp.) using fe (ii) as electron donor. Ann. Microbiol. 2015, 65, 1069–1078. [Google Scholar] [CrossRef]
- Cunningham, J.A.; Rahme, H.; Hopkins, G.D.; Lebron, C.; Reinhard, M. Enhanced in situ bioremediation of btex-contaminated groundwater by combined injection of nitrate and sulfate. Environ. Sci. Technol. 2001, 35, 1663–1670. [Google Scholar] [CrossRef]
- Liu, Y.; Whitman, W.B. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 2008, 1125, 171–189. [Google Scholar] [CrossRef]
- Jarrell, K.F. Extreme oxygen sensitivity in methanogenic archaebacteria. Bioscience 1985, 35, 298–302. [Google Scholar] [CrossRef]
- Alvarez, P.; Vogel, T. Degradation of btex and their aerobic metabolites by indigenous microorganisms under nitrate reducing conditions. Water Sci. Technol. 1995, 31, 15–28. [Google Scholar] [CrossRef]
- Knowles, R. Denitrification. Microbiol. Rev. 1982, 46, 43. [Google Scholar] [PubMed]
- Lovley, D. Potential for anaerobic bioremediation of btex in petroleum-contaminated aquifers. J. Ind. Microbiol. Biotechnol. 1997, 18, 75–81. [Google Scholar] [CrossRef]
- Schroth, M.H.; Istok, J.D.; Conner, G.T.; Hyman, M.R.; Haggerty, R.; O’Reilly, K.T. Spatial variability in in situ aerobic respiration and denitrification rates in a petroleum-contaminated aquifer. Groundwater 1998, 36, 924–937. [Google Scholar] [CrossRef]
- Schürmann, A.; Schroth, M.; Saurer, M.; Bernasconi, S.; Zeyer, J. Nitrate-consuming processes in a petroleum-contaminated aquifer quantified using push–pull tests combined with 15n isotope and acetylene-inhibition methods. J. Contam. Hydrol. 2003, 66, 59–77. [Google Scholar] [CrossRef]
- Smith, R.L.; Duff, J.H. Denitrification in a sand and gravel aquifer. Appl. Environ. Microbiol. 1988, 54, 1071–1078. [Google Scholar]
- Myhr, S.; Torsvik, T. Denitrovibrio acetiphilus, a novel genus and species of dissimilatory nitrate-reducing bacterium isolated from an oil reservoir model column. Int. J. Syst. Evol. Microbiol. 2000, 50, 1611–1619. [Google Scholar] [CrossRef]
- Tiedje, J.M. Ecology of denitrification and dissimilatory nitrate reduction to ammonium. Biol. Anaerob. Microorg. 1988, 717, 179–244. [Google Scholar]
- Stenstrom, M.K.; Poduska, R.A. The effect of dissolved oxygen concentration on nitrification. Water Res. 1980, 14, 643–649. [Google Scholar] [CrossRef]
- Black, E.M.; Chimenti, M.S.; Just, C.L. Metagenomic analysis of nitrogen-cycling genes in upper mississippi river sediment with mussel assemblages. MicrobiologyOpen 2019, 8, e00739. [Google Scholar] [CrossRef]
- Gallon, J. The oxygen sensitivity of nitrogenase: A problem for biochemists and micro-organisms. Trends Biochem. Sci. 1981, 6, 19–23. [Google Scholar] [CrossRef]
- Herrero, A.; Muro-Pastor, A.M.; Flores, E. Nitrogen control in cyanobacteria. J. Bacteriol. 2001, 183, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.S.; Pardue, J.H.; Jackson, W.A. Oxygen demand and sulfate reduction in petroleum hydrocarbon contaminated salt marsh soils. Water Res. 2000, 34, 1345–1353. [Google Scholar] [CrossRef]
