Distribution and Antimicrobial Resistance of Salmonella Isolated from Pigs with Diarrhea in China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation of Strains
2.3. Biochemical Tests and Serotyping
2.4. Antimicrobial Susceptibility Testing
2.5. Molecular Typing
2.6. Pathogenicity Analysis
2.7. Heavy Metal Tolerance
2.8. Whole-Genome Sequencing and SNP Based Phylogeny
2.9. Statistical Analysis
3. Results
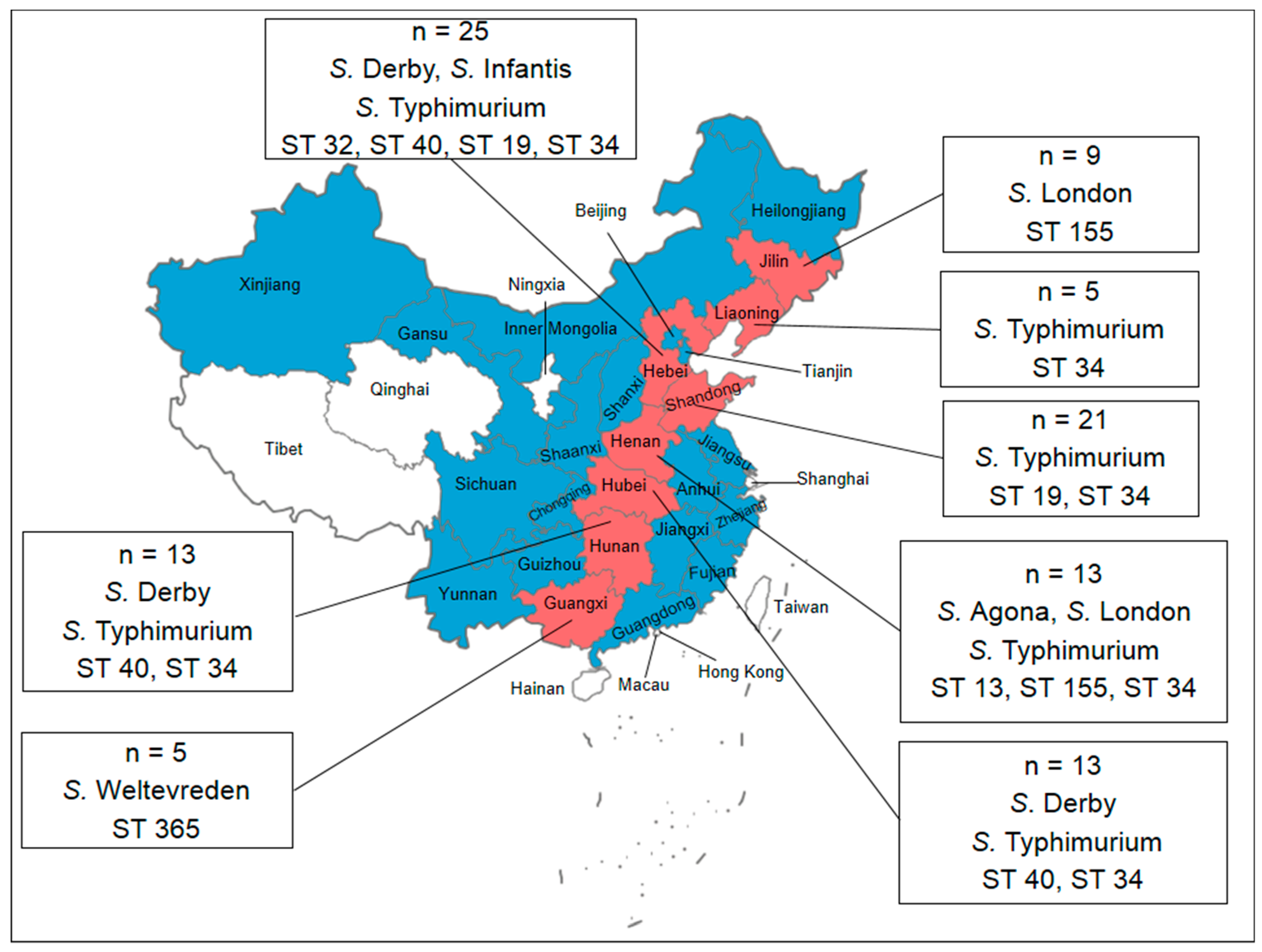
3.1. Prevalence and Distribution of Salmonella
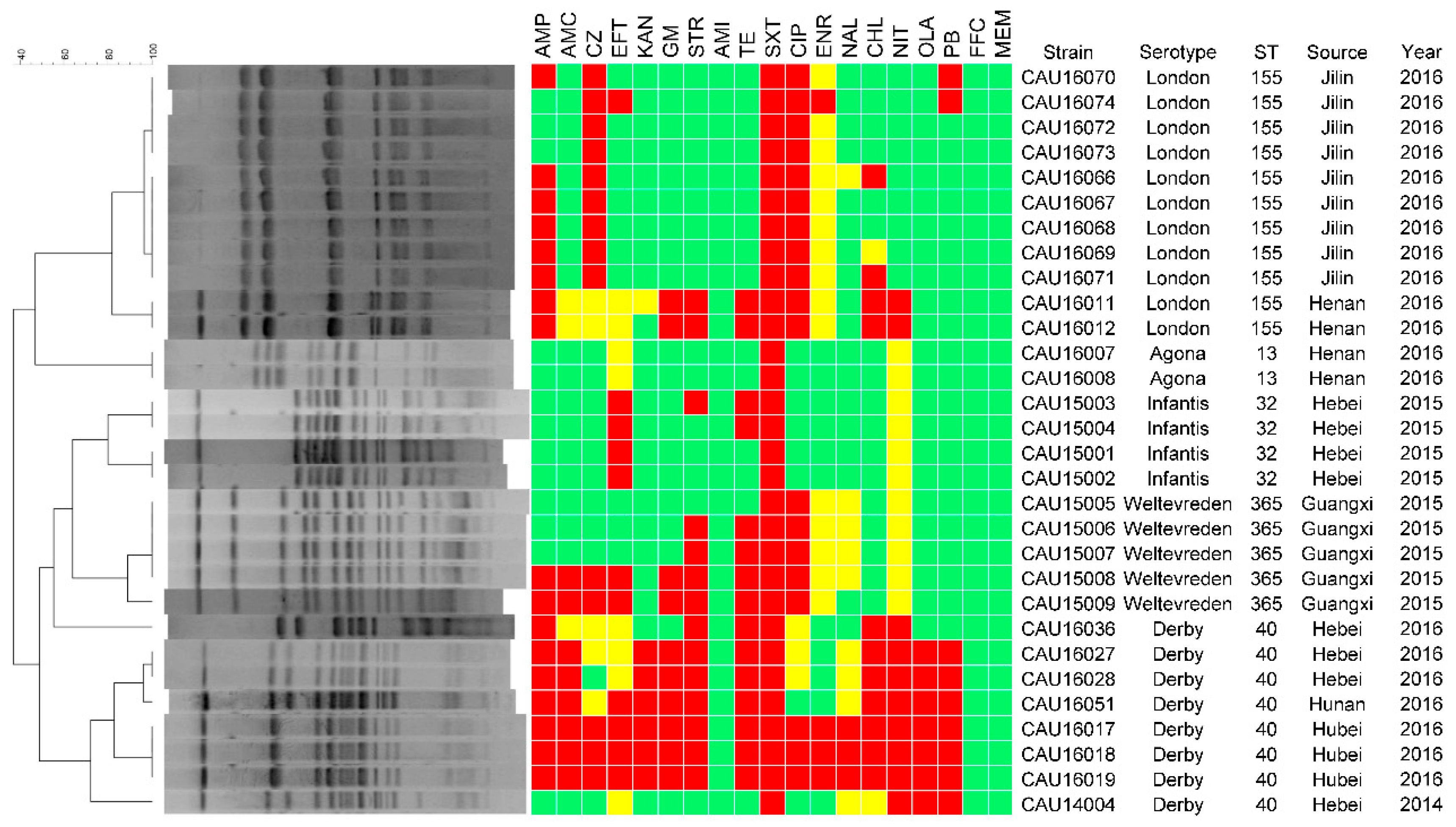
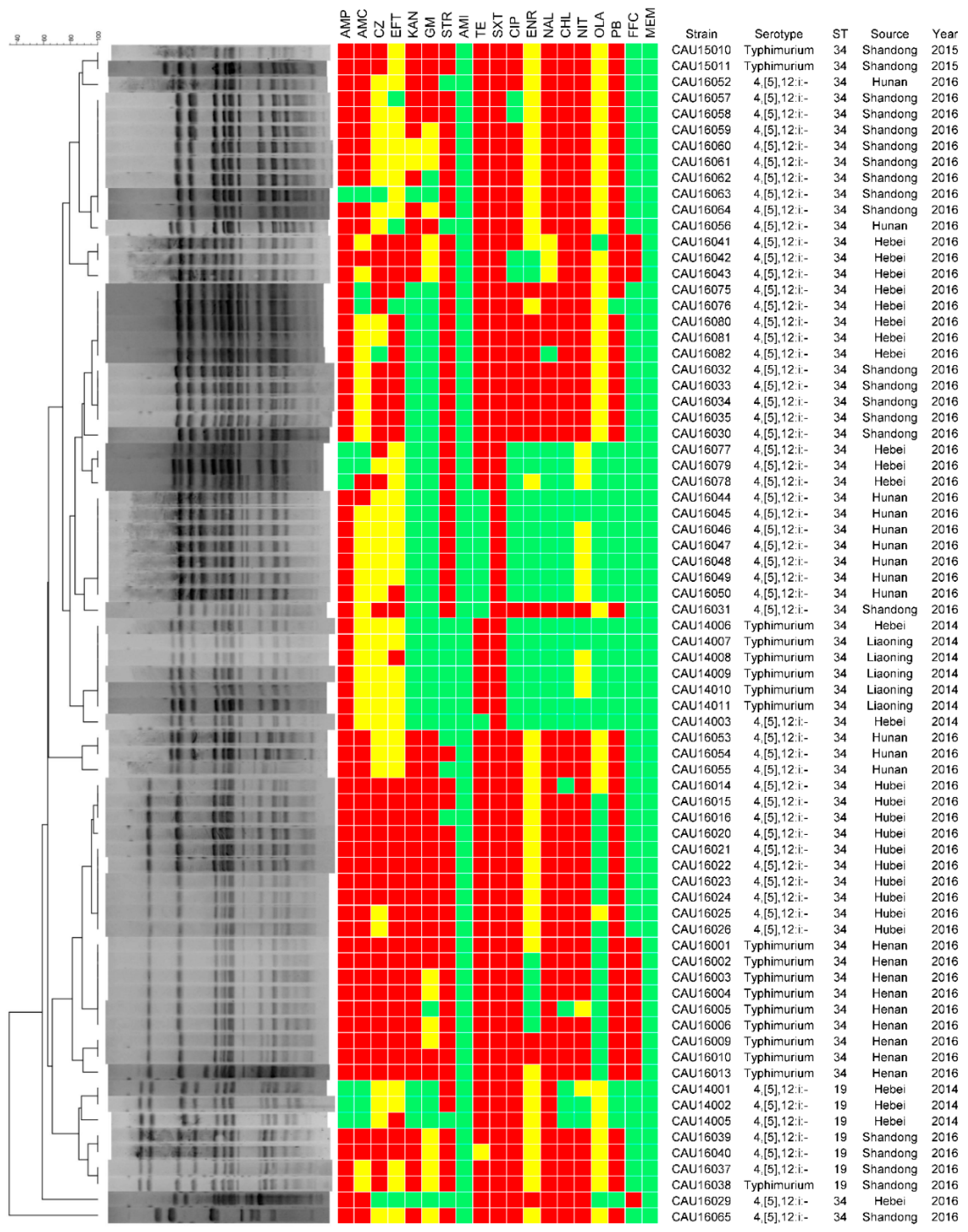
3.2. Serotyping
3.3. Antimicrobial Resistance
3.4. Molecular Typing
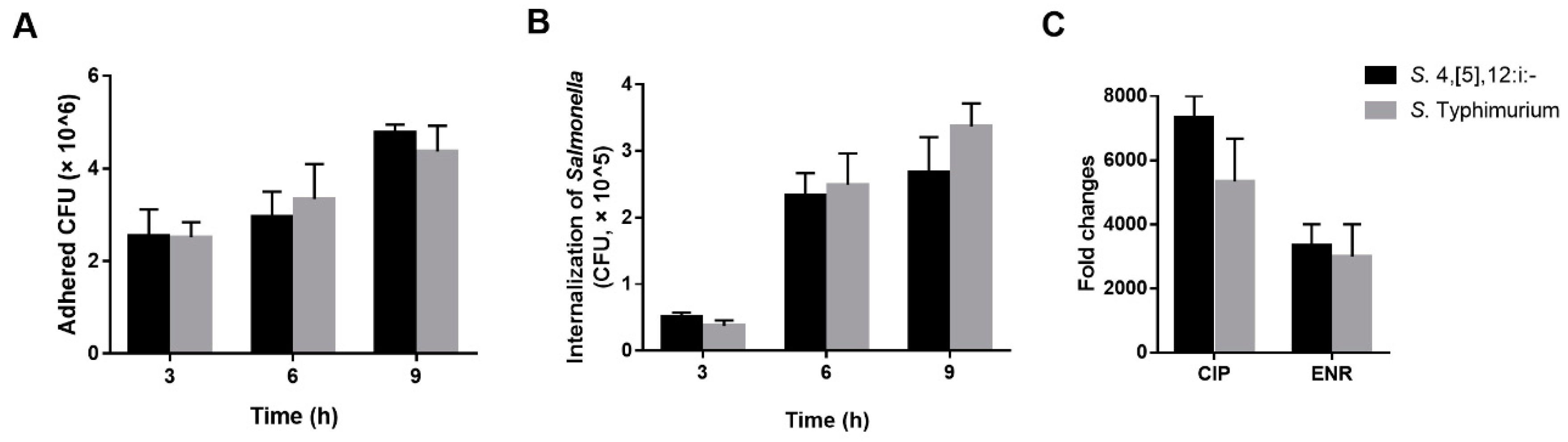
3.5. Pathogenicity Analysis
3.6. Heavy Metal Tolerance
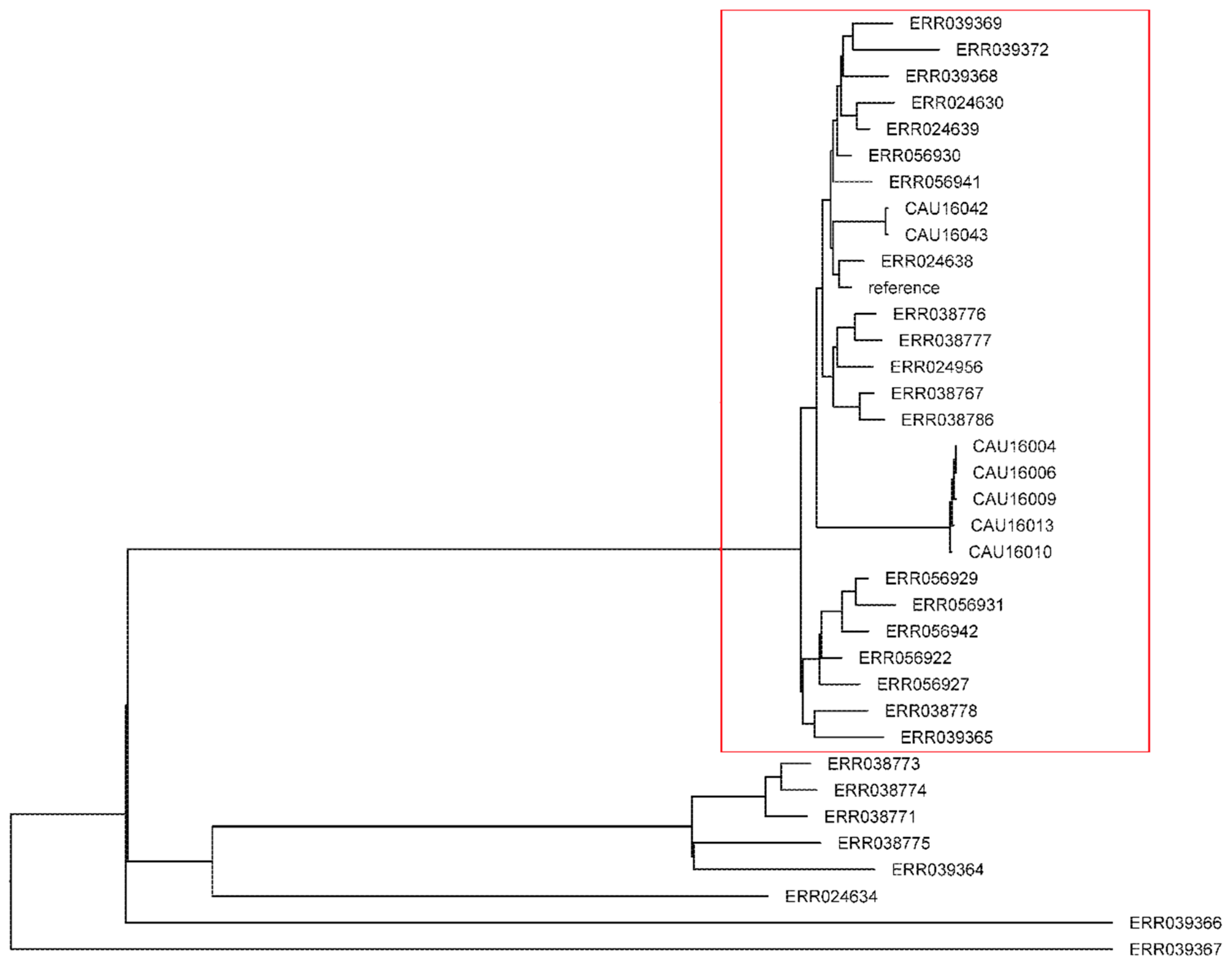
3.7. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kim, H.B.; Isaacson, R.E. Salmonella in swine: Microbiota interactions. Annu. Rev. Anim. Biosci. 2017, 5, 43–63. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Xia, X.; Cui, Y.; Hu, Y.; Xi, M.; Wang, X.; Shi, X.; Wang, D.; Meng, J.; Yang, B. Prevalence of extended-spectrum b-lactamase-producing Salmonella on retail chicken in six provinces and two national cities in the People’s Republic of China. J. Food Protect. 2013, 76, 2040–2044. [Google Scholar] [CrossRef] [PubMed]
