Variability in Characterizing Escherichia coli from Cattle Feces: A Cautionary Tale
Abstract
1. Introduction
2. Materials and Methods
2.1. PCR Confirmation of Serogroup and stx at Initial Isolation by Laboratory A (LAB A)
2.2. PCR Methodology Used for Detection of stx Genes by Laboratory B (LAB B)
2.3. Serotyping of Isolates and Detection of stx Genes by Laboratory C (LAB C)
2.4. Pulsed-Field Gel Electrophoresis (PFGE)
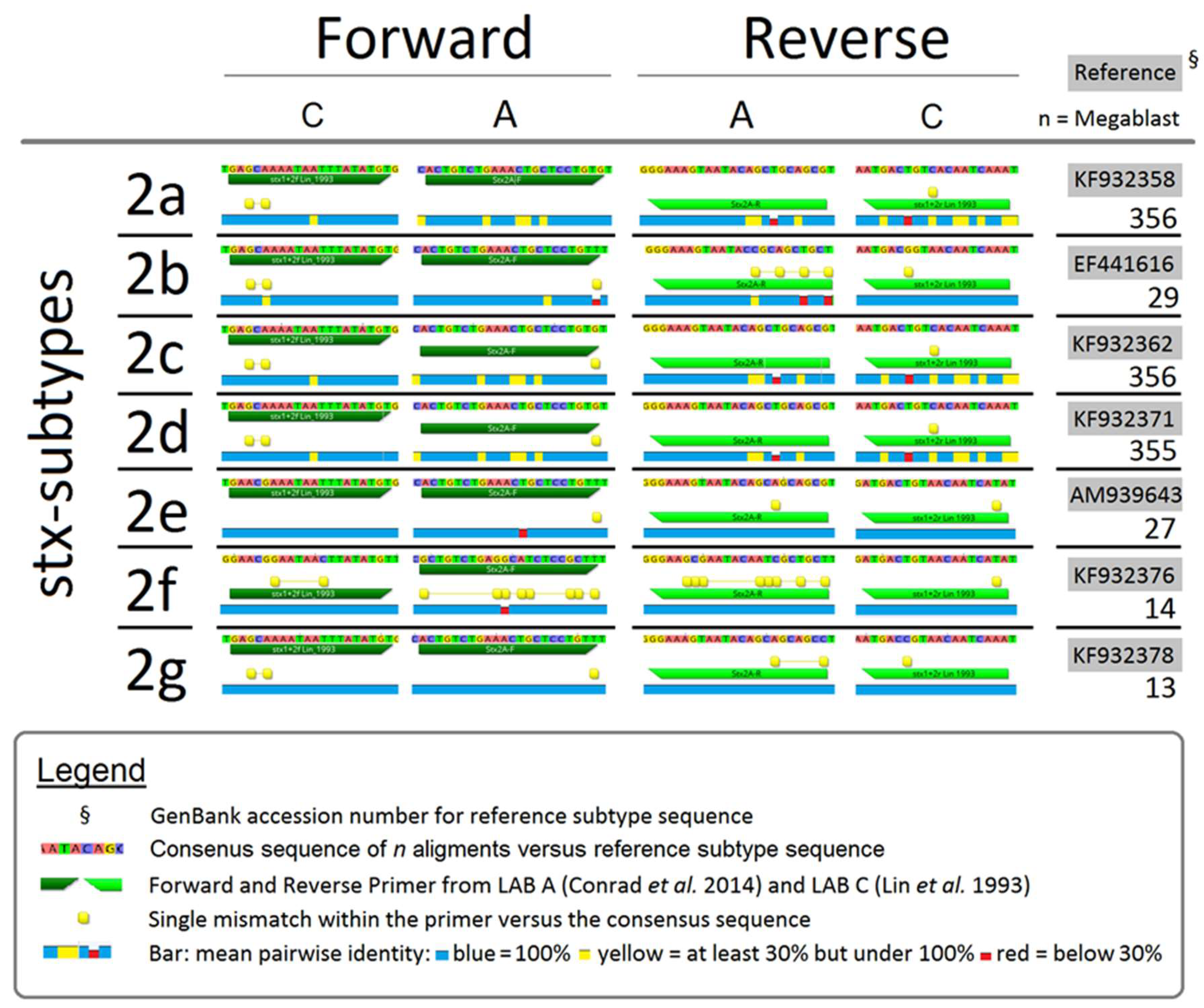
2.5. Comparing Consensus Sequences of PCR Primers and stx
2.6. Statistical Analyses
3. Results
3.1. O Group as Determined by PCR and TS
3.2. Serotypes of Isolates Where PCR O Group Agreed with TS
3.3. Comparison of PFGE and TS for O26, O103, O111 and O157
3.4. Inconsistent Detection of stx1 and stx2
4. Discussion
4.1. Determination of O Group by PCR and TS
4.2. Serotypes of Top 7 Isolated from Cattle Feces
4.3. Impacts of Serogroup and Serotype on PFGE Analyses
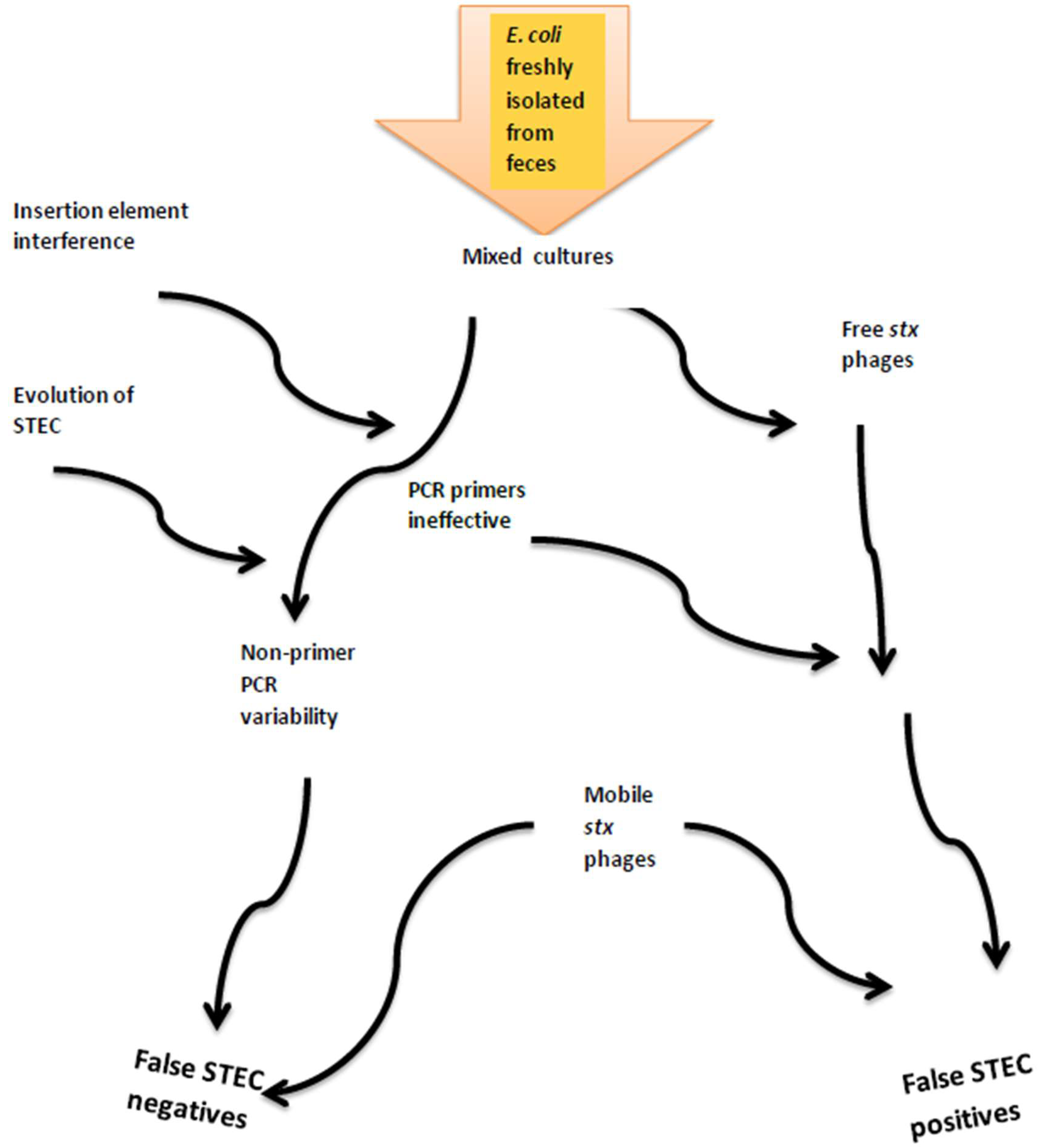
4.4. Inconsistent Detection of stx1 and stx2
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bosilevac, J.M.; Koohmaraie, M. Prevalence and characterization of non-O157 Shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl. Environ. Microbiol. 2011, 77, 2103–2112. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, M.W.; Gay, J.M.; Hancock, D.D.; Gay, C.C.; Fox, L.K.; Besser, T.E. Sensitivity of bacteriologic culture for detection of Escherichia coli O157:H7 in bovine feces. J. Clin. Microbiol. 1995, 3, 2616–2619. [Google Scholar]
- Cooley, M.B.; Jay-Russell, M.; Atwill, E.R.; Carychao, D.; Nguyen, K.; Quinones, B.; Patel, R.; Walker, S.; Swimley, M.; Pierre-Jerome, E.; et al. Development of a robust method for isolation of Shiga toxin-positive Escherichia coli (STEC) from fecal, plant, soil and water samples from a leafy greens production region in California. PLoS ONE 2013, 8, e65716. [Google Scholar] [CrossRef] [PubMed]
- Noll, L.W.; Shridhar, P.B.; Dewsbury, D.M.; Shi, X.; Cernicchiaro, N.; Renter, D.G.; Nagaraja, T.G. A Comparison of Culture- and PCR-Based Methods to Detect Six Major Non-O157 Serogroups of Shiga Toxin-Producing Escherichia coli in Cattle Feces. PLoS ONE 2015, 10, e0135446. [Google Scholar] [CrossRef] [PubMed]
- Hallewell, J.; Alexander, T.; Reuter, T.; Stanford, K. Limitations of immunomagnetic separation for detection of the top seven serogroups of Shiga-toxin producing Escherichia coli. J. Food Prot. 2017, 80, 598–603. [Google Scholar] [CrossRef] [PubMed]
- Gannon, V.P.J.; King, R.K.; Kim, J.Y.; Golsteyn Thomas, E.J. Rapid and sensitive method for detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Appl. Environ. Microbiol. 1992, 58, 3809–3815. [Google Scholar] [PubMed]
