A Novel Quinolone JH62 (E-2-(Tridec-4-en-1-yl)-quinolin-4(1H)-one) from Pseudomonas aeruginosa Exhibits Potent Anticancer Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Pa Strains
2.3. Mice
2.4. Reagents
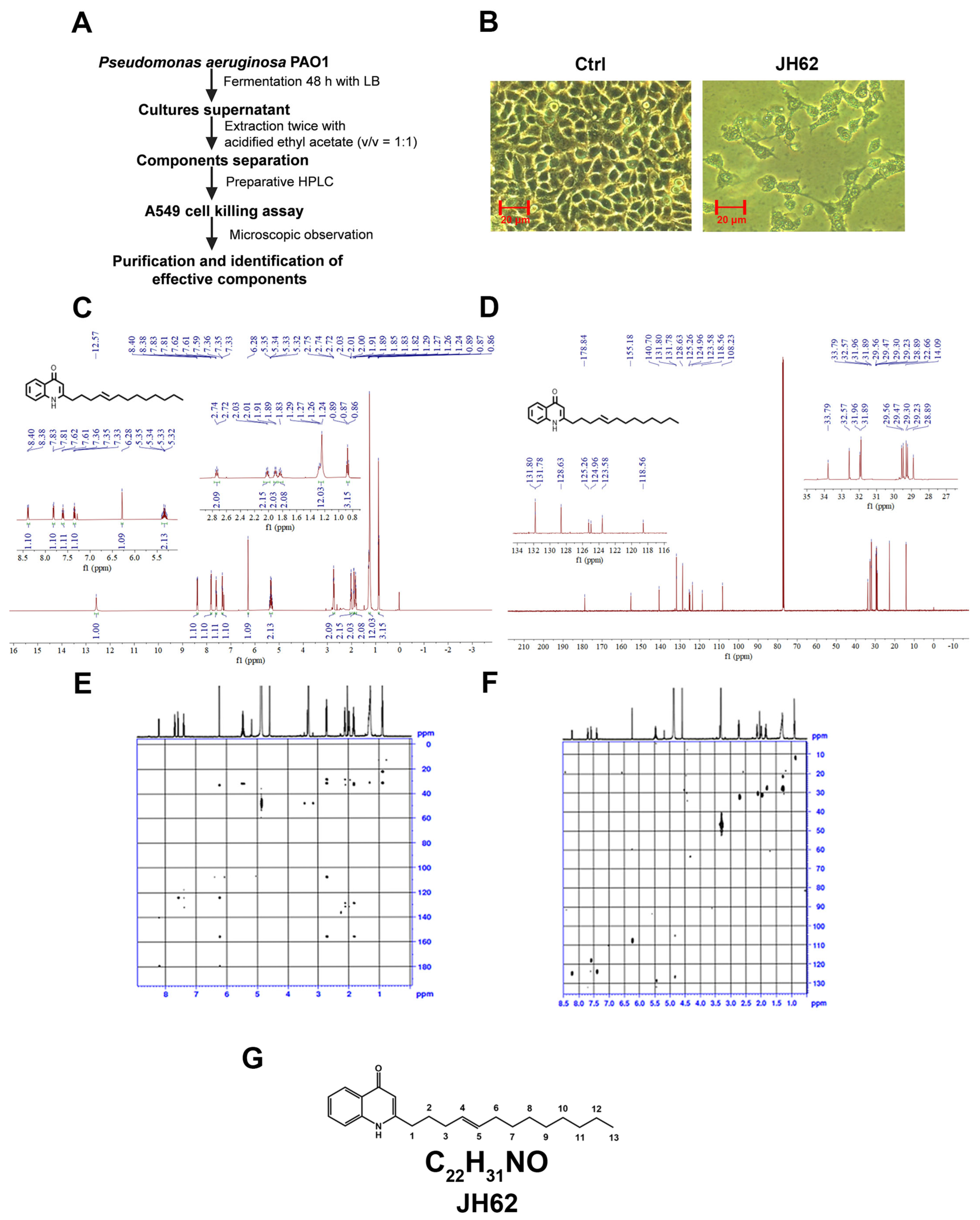
2.5. Preparation of Bacterial Supernatants and Purification of Inhibitory Compound
2.6. Chemical Structural Analysis
2.7. Cell Viability
2.8. Drug Combination Study
2.9. Tumorigenicity Assay
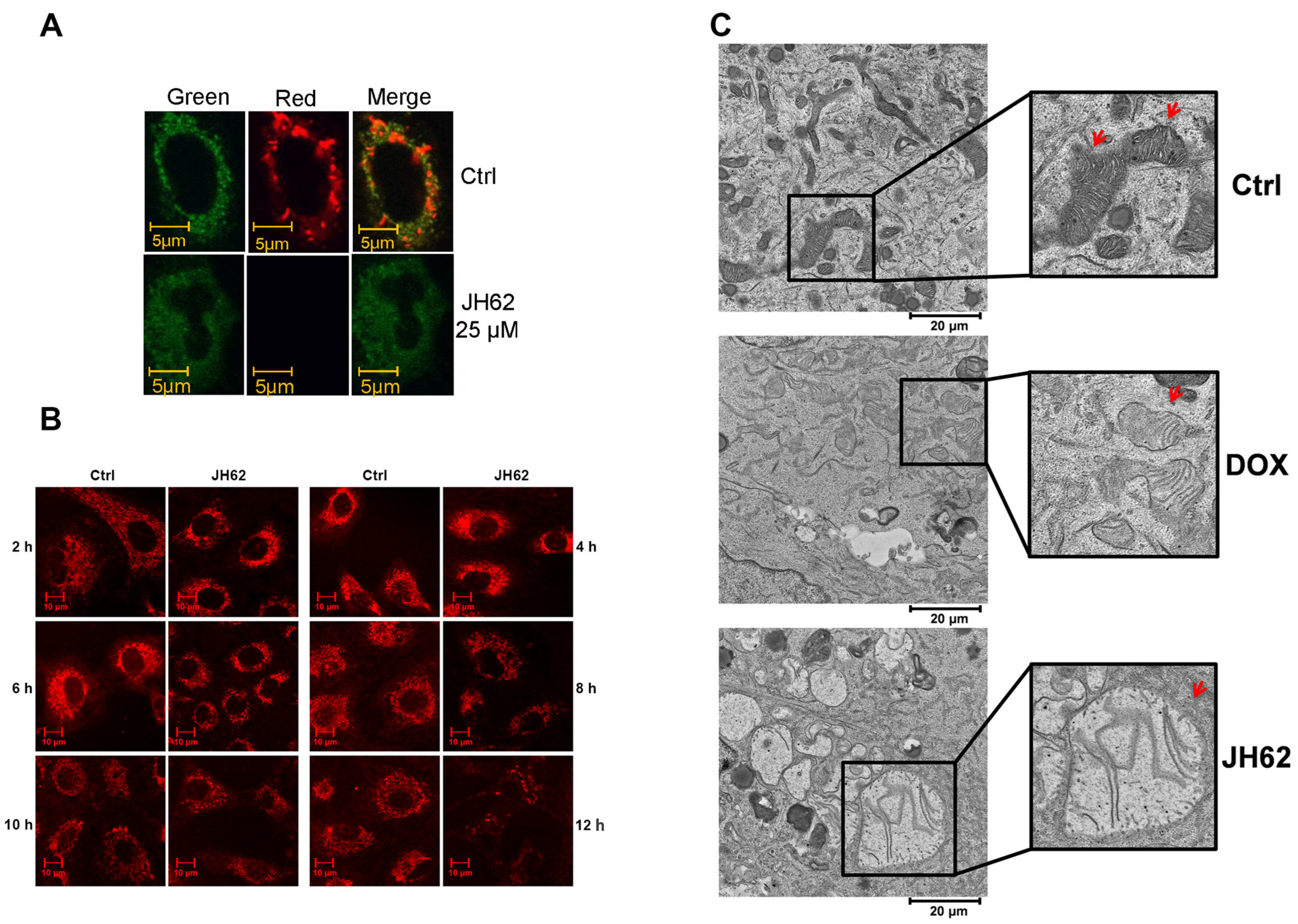
2.10. Mitochondria JC-1 and MitoTracker Staining
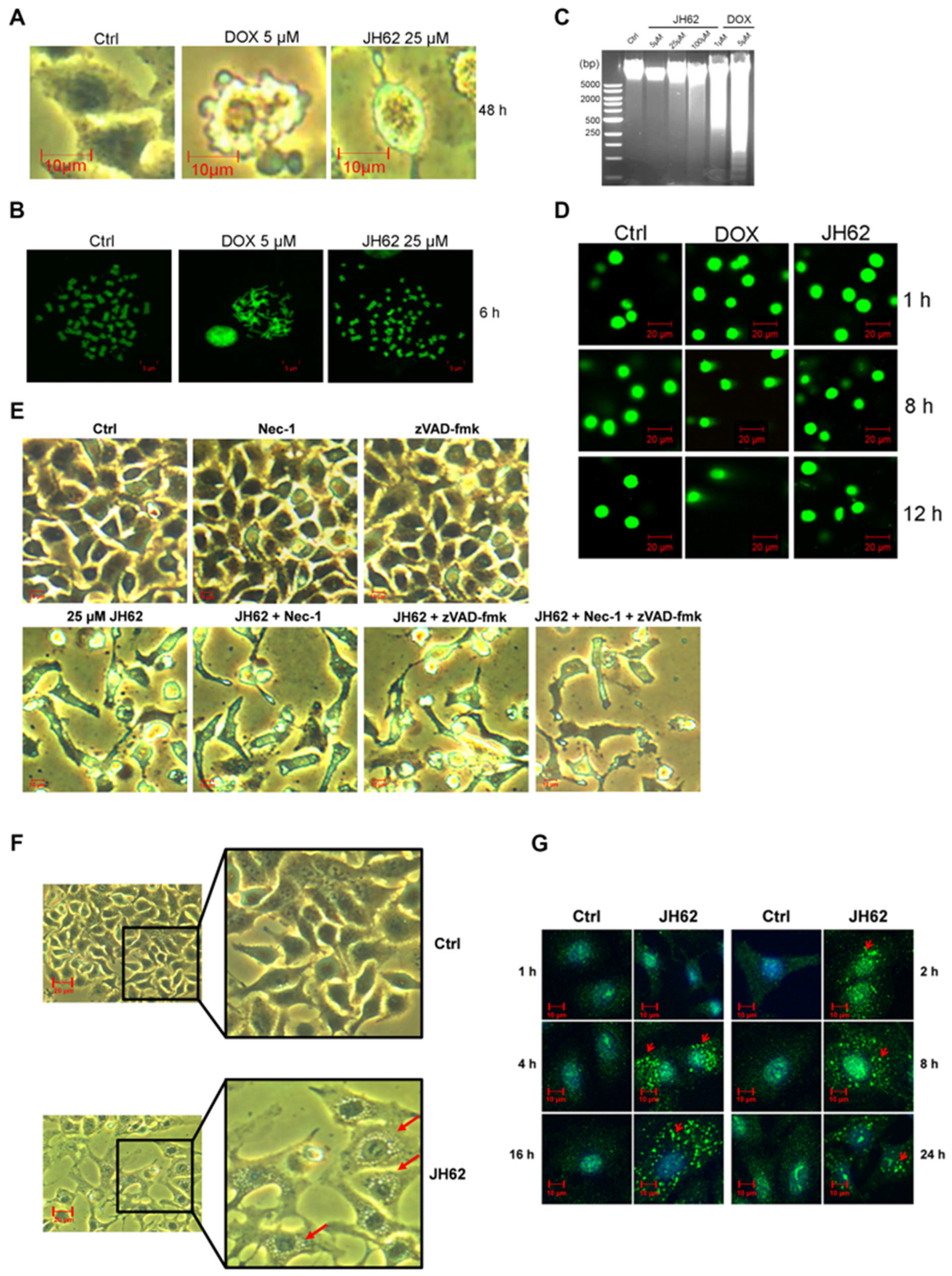
2.11. Chromosome Staining Experiment
2.12. DNA Fragmentation Analysis
2.13. Comet Assay
2.14. JH62 Extraction and Quantitative Analysis
2.15. Construction of Pa Deletion Mutant and Complemented Strains
2.16. ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) Analysis
2.17. Data Analysis
2.18. Ethics Statement
3. Results
3.1. A Novel Quinolone Compound JH62 Isolated from Pa Inhibits Cancer Cell Viability
3.2. JH62 Induces A549 Cell Death and Inhibits Tumor Growth
3.3. JH62 Induces Mitochondrial Dysfunction and Structural Disruption in A549 Lung Cancer Cells
3.4. JH62 Induces Autophagic Cell Death Distinct from DOX-Mediated Apoptosis
3.5. The pqs Gene Cluster Mediates JH62 Biosynthesis in Pa
3.6. ADMET Evaluation Identifies JH62 as a Promising Lead Compound for Drug Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AQNO | 2-alkyl-4-hydroxyquinoline N-oxide |
| DHQ | 2, 4-dihydroxyquinoline |
| DMSO | Dimethyl sulfoxide |
| DOX | Doxorubicin |
| HHQ | 2-heptyl-4-hydroxyquinoline |
| HNQ | 2-nonyl-4-hydroxyquinoline |
| HPLC | High Performance Liquid Chromatography |
| HQNO | 2-heptyl-4-hydroxyquinoline N-oxide |