- Vo, A.T.; van Duijkeren, E.; Fluit, A.C.; Heck, M.E.; Verbruggen, A.; Maas, H.M.; Gaastra, W. Distribution of Salmonella enterica serovars from humans, livestock and meat in vietnam and the dominance of Salmonella typhimurium phage type 90. Vet. Microbiol. 2006, 113, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Meurens, F.; Berri, M.; Auray, G.; Melo, S.; Levast, B.; Virlogeux-Payant, I.; Chevaleyre, C.; Gerdts, V.; Salmon, H. Early immune response following Salmonella enterica subspecies enterica serovar Typhimurium infection in porcine jejunal gut loops. Vet. Res. 2009, 40, 5. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.; Rajora, V.R.; Arora, N. Prevalence of Salmonella in pigs and broilers in the tarai region of uttarakhand, India. Indian J. Med. Microbiol. 2014, 32, 99–101. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Sanz, R.; Herrera-Leon, S.; de la Fuente, M.; Arroyo, M.; Echeita, M.A. Emergence of extended-spectrum beta-lactamases and ampc-type beta-lactamases in human Salmonella isolated in Spain from 2001 to 2005. J. Antimicrob. Chemoth. 2009, 64, 1181–1186. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liao, X.; Yang, Y.; Sun, J.; Li, L.; Liu, B.; Yang, S.; Ma, J.; Li, X.; Zhang, Q.; et al. Spread of oqxab in Salmonella enterica serotype Typhimurium predominantly by IncHI2 plasmids. J. Antimicrob. Chemoth. 2013, 68, 2263–2268. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.H.; Chan, E.W.; Liu, L.Z.; Chen, S. PMQR genes oqxAB and aac(6′)Ib-cr accelerate the development of fluoroquinolone resistance in Salmonella Typhimurium. Front. Microbiol. 2014, 5, 521. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Catry, B.; Cavaleri, M.; Baptiste, K.; Grave, K.; Grein, K.; Holm, A.; Jukes, H.; Liebana, E.; Navas, A.L.; Mackay, D.; et al. Use of colistin-containing products within the European Union and European Economic Area (EU/EEA): Development of resistance in animals and possible impact on human and animal health. Int J. Antimicrob. Ag. 2015, 46, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Machado, J.; Bernardo, F. Prevalence of Salmonella in chicken carcasses in portugal. J. Appl. Microbiol. 1990, 69, 477–480. [Google Scholar] [CrossRef]