- Lin, Z.; Kurazono, H.; Yamasaki, S.; Takeda, Y. Detection of various variant verotoxin genes in Escherichia coli by polymerase chain reaction. Microbiol. Immunol. 1993, 37, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Paton, A.W.; Paton, J.C. Detection and characterization of Shiga toxigenic Escherichia coli by using Multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hylA, rfbO111, and rfbO157. J. Clin. Microbiol. 1998, 36, 598–602. [Google Scholar]
- Bai, J.; Paddock, Z.D.; Shi, X.; Li, S.; An, B.; Nagaraja, T.G. Applicability of a multiplex PCR to detect the seven major Shiga toxin-producing Escherichia coli based on genes that code for serogroup-specific O-antigens and major virulence factors in cattle feces. Foodborne Pathog. Dis. 2012, 9, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Conrad, C.C.; Stanford, K.; McAllister, T.A.; Thomas, J.; Reuter, T. Further development of sample preparation and detection methods for O157 and the top 6 non-O157 STEC serogroups in cattle feces. J. Microbiol. Methods 2014, 105, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Goji, N.; Mathews, A.; Huszczynski, G.; Laing, C.R.; Gannon, V.P.J.; Graham, M.R.; Amoako, K.K. A new pyrosequencing assay for rapid detection and genotyping of Shiga toxin, intimin and O157-specific rfbE genes of Escherichia coli. J. Microbiol. Meth. 2015, 109, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Ziebell, K.A.; Read, S.C.; Johnson, R.P.; Gyles, C.L. Evaluation of PCR and PCR-RFLP protocols for identifying Shiga toxins. Res. Microbiol. 2002, 153, 289–300. [Google Scholar] [CrossRef]
- Wells, J.G.; Davis, B.; Wachsmuth, K.; Riley, L.W.; Remis, R.; Sokolow, R.; Morris, G.K. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J. Clin. Microbiol. 1983, 18, 512–520. [Google Scholar] [PubMed]
- DeBroy, C.; Frantamico, P.M.; Roberts, E.; Davis, M.A.; Liu, Y. Development of PCR assays targeting genesin O-antigen gene clusters for detection and identification of Escherichia coli O45 and O55 serogroups. Appl. Environ. Microbiol. 2005, 71, 4919–4924. [Google Scholar] [CrossRef] [PubMed]
- Frantamico, P.M.; Debroy, C.; Liu, Y.; Needleman, D.S.; Barazoni, G.M.; Feng, P. Advances in molecular serotyping and subtyping Escherichia coli. Front. Microbiol. 2016, 7, 644. [Google Scholar] [CrossRef]
- DeBroy, C.; Fratamico, P.M.; Yan, X.; Baranzoni, G.M.; Liu, Y.; Needleman, D.S.; Tebbs, R.; O’Connell, C.D.; Allred, A.; Swimley, M.; et al. Comparison of O-antigen gene clusters of all O-serogroups of Escherichia coli and proposal for adopting a new nomenclature for O-typing. PLoS ONE 2016, 11, e0147434. [Google Scholar]
- Shridhar, P.B.; Noll, L.W.; Shi, X.; Cernicchiaro, N.; Renter, D.G.; Bai, J.; Nagaraja, T.G. Escherichia coli O104 in feedlot cattle feces: Prevalence, isolation and characterization. PLoS ONE 2016, 11, e0152101. [Google Scholar] [CrossRef] [PubMed]
- Duda, K.A.; Linder, B.; Brade, H.; Leimback, A.; Brzuskiewicz, E.; Dobindt, U.; Holst, O. The lipopolysaccharide of the mastitis isolate Escherichia coli strain 1303 comprises a novel O antigen and the rare K-12 core type. Microbiology 2011, 157, 1750–1760. [Google Scholar] [CrossRef] [PubMed]