| IC50 | Half maximal inhibitory concentration |
| LC-MS | Liquid chromatography-mass spectrometry |
| NMR | Nuclear magnetic resonance |
| NQNO | 2-nonyl-4-hydroxyquinoline N-oxide |
| Pa | Pseudomonas aeruginosa |
| PPB | Plasma protein binding |
| PQS | 2-heptyl-3-hydroxy-4(1H)-quinolone |
| UQNO | 2-undecyl-4-hydroxyquinoline N-oxide |
| WT | Wild type |
References
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef]
- Elbehiry, A.; Marzouk, E.; Aldubaib, M.; Abalkhail, A.; Anagreyyah, S.; Anajirih, N.; Almuzaini, A.M.; Rawway, M.; Alfadhel, A.; Draz, A.; et al. Helicobacter pylori infection: Current status and future prospects on diagnostic, therapeutic and control challenges. Antibiotics 2023, 12, 191. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Li, S.; Gan, R.-Y.; Zhou, T.; Xu, D.-P.; Li, H.-B. Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef]
- Coley, W.B. The treatment of malignant tumors by repeated inoculations of erysipelas. With a report of ten original cases. 1893. Clin. Orthop. Relat. Res. 1991, 98, 563–577. [Google Scholar]
- Niu, H.; Zhou, M.; Zogona, D.; Xing, Z.; Wu, T.; Chen, R.; Cui, D.; Liang, F.; Xu, X. Akkermansia muciniphila: A potential candidate for ameliorating metabolic diseases. Front. Immunol. 2024, 15, 1370658. [Google Scholar] [CrossRef]
- Di Gioia, D.; Aloisio, I.; Mazzola, G.; Biavati, B. Bifidobacteria: Their impact on gut microbiota composition and their applications as probiotics in infants. Appl. Microbiol. Biotechnol. 2014, 98, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Hazan, S. Rapid improvement in Alzheimer’s disease symptoms following fecal microbiota transplantation: A case report. J. Int. Med. Res. 2020, 48, 0300060520925930. [Google Scholar] [CrossRef] [PubMed]