- Mastrorilli, E.; Pietrucci, D.; Barco, L.; Ammendola, S.; Petrin, S.; Longo, A.; Mantovani, C.; Battistoni, A.; Ricci, A.; Desideri, A.; et al. A comparative genomic analysis provides novel insights into the ecological success of the monophasic Salmonella serovar 4,[5],12:I. Front. Microbiol. 2018, 9, 715. [Google Scholar] [CrossRef] [PubMed]
- Petrovska, L.; Mather, A.E.; AbuOun, M.; Branchu, P.; Harris, S.R.; Connor, T.; Hopkins, K.L.; Underwood, A.; Lettini, A.A.; Page, A.; et al. Microevolution of monophasic Salmonella Typhimurium during epidemic, United Kingdom, 2005–2010. Emerg Infect. Dis. 2016, 22, 617–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barco, L.; Mancin, M.; Ruffa, M.; Saccardin, C.; Minorello, C.; Zavagnin, P.; Lettini, A.A.; Olsen, J.E.; Ricci, A. Application of the random forest method to analyse epidemiological and phenotypic characteristics of Salmonella 4,[5],12:I:- and Salmonella Typhimurium strains. Zoonoses Public Health 2012, 59, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-fifth Informational Supplement; M100-s25; Clinical and laboratory standards institute: Wayne, PA, USA, 2015. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 4th ed.; Clinical and laboratory standards institute: Wayne, PA, USA, 2013. [Google Scholar]