- Joris, M.A.; Verstraete, K.; De Reu, K.; De Zutter, L. Loss of vtx genes after the first sub-cultivation step of verocytoxigenic Escherichia coli O157 and non-O157 during isolation from naturally contaminated fecal samples. Toxins 2011, 3, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Loftsdottir, H.; Soderlund, R.; Jinnerot, T.; Eriksson, E.; Bongcam-Rudolff, E.; Aspan, A. Dynamics of insertion sequence element IS629 inactivation of verotoxin 2 genes in Escherichia coli O157:H7. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed]
- Quiros, P.; Martinez-Castillo, A.; Muniesa, M. Improving detection of Shiga toxin-producing Escherichia coli by molecular methods by reducing the interferences of free Shiga toxin-encoding bacteriophages. Appl. Environ. Microbiol. 2015, 81, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Stanford, K.; Johnson, R.P.; Alexander, T.; McAllister, T.; Reuter, T. Influence of season and feedlot location on prevalence and virulence of Escherichia coli from feces of western Canadian slaughter cattle. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Ribot, E.M.; Fair, M.A.; Gautom, R.; Cameron, D.N.; Hunter, S.B.; Swaminathan, B.; Barrett, T.J. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella and Shigella for PulseNet. Foodborne Pathog. Dis. 2006, 3, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Scheutz, F.; Beutin, L.; Pierard, D.; Buvens, G.; Karch, H.; Mellmann, A.; Caprioli, A.; Tozzoli, R.; Morabito, S.; Strockbine, N.A.; et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 2012, 50, 2951–2963. [Google Scholar] [CrossRef] [PubMed]
- Iguchi, A.; Iyoda, S.; Kikuchi, T.; Ogura, Y.; Katsura, K.; Ohnishi, M.; Hayashi, T.; Thomson, N.R. A complete view of the genetic diversity of the Escherichia coli O-antigen biosynthesis gene cluster. DNA Res. 2015, 22, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Geue, L.; Scgares, S.; Mintel, B.; Conraths, F.J.; Muller, E.; Ehricht, R. Rapid microarray-based genotyping of Escherichia coli serotype O156:H25/H-/Hnt isolates from cattle and clonal relationship analysis. Appl. Environ. Microbiol. 2010, 76, 5510–5519. [Google Scholar] [CrossRef] [PubMed]
- Kerr, P.; Ball, H.; China, B.; Mainil, J.; Finlay, D.; Pollock, D.; Wilson, I.; Mackie, D. Use of a monoclonal antibody against an Escherichia coli O26 surface protein for detection of enteropathogenic and enterohemorrhagic strains. Clin. Diagn. Lab. Immunol. 1999, 6, 610–614. [Google Scholar] [PubMed]
- Yonekita, T.; Fukimura, T.; Morishita, N.; Matsumoto, T.; Morimatsu, F. Simple, rapid and reliable detection of Escherichia coli O26 using immunochromatography. J. Food Prot. 2013, 76, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Delannoy, S.; Beutin, L.; Fach, P. Use of clustered regularly interspaced short palindromic repeat sequence polymorphisms for specific detection of enterohemorrhagic Escherichia coli strains of serotypes O26:H11, O45:H2, O103:H2, O111:H8, O121:H19, O145:H28 and O157:H7 by real-time PCR. J. Clin. Microbiol. 2012, 50, 4035–4040. [Google Scholar] [CrossRef] [PubMed]
- Jacob, M.E.; Almes, K.M.; Shi, X.; Sargeant, J.M.; Nagaraja, T.G. Escherichia coli O157:H7 genetic diversity in bovine fecal samples. J. Food Prot. 2011, 74, 1186–1188. [Google Scholar] [CrossRef] [PubMed]
- Strachan, N.J.C.; Rotariu, O.; Lopes, B.; MacRae, M.; Fairly, S.; Laing, C.; Gannon, V.; Allison, L.J.; Hanson, M.F.; Dallman, T.; et al. Whole genome sequencing demonstrates that geographic variation of Escherichia coli O157 genotypes dominates host association. Sci. Rep. 2015, 5, 14145. [Google Scholar] [CrossRef] [PubMed]
- Dewsbury, D.M.; Renter, D.G.; Shridhar, P.B.; Noll, L.W.; Shi, X.; Nagaraja, T.G. Summer and winter prevalence of Shiga toxin-producing Escherichia coli (STEC) O26, O45, O103, O111, O121, O145 and O157 in feces of feedlot cattle. Foodborne Pathog. Dis. 2015, 12, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Beutin, L.; Kaulfuss, S.; Herold, S.; Oswald, E.; Schmidt, H. Generic analysis of enteropathogenic and enterohemorrhagic Escherichia coli serogroup O103 strains by molecular typing of virulence and housekeeping genes and pulsed-field gel electrophoresis. J. Clin. Microbiol. 2005, 43, 1552–1563. [Google Scholar] [CrossRef] [PubMed]
- Seske, C.; Sunde, M.; Hopp, P.; Brulheim, T.; Sewornu Cudjoe, K.; Kvitle, B.; Urdahl, A.M. Occurrence of potentially human-pathogenic Escherichia coli O103 in Norwegian sheep. Appl. Environ. Microbiol. 2013, 79, 7502–7509. [Google Scholar]
- Eichhorn, H.; Heidemanns, K.; Semmler, T.; Kinnemann, B.; Mellmann, A.; Harmsen, D.; Anjum, M.; Schmidt, H.; Fruth, A.; Valentin-Weigand, P.; et al. Highly virulent non-O157 enterohemorrhagic Escherichia coli (EHEC) serotypes reflect similar phylogenetic lineages, providing new insights into the evolution of EHEC. Appl. Environ. Microbiol. 2015, 81, 7041–7047. [Google Scholar] [CrossRef] [PubMed]
- Beutin, L.; Delannoy, S.; Fach, P. Genetic diversity of the fliC genes encoding the flagellar antigen H19 or Escherichia coli and applications to the specific identification of enterohemorrhagic E. coli O121:H19. Appl. Environ. Microbiol. 2015, 81, 4224–4230. [Google Scholar] [CrossRef] [PubMed]
- Cooper, K.K.; Mandrell, R.E.; Louie, J.W.; Korlach, J.; Clark, T.A.; Parker, C.T.; Huynh, S.; Chain, P.S.; Ahmen, S.; Carter, M.Q. Comparative genomics of enterohemorrhagic Escherichia coli O145:H28 demonstrates a common evolutionary lineage with Escherichia coli O157:H7. BMC Genom. 2014, 15, 17. [Google Scholar] [CrossRef] [PubMed]
- Delannoy, S.; Mariani-Kurkdjian, P.; Bonacorsi, S.; Liguori, S.; Fach, P. Characteristics of emerging human pathogenic Escherichia coli O26:H11 strains isolated in France between 2010 and 2013 and carrying the stx2d gene only. J. Clin. Microbiol. 2015, 53, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Fields, P.I.; Swaminathan, B.; Whittam, T.S. Characterization of non-motile variants of Escherichia coli O157 and other serotypes by using an antiflagellin monoclonal antibody. J. Clin. Microbiol. 1996, 34, 2856–2859. [Google Scholar] [PubMed]
- Tominaga, A.M.; Mahmoud, A.-H.; Mukaihara, T.; Enomoto, M. Molecular characterization of intact, but cryptic flagellin genes in the genus Shigella. Mol. Microbiol. 1994, 12, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Conrad, C.C.; Stanford, K.; McAllister, T.A.; Thomas, J.; Reuter, T. Competition during enrichment of pathogenic Escherichia coli may result in culture bias. Facets 2016, 1, 114–126. [Google Scholar] [CrossRef]
- Booth, C.S.; Pienaar, E.; Termaat, J.R.; Whitney, S.E.; Louw, T.M.; Viljoen, H.J. Efficiency of the polymerase chain reaction. Chem. Eng. Sci. 2010, 65, 4996–5006. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Dobrindt, U.; Hacker, J.; Hasnain, S.E. Genomic fluidity and pathogenic bacteria: applications in diagnostics, epidemiology and intervention. Nat. Rev. Microbiol. 2008, 6, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, N.; Tarr, P.I. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations and evolutionary implications. J. Bacteriol. 2003, 189, 6645–6654. [Google Scholar]
- Mellmann, A.; Lu, S.; Karch, H.; Xu, J.-G.; Harmsen, D.; Schmidt, M.A.; Bielaszewska, M. Recycling of Shiga toxin 2 genes in sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl. Environ. Microbiol. 2008, 74, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.-T.; Qintela, I.A.; Nguyen, K.; Salvador, A.; Cooley, M.B.; Wu, V.C.H. Investigation of prevalence offree Shiga-toxin producing Escherichia coli (STEC)-specific bacteriophages and its correlation with STEC bacterial hosts in a produce-growing area in Salinas California. PLoS ONE 2018, 13, e0190534. [Google Scholar] [CrossRef] [PubMed]
- Rooks, D.J.; Yan, Y.; McDonald, J.E.; Woodward, M.J.; McCarthy, A.J.; Alison, H.E. Development and validation of a qPCR-based method for quantifying Shiga toxin-encoding and other lambdoid bacteriophages. Environ. Microbiol. 2010, 12, 1194–1204. [Google Scholar] [CrossRef] [PubMed]
- Hartland, E.L.; Leong, J.M. Enteropathogenic and enterohemorrhagic E. coli: Ecology, pathogenesis, and evolution. Front. Cell. Infect. Microbiol. 2013, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, M.; Zhou, K.; Mellmann, A.; Morabito, S.; Croughs, P.D.; de Boer, R.F.; Koolstra-Smid, A.M.D.; Rossen, J.W.A.; Friedrich, A.W. Is Shiga toxin-negative Escherichia coli O157:H7 enteropathogenic or enterohemorrhagic Escherichia coli? Comprehensive molecular analysis using whole-genome sequencing. J. Clin. Microbiol. 2015, 53, 3530–3538. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Eydallin, G.; Maharajan, R.P.; Feng, L.; Wang, L.; Ferenci, T. Natural Escherichia coli isolates rapidly acquire genetic changes upon laboratory domestication. Microbiology 2017, 163, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Toth, I.; Schmidt, H.; Kardos, G.; Lancz, Z.; Creuzburg, K.; Damjanova, I.; Paszti, J.; Beutin, L.; Nagy, B. Virulence genes and molecular typing of different groups of Escherichia coli O157 strains in cattle. Appl. Environ. Microbiol. 2009, 75, 6282–6291. [Google Scholar] [CrossRef] [PubMed]
- Sanjar, F.; Rusconi, B.; Hazen, T.H.; Koenig, S.S.K.; Hammel, M.K.; Feng, P.C.H.; Rasko, D.A.; Eppinger, M. Characterization of the pathogenome and phylogenomic classification of enteropathogenic Escherichia coli of the O157: Non-H7 serotypes. FEMS Pathog. Dis. 2015, 73. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, A.W.; Bielaszewska, M.; Zhang, W.; Pulz, M.; Kuczius, T.; Ammon, A.; Karch, H. Escherichia coli harboring Shiga toxin 2 gene variants: Frequency and association with clinical symptoms. J. Infect. Dis. 2002, 185, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Possé, B.; de Zutter, L.; Heyndrickx, M.; Herman, L. Quantitative isolation efficiency of O26, O103, O111, O145 and O157 STEC serotypes from artificially-contaminated food and cattle faeces samples using a new isolation protocol. J. Appl. Microbiol. 2008, 105, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Bielaszewska, M.; Karch, H. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 1999, 65, 3855–3861. [Google Scholar] [PubMed]
- Stromberg, Z.R.; Lewis, G.L.; Moxley, R.A. Comparison of agar media for detection and quantification of Shiga toxin-producing Escherichia coli from cattle feces. J. Food Prot. 2016, 79, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Creuzburg, K.; Kohler, B.; Hempel, H.; Schreier, P.; Jacobs, E.; Schmidt, H. Genetic structure and chromosomal integration site of the cryptic prophage CP-1639 encoding Shiga toxin 1. Microbiology 2005, 151, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Asadulghani, M.; Ogura, Y.; Ooka, T.; Itoh, T.; Sawaguchi, A.; Iguchi, A.; Nakayama, K.; Hatashi, T. The defective prophage pool of Escherichia coli O157: Prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 2009, 5, e10000408. [Google Scholar] [CrossRef] [PubMed]
- Watahiki, M.; Isobe, J.; Limata, K.; Shima, T.; Kanatani, J.; Shimizzu, M.; Nagata, A.; Kawakami, K.; Yamada, M.; Izumiya, H.; et al. Characterization of enterohemorrhagic Escherichia coli O111 and O157 strains isolated from outbreak patients in Japan. J. Clin. Microbiol. 2014, 52, 2757–2763. [Google Scholar] [CrossRef] [PubMed]
- Morton, V.; Cheng, J.M.; Sharma, D.; Kearney, A. Notes from the field: an outbreak of Shiga-toxin producing Escherichia coli infections associated with flour, Canada 2016–2017. MMWR Morb. Mortal. Wkly. Rep. 2017, 66, 705–706. [Google Scholar] [CrossRef] [PubMed]
| PCR-O Group | #Isolates Serotyped | Isolates PCR = TS O Group (%) | SEM 1 | Predominant Mismatches (#of Isolates) | #Mismatched O Groups by Serotyping | #H Groups by TS of Mismatched O Groups |