- Deo, S.V.S.; Sharma, J.; Kumar, S. GLOBOCAN 2020 report on global cancer burden: Challenges and opportunities for surgical oncologists. Ann. Surg. Oncol. 2022, 29, 6497–6500. [Google Scholar] [CrossRef]
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer treatment and survivorship statistics, 2022. CA A Cancer J. Clin. 2022, 72, 409–436. [Google Scholar] [CrossRef]
- Hashem, S.; Ali, T.A.; Akhtar, S.; Nisar, S.; Sageena, G.; Ali, S.; Al-Mannai, S.; Therachiyil, L.; Mir, R.; Elfaki, I.; et al. Targeting cancer signaling pathways by natural products: Exploring promising anti-cancer agents. Biomed. Pharmacother. 2022, 150, 113054. [Google Scholar] [CrossRef]
- Ji, G.; Zhao, J.; Si, X.; Song, W. Targeting bacterial metabolites in tumor for cancer therapy: An alternative approach for targeting tumor-associated bacteria. Adv. Drug Deliv. Rev. 2024, 211, 115345. [Google Scholar] [CrossRef]
- Cort, A.; Timur, M.; Ozdemir, E.; Ozben, T. Effects of curcumin on bleomycin-induced apoptosis in human malignant testicular germ cells. J. Physiol. Biochem. 2013, 69, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Martinez, E.D.; MacMillan, J.B. Anthraquinones from a marine-derived Streptomyces spinoverrucosus. J. Nat. Prod. 2012, 75, 1759–1764. [Google Scholar] [CrossRef] [PubMed]
- Tuon, F.F.; Dantas, L.R.; Suss, P.H.; Tasca Ribeiro, V.S. Pathogenesis of the Pseudomonas aeruginosa biofilm: A review. Pathogens 2022, 11, 300. [Google Scholar] [CrossRef]
- Abraham, E.H.; Vos, P.; Kahn, J.; Grubman, S.A.; Jefferson, D.M.; Ding, I.; Okunieff, P. Cystic fibrosis hetero- and homozygosity is associated with inhibition of breast cancer growth. Nat. Med. 1996, 2, 593–596. [Google Scholar] [CrossRef]