- Chen, S.; Zhao, S.H.; White, D.G.; Schroeder, C.M.; Lu, R.; Yang, H.C.; McDermott, P.F.; Ayers, S.; Meng, J.H. Characterization of multiple-antimicrobial-resistant Salmonella serovars isolated from retail meats. Appl. Environ. Microb. 2004, 70, 1–7. [Google Scholar] [CrossRef]
- Sorensen, A.H.; Hansen, L.H.; Johannesen, E.; Sorensen, S.J. Conjugative plasmid conferring resistance to olaquindox. Antimicrob. Agents Ch. 2003, 47, 798–799. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of mics and zone diameters version 7.1, valid from 2017-03-10. 2017. [Google Scholar]
- Centers for Disease Control Prevention. Standard Operating Procedure for Pulsenet Pfge of Escherichia coli o157: H7, Escherichia coli non-o157 (stec), Salmonella serotypes, Shigella sonnei and Shigella flexneri; Centers for Disease Control and Prevention: Atlanta, Georgia, 2013.
- Rosselin, M.; Virlogeux-Payant, I.; Roy, C.; Bottreau, E.; Sizaret, P.Y.; Mijouin, L.; Germon, P.; Caron, E.; Velge, P.; Wiedemann, A. Rck of Salmonella enterica, subspecies enterica serovar enteritidis, mediates zipper-like internalization. Cell. Res. 2010, 20, 647–664. [Google