|---|---|---|---|---|---|---|
| O26 | 115 | 62.6 a | 4.5 | NA 2 | 34 | 22 |
| O45 | 116 | 69.8 a | 4.3 | O110:H31 (13) | 21 | 16 |
| O103 | 116 | 89.0 b | 2.9 | NA | 8 | 6 |
| O111 | 39 | 95.0 b | 3.5 | NA | 1 | 1 |
| O121 | 115 | 91.4 b | 8.6 | NA | 9 | 9 |
| O145 | 79 | 92.4 b | 7.6 | NA | 6 | 5 |
| O157 | 109 | 83.9 b | 3.5 | O71: H32 (3) | 13 | 9 |
| Total | 689 | 83.0 | − | NA | 92 | 68 |
| O Group | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| O26 | O45 | O103 | O111 | O121 | O145 | O157 | TOTAL | ||
| H group by traditional serotyping | NM 1 | 42 | 3 | 23 | 21 | 6 | 64 | 11 | 170 |
| 7 | 4 | 54 | 55 | 126 | |||||
| 4 | 34 | 34 | |||||||
| 19 | 6 | 2 | 20 | 28 | |||||
| 2 | 26 | 26 | |||||||
| 21 | 18 | 1 | 19 | ||||||
| 46 | 1 | 16 | 17 | ||||||
| 9 | 5 | 8 | 13 | ||||||
| 11 | 9 | 1 | 2 | 12 | |||||
| 12 | 11 | 11 | |||||||
| 10 | 7 | 3 | 10 | ||||||
| 38 | 1 | 9 | 10 | ||||||
| 29 | 8 | 8 | |||||||
| 14 | 2 | 4 | 1 | 7 | |||||
| unknown | 2 | 2 | 2 | 1 | 7 | ||||
| 8 | 1 | 5 | 13 | 6 | |||||
| 32 | 3 | 1 | 4 | ||||||
| 16 | 1 | 2 | 3 | ||||||
| 25 | 1 | 2 | 3 | ||||||
| 6 | 2 | 2 | |||||||
| 18 | 2 | 2 | |||||||
| 30 | 2 | 2 | |||||||
| 34 | 2 | 2 | |||||||
| 43 | 1 | 1 | |||||||
| 45 | 1 | 1 | |||||||
| 52 | 1 | 1 | |||||||
| TOTAL | 62 | 71 | 102 | 34 | 104 | 67 | 85 | 525 | |
| Serogroup | #Clusters 1 | #(%) NT 2 Isolates | #(%) PCR Mismatch TS Serogroup | #PCR and TS Matching Isolates | Isolates Where PCR and Traditional Serotyping Matched Serogroup #Isolates Within Serogroup-Specific Clusters #Unique Isolates | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O26:NM | O26:H9 | O26:H11 | O26:H32 | |||||||||
| O26 | 12 | 5(7.0) | 29 (40.8) | 42 | 11/23 | 1/3 | 2/6 | 1/3 | 17 (40.4) | |||
| O103:NM | O103:H2 | O103:H6 3 | O103:H7 | O103:H8 | O103:H14 | O103:H19 | ||||||
| O103 | 12 | 2(3.2) | 2 (3.2) | 58 | 4/14 | 8/11 | 1/1 | 0/3 | 4/4 | 2/4 | 0/1 | 22 (37.9) |
| O103:H21 | O103:H25 | O103:H38 | O103:H43 | |||||||||
| 6/10 | 0/1 | 2/6 | 0/1 | |||||||||
| O111:NM | O111:H8 | |||||||||||
| O111 | 4 | 3(17.6) | 0 (0) | 14 | 5/6 | 8/8 | 0 (0) | |||||
| O157:NM | O157:H7 | O157:H12 | O157:H29 | |||||||||
| O157 | 7 | 2(4.0) | 6(12.0) | 42 | 3/5 | 24/29 | 0/2 | 0/2 | 9 (21.4) | |||
| O Group | #Isolates Labs A and C (LAB B) | % STEC 1 LAB A | % STEC LAB B | % STEC LAB C | %stx1 + Labs A&B | % stx1 + Labs A&C | %stx2 + Labs A&B | % stx2 + Labs A&C | H Types STEC in Labs A& C (n) |
|---|---|---|---|---|---|---|---|---|---|
| O26 | 62 (52) | 71.5 a ± 3.2 | 30.0 a ± 3.2 | 8.3 a ± 3.0 | 25.5 a ± 2.9 | 8.3 b ± 3.3 | 3.6 a ± 1.3 | 0.0 a ± 0.0 | H11 (7/9) |
| O45 | 71 (0) | 80.6 ab ± 2.8 | NE 1 | 0.0 a ± 0.0 | NE | 0.0 a ± 0.0 | NE | 0.0 a ± 0.0 | None |
| O103 | 102 (31) | 69.8 a ± 3.0 | 41.9 a + 7.5 | 2.8 a ± 1.5 | 39.5 b ± 2.3 | 2.8 a ± 1.6 | 0.0 a ± 0.0 | 0.0 a ± 0.0 | H11 (2/2), H25 (1/1) |