- Maisonneuve, P.; Marshall, B.C.; Knapp, E.A.; Lowenfels, A.B. Cancer risk in cystic fibrosis: A 20-year nationwide study from the United States. JNCI J. Natl. Cancer Inst. 2013, 105, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Goto, M.; Punj, V.; Zaborina, O.; Kimbara, K.; Das Gupta, T.K.; Chakrabarty, A.M. The bacterial redox protein azurin induces apoptosis in J774 macrophages through complex formation and stabilization of the tumor suppressor protein p53. Infect. Immun. 2002, 70, 7054–7062. [Google Scholar] [CrossRef]
- Qi, J.-L.; He, J.-R.; Jin, S.-M.; Yang, X.; Bai, H.-M.; Liu, C.-B.; Ma, Y.-B. P. aeruginosa mediated necroptosis in mouse tumor cells induces long-lasting systemic antitumor immunity. Front. Oncol. 2021, 10, 610651. [Google Scholar] [CrossRef]
- Lv, F.; Cao, J.; Liu, Z.; Wang, Z.; Zhang, J.; Zhang, S.; Wang, L.; Zhao, X.; Shao, Z.; Wang, B.; et al. Phase II study of Pseudomonas aeruginosa-mannose-sensitive hemagglutinin in combination with capecitabine for Her-2-negative metastatic breast cancer pretreated with anthracycline and taxane. PLoS ONE 2015, 10, e0118607. [Google Scholar] [CrossRef]
- Wolf, P.; Elsaesser-Beile, U. Pseudomonas exotoxin A: From virulence factor to anti-cancer agent. Int. J. Med. Microbiol. 2009, 299, 161–176. [Google Scholar] [CrossRef]
- Huang, F.; Shu, Q.; Qin, Z.; Tian, J.; Su, Z.; Huang, Y.; Gao, M. Anticancer actions of azurin and its derived peptide p28. Protein J. 2020, 39, 182–189. [Google Scholar] [CrossRef]
- Ghssein, G.; Ezzeddine, Z. A review of Pseudomonas aeruginosa metallophores: Pyoverdine, pyochelin and pseudopaline. Biology 2022, 11, 1711. [Google Scholar] [CrossRef]