Scholar] [CrossRef] [PubMed]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, J.; Wu, C.M.; Wu, C.B.; Qi, J.; Wang, Y.; Wang, H.Y.; Liu, Y.Q.; Shen, J.Z. Serotype distribution and antibiotic resistance of Salmonella in food-producing animals in Shandong province of China, 2009 and 2012. Int. J. Food Microbiol. 2014, 180, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Kuang, X.H.; Hao, H.H.; Dai, M.H.; Wang, Y.L.; Ahmad, I.; Liu, Z.L.; Yuan, Z.H. Serotypes and antimicrobial susceptibility of Salmonella spp. Isolated from farm animals in China. Front. Microbiol. 2015, 6, 602. [Google Scholar] [CrossRef] [PubMed]
- Li, R.C.; Lai, J.; Wang, Y.; Liu, S.L.; Li, Y.; Liu, K.Y.; Shen, J.Z.; Wu, C.M. Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan province, China. Int. J. Food. Microbiol. 2013, 163, 14–18. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority. The European union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 2015, 18, 45. [Google Scholar]
- National Antimicrobial Resistance Monitoring System. Animal Arm Annual Report. Available online: Https://www.Ars.Usda.Gov/arsuserfiles/60401020/narms/narms2011/narms%20usda%202011%20report.Pdf (accessed on 18 November 2018).