- Cardoso, R.B.; Sierra-Alvarez, R.; Rowlette, P.; Flores, E.R.; Gómez, J.; Field, J.A. Sulfide oxidation under chemolithoautotrophic denitrifying conditions. Biotechnol. Bioeng. 2006, 95, 1148–1157. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Lü, C.; Hou, N.; Xin, Y.; Liu, J.; Liu, H.; Xun, L. Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. Isme J. 2017, 11, 2754. [Google Scholar] [CrossRef] [PubMed]
- Schiff, J.; Fankhauser, H. Assimilatory sulfate reduction. In Biology of Inorganic Nitrogen and Sulfur; Springer: Berlin, Germany, 1981; pp. 153–168. [Google Scholar]
- Ioki, M.; Baba, M.; Bidadi, H.; Suzuki, I.; Shiraiwa, Y.; Watanabe, M.M.; Nakajima, N. Modes of hydrocarbon oil biosynthesis revealed by comparative gene expression analysis for race a and race b strains of botryococcus braunii. Bioresour. Technol. 2012, 109, 271–276. [Google Scholar] [CrossRef][Green Version]
- Ollivier, B.; Magot, M. Petroleum Microbiology; ASM Press: Washington, DC, USA, 2005. [Google Scholar]
- Sundberg, K.; Johansson, A.-S.; Stenberg, G.; Widersten, M.; Seidel, A.; Mannervik, B.; Jernström, B. Differences in the catalytic efficiencies of allelic variants of glutathione transferase p1-1 towards carcinogenic diol epoxides of polycyclic aromatic hydrocarbons. Carcinogenesis 1998, 19, 433–436. [Google Scholar] [CrossRef]
- Harayama, S.; Kishira, H.; Kasai, Y.; Shutsubo, K. Petroleum biodegradation in marine environments. J. Mol. Microbiol. Biotechnol. 1999, 1, 63–70. [Google Scholar]
- Vomberg, A.; Klinner, U. Distribution of alkb genes within n-alkane-degrading bacteria. J. Appl. Microbiol. 2000, 89, 339–348. [Google Scholar] [CrossRef]
- Zylstra, G.J.; Gibson, D.T. Aromatic hydrocarbon degradation: A molecular approach. In Genetic Engineering; Springer: Berlin, Germany, 1991; pp. 183–203. [Google Scholar]
- Tan, B.; Dong, X.; Sensen, C.W.; Foght, J. Metagenomic analysis of an anaerobic alkane-degrading microbial culture: Potential hydrocarbon-activating pathways and inferred roles of community members. Genome 2013, 56, 599–611. [Google Scholar] [CrossRef]
- Foght, J. Anaerobic biodegradation of aromatic hydrocarbons: Pathways and prospects. J. Mol. Microbiol. Biotechnol. 2008, 15, 93–120. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, E.; Ahmed, S.I.; Devol, A.H. Aerobic and anaerobic decomposition of organic matter in marine sediment: Which is fastest? Limnol. Oceanogr. 1995, 40, 1430–1437. [Google Scholar] [CrossRef]




| Well | MW14 | MW4 | NA125 | PM7 | PM3 | MW3 | NA7 | NA68 | MW6 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Contamination indices | COD (mg·L−1) | 0 | 0 | 0 | 17 | 27 | 131 | 482 | 295 | 50 |
| TPH (μg·L−1) | 640.6 | 659.8 | 619.1 | 6433.7 | 1872.6 | 4329 | 659 | 13,558 | 15,280.9 | |
| VOCs (μg·L−1) | 5.7 | 3 | 3.5 | 4.1 | 2.8 | 33 | 1848.7 | 2807.1 | 1699.5 | |
| BTEX (μg·L−1) | 2.6 | 1.7 | 1.5 | 1.2 | 1.4 | 8 | 1703.7 | 1493.5 | 1461.9 | |