- Si, T.; Wang, A.; Yan, H.; Kong, L.; Guan, L.; He, C.; Ma, Y.; Zhang, H.; Ma, H. Progress in the study of natural antimicrobial active substances in Pseudomonas aeruginosa. Molecules 2024, 29, 4400. [Google Scholar] [CrossRef]
- Wells, I.C.; Elliott, W.H.; Thayer, S.A.; Doisy, E.A. Ozonization of some antibiotic substances produced by Pseudomonas aeruginosa. J. Biol. Chem. 1952, 196, 321–330. [Google Scholar] [CrossRef]
- Pesci, E.C.; Milbank, J.B.; Pearson, J.P.; McKnight, S.; Kende, A.S.; Greenberg, E.P.; Iglewski, B.H. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1999, 96, 11229–11234. [Google Scholar] [CrossRef] [PubMed]
- Bredenbruch, F.; Nimtz, M.; Wray, V.; Morr, M.; Müller, R.; Häussler, S. Biosynthetic pathway of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines. J. Bacteriol. 2005, 187, 3630–3635. [Google Scholar] [CrossRef] [PubMed]
- Lépine, F.; Déziel, E. Liquid chromatography/mass spectrometry for the detection and quantification of N-acyl-L-homoserine lactones and 4-hydroxy-2-alkylquinolines. Methods Mol. Biol. 2011, 692, 61–69. [Google Scholar] [CrossRef]
- Lin, J.; Cheng, J.; Wang, Y.; Shen, X. The Pseudomonas quinolone signal (PQS): Not just for quorum sensing anymore. Front. Cell. Infect. Microbiol. 2018, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Legendre, C.; Reen, F.J.; Mooij, M.J.; McGlacken, G.P.; Adams, C.; O’Gara, F. Pseudomonas aeruginosa Alkyl quinolones repress hypoxia-inducible factor 1 (HIF-1) signaling through HIF-1α degradation. Infect. Immun. 2012, 80, 3985–3992. [Google Scholar] [CrossRef]
- Jayaseelan, S.; Ramaswamy, D.; Dharmaraj, S. Pyocyanin: Production, applications, challenges and new insights. World J. Microbiol. Biotechnol. 2014, 30, 1159–1168. [Google Scholar] [CrossRef]