- National Bureau of Statistics of China. China Statistical Yearbook; National Bureau of Statistics of China: Beijing, China, 2016.
- Doumith, M.; Godbole, G.; Ashton, P.; Larkin, L.; Dallman, T.; Day, M.; Day, M.; Muller-Pebody, B.; Ellington, M.J.; de Pinna, E.; et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J. Antimicrob. Chemoth. 2016, 71, 2300–2305. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.S.; Xu, M.; Zhu, C.H.; Miao, J.F.; Liu, X.X.; Xu, B.; Zhang, J.Q.; Yu, Y.; Jia, X.B. Antimicrobial resistance, presence of integrons and biofilm formation of Salmonella pullorum isolates from eastern China (1962-2010). Avian Pathol. 2013, 42, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Slade, R.D.; Kyriazakis, I.; Carroll, S.M.; Reynolds, F.H.; Wellock, I.J.; Broom, L.J.; Miller, H.M. Effect of rearing environment and dietary zinc oxide on the response of group-housed weaned pigs to enterotoxigenic Escherichia coli O149 challenge. Animal 2011, 5, 1170–1178. [Google Scholar] [CrossRef] [PubMed]
- Wales, A.D.; Davies, R.H. Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics 2015, 4, 567–604. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.L.; Ran, L.; Wu, S.Y.; Ke, B.X.; He, D.M.; Yang, X.F.; Zhang, Y.H.; Ke, C.W.; Klena, J.D.; Yan, M.Y.; et al. Laboratory-based surveillance of non-typhoidal Salmonella infections in Guangdong province, China. Foodborne Pathog. Dis. 2012, 9, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Wasyl, D.; Hoszowski, A. Occurrence and characterization of monophasic Salmonella enterica serovar Typhimurium (4,[5], 12:i:-) of non-human origin in Poland. Foodborne Pathog. Dis. 2012, 9, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
| Resistance Pattern | Guangxi | Liaoning | Jilin | Henan | Hubei | Hunan | Shandong | Hebei |
|---|---|---|---|---|---|---|---|---|
| 3 | 0 | 5 | 2 | 0 | 0 | 0 | 0 | 1 |
| 4 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 2 |
| 5 | 2 | 0 | 0 | 0 | 3 | 6 | 11 | 4 |
| 6 | 0 | 0 | 0 | 1 | 6 | 0 | 3 | 2 |
| 7 | 0 | 0 | 0 | 8 | 1 | 0 | 6 | 4 |
| 8 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| Total | 2 | 5 | 2 | 11 | 13 | 6 | 21 | 13 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, J.-H.; Zhu, Y.-H.; Ren, T.-Y.; Guo, L.; Yang, G.-Y.; Jiao, L.-G.; Wang, J.-F. Distribution and Antimicrobial Resistance of Salmonella Isolated from Pigs with Diarrhea in China. Microorganisms 2018, 6, 117. https://doi.org/10.3390/microorganisms6040117
Su J-H, Zhu Y-H, Ren T-Y, Guo L, Yang G-Y, Jiao L-G, Wang J-F. Distribution and Antimicrobial Resistance of Salmonella Isolated from Pigs with Diarrhea in China. Microorganisms. 2018; 6(4):117. https://doi.org/10.3390/microorganisms6040117
Chicago/Turabian StyleSu, Jin-Hui, Yao-Hong Zhu, Tian-Yi Ren, Liang Guo, Gui-Yan Yang, Lian-Guo Jiao, and Jiu-Feng Wang. 2018. "Distribution and Antimicrobial Resistance of Salmonella Isolated from Pigs with Diarrhea in China" Microorganisms 6, no. 4: 117. https://doi.org/10.3390/microorganisms6040117
APA StyleSu, J.-H., Zhu, Y.-H., Ren, T.-Y., Guo, L., Yang, G.-Y., Jiao, L.-G., & Wang, J.-F. (2018). Distribution and Antimicrobial Resistance of Salmonella Isolated from Pigs with Diarrhea in China. Microorganisms, 6(4), 117. https://doi.org/10.3390/microorganisms6040117