| Electron acceptors | DO (mg·L−1) | 4.69 | 1.89 | 2.16 | 1.58 | 2.42 | 2.21 | 1.02 | 1.79 | 1.44 |
| SO42- (mg·L−1) | 237.5 | 269 | 123.6 | 154.9 | 155.9 | 107.7 | 24.68 | 49.95 | 51.06 | |
| NO3- (mg·L−1) | 116.4 | 87.6 | 17.33 | 2.42 | 3.54 | 10.61 | 33.21 | 64.97 | 8.13 | |
| Metabolic byproducts | Fe2+ (mg·L−1) | <0.01 | 0.011 | <0.01 | <0.01 | 0.962 | 1.512 | 5.164 | 0.019 | 3.099 |
| Mn2+ (mg·L−1) | 0.022 | 0.013 | 0.271 | 1.661 | 2.182 | 2.145 | 2.711 | 0.856 | 3.139 | |
| Other parameters | DIC (mg·L−1) | 248 | 274 | 146 | 187 | 212 | 249 | 292 | 285 | 319 |
| K+ (mg·L−1) | 4.41 | 4.83 | 2.6 | 4.32 | 4.35 | 4.27 | 4.06 | 8.54 | 3.1 | |
| Na+ (mg·L−1) | 115 | 137.7 | 125.3 | 152.1 | 143.3 | 155.1 | 147.4 | 190.5 | 123.7 | |
| Ca2+ (mg·L−1) | 240 | 246.4 | 144.8 | 165 | 157.3 | 122 | 135.3 | 87.32 | 192.9 | |
| Mg2+ (mg·L−1) | 83 | 89 | 51 | 67 | 69 | 56 | 62 | 66 | 77 | |
| NH4+ (mg·L−1) | 0 | 0 | 0 | 0 | 0 | 27.5 | 0 | 300 | 0 | |
| Cl– (mg·L−1) | 167 | 202 | 229 | 246 | 202 | 185 | 167 | 475 | 241 | |
| ORP (mv) | 130 | 203.1 | 115.8 | −35.8 | −78.6 | −51.9 | −98.2 | −55.5 | −92.7 | |
| pH | 6.82 | 6.75 | 6.98 | 6.89 | 6.90 | 6.81 | 6.78 | 7.20 | 6.72 | |
| Domain | MW14 | MW4 | NA125 | PM7 | PM3 | MW3 | NA7 | NA68 | MW6 |
|---|---|---|---|---|---|---|---|---|---|
| Bacteria | 98.74 | 98.56 | 98.97 | 97.09 | 99.20 | 99.34 | 98.43 | 99.50 | 98.55 |
| Archaea | 0.72 | 0.55 | 0.24 | 2.01 | 0.44 | 0.07 | 0.17 | 0.19 | 0.10 |
| Eukaryota | 0.26 | 0.49 | 0.29 | 0.49 | 0.25 | 0.21 | 0.44 | 0.17 | 1.14 |
| Viruses | 0.20 | 0.34 | 0.45 | 0.37 | 0.08 | 0.36 | 0.92 | 0.11 | 0.20 |
| Unclassified | 0.08 | 0.06 | 0.05 | 0.04 | 0.03 | 0.03 | 0.03 | 0.03 | 0.02 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, P.; Ning, Z.; Zhang, N.; Zhang, M.; Guo, C.; Niu, M.; Shi, J. Insights into Biodegradation Related Metabolism in an Abnormally Low Dissolved Inorganic Carbon (DIC) Petroleum-Contaminated Aquifer by Metagenomics Analysis. Microorganisms 2019, 7, 412. https://doi.org/10.3390/microorganisms7100412
Cai P, Ning Z, Zhang N, Zhang M, Guo C, Niu M, Shi J. Insights into Biodegradation Related Metabolism in an Abnormally Low Dissolved Inorganic Carbon (DIC) Petroleum-Contaminated Aquifer by Metagenomics Analysis. Microorganisms. 2019; 7(10):412. https://doi.org/10.3390/microorganisms7100412
Chicago/Turabian StyleCai, Pingping, Zhuo Ning, Ningning Zhang, Min Zhang, Caijuan Guo, Manlan Niu, and Jiansheng Shi. 2019. "Insights into Biodegradation Related Metabolism in an Abnormally Low Dissolved Inorganic Carbon (DIC) Petroleum-Contaminated Aquifer by Metagenomics Analysis" Microorganisms 7, no. 10: 412. https://doi.org/10.3390/microorganisms7100412
APA StyleCai, P., Ning, Z., Zhang, N., Zhang, M., Guo, C., Niu, M., & Shi, J. (2019). Insights into Biodegradation Related Metabolism in an Abnormally Low Dissolved Inorganic Carbon (DIC) Petroleum-Contaminated Aquifer by Metagenomics Analysis. Microorganisms, 7(10), 412. https://doi.org/10.3390/microorganisms7100412