- Vilaplana, L.; Marco, M.P. Phenazines as potential biomarkers of Pseudomonas aeruginosa infections: Synthesis regulation, pathogenesis and analytical methods for their detection. Anal. Bioanal. Chem. 2020, 412, 5897–5912. [Google Scholar] [CrossRef]
- Chong, H.; Li, Q. Microbial production of rhamnolipids: Opportunities, challenges and strategies. Microb. Cell Fact. 2017, 16, 137. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Revtovich, A.V.; Chen, Q.; Shah, K.N.; Cannon, C.L.; Kirienko, N.V. Pyoverdine-dependent virulence of Pseudomonas aeruginosa isolates from cystic fibrosis patients. Front. Microbiol. 2019, 10, 2048. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.R.; Musaev, D.G.; Wuest, W.M. Pyochelin biosynthetic metabolites bind iron and promote growth in Pseudomonads demonstrating siderophore-like activity. ACS Infect. Dis. 2021, 7, 544–551. [Google Scholar] [CrossRef]
- Smith, D.; Spaněl, P.; Gilchrist, F.J.; Lenney, W. Hydrogen cyanide, a volatile biomarker of Pseudomonas aeruginosa infection. J. Breath Res. 2013, 7, 044001. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef]
- Diggle, S.P.; Cornelis, P.; Williams, P.; Cámara, M. 4-quinolone signalling in Pseudomonas aeruginosa: Old molecules, new perspectives. Int. J. Med. Microbiol. IJMM 2006, 296, 83–91. [Google Scholar] [CrossRef]
- Leid, J.G.; Willson, C.J.; Shirtliff, M.E.; Hassett, D.J.; Parsek, M.R.; Jeffers, A.K. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 2005, 175, 7512–7518. [Google Scholar] [CrossRef]
- Marmont, L.S.; Rich, J.D.; Whitney, J.C.; Whitfield, G.B.; Almblad, H.; Robinson, H.; Parsek, M.R.; Harrison, J.J.; Howell, P