| O111 | 34 (15) | 100.0 c ± 0.0 | 100.0 b + 0.0 | 89.5 c ± 4.5 | 100.0 c ± 0.0 | 89.5 d ± 4.5 | 0.0 a ± 0.0 | 0.0 a ± 0.0 | H8 (21/21), NM (9/13) |
| O121 | 104 (0) | 85.2 b ± 2.7 | NE | 33.0 b ± 4.2 | NE | 15.1 b ± 3.5 | NE | 16.0 b ± 3.6 | H7 (16/54), H10 (2/3) H19 (15/20), H32 (1/1) |
| O145 | 67 (0) | 80.8 ab ± 4.5 | NE | 49.3 b + 5.3 | NE | 49.3 c ± 5.9 | NE | 2.7 a ± 1.9 | NM (32/64) |
| O157:H12/H29 | 19 (19) | 93.2 c ± 2.2 | 44.2 a ± 7.6 | 0.0 a ± 0.0 | 42.9 b ± 7.7 | 0.0 a ± 0.0 | 21.4 b ± 5.9 | 0.0 a ± 0.0 | None |
| O157:H7/NM | 66 (54) | 98.7 c ± 1.4 | 98.8 b ± 1.2 | 91.4 c + 3.0 | 81.6 c ± 3.0 | 80.0 d ± 4.8 | 84.3 c ± 2.7 | 90.7 c ± 4.2 | H7 (54/55), NM (7/11) |
| #Isolates Sharing PCR Profile | PCR | Year of PCR | LAB | PCR stx1 | Detection 2 stx2 | Comments |
|---|---|---|---|---|---|---|
| 6 | First 1 | 2013 | A | + | + | LAB A PCRs are the same over years, LAB C negative for stx1 |
| Second | 2015 | C | - | + | ||
| Third | 2017 | A | + | + | ||
| 2 | First | 2013 | A | - | + | All three PCRs are the same never stx1 |
| Second | 2015 | C | - | + | ||
| Third | 2017 | A | - | + | ||
| 2 | First | 2013 | A | - | - | LAB A and LAB C initially the same, stx1 positive in second LAB A analysis |
| Second | 2015 | C | - | - | ||
| Third | 2017 | A | + | - | ||
| 2 | First | 2013 | A | + | + | Second LAB A analysis negative for stx1 and now agrees with LAB C |
| Second | 2016 | C | - | + | ||
| Third | 2017 | A | - | + |
| #Isolates Sharing PCR Profile | PCR | Year of PCR | LAB | PCR stx1 | Detection 2 stx2 | Comments |
|---|---|---|---|---|---|---|
| 4 | First 1 | 2013 | A | + | − | stx2 never detected by any laboratory |
| SecondThird | 2014 2014 | B C | + + | − − | ||
| Fourth | 2017 | A | + | − | ||
| 2 | First Second | 2013 2014 | A B | + + | + − | stx2 present in initial isolation PCR only |
| Third | 2014 | C | + | − | ||
| Fourth | 2017 | A | + | − |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stanford, K.; Reuter, T.; Hallewell, J.; Tostes, R.; Alexander, T.W.; McAllister, T.A. Variability in Characterizing Escherichia coli from Cattle Feces: A Cautionary Tale. Microorganisms 2018, 6, 74. https://doi.org/10.3390/microorganisms6030074
Stanford K, Reuter T, Hallewell J, Tostes R, Alexander TW, McAllister TA. Variability in Characterizing Escherichia coli from Cattle Feces: A Cautionary Tale. Microorganisms. 2018; 6(3):74. https://doi.org/10.3390/microorganisms6030074
Chicago/Turabian StyleStanford, Kim, Tim Reuter, Jennyka Hallewell, Renata Tostes, Trevor W. Alexander, and Tim A. McAllister. 2018. "Variability in Characterizing Escherichia coli from Cattle Feces: A Cautionary Tale" Microorganisms 6, no. 3: 74. https://doi.org/10.3390/microorganisms6030074
APA StyleStanford, K., Reuter, T., Hallewell, J., Tostes, R., Alexander, T. W., & McAllister, T. A. (2018). Variability in Characterizing Escherichia coli from Cattle Feces: A Cautionary Tale. Microorganisms, 6(3), 74. https://doi.org/10.3390/microorganisms6030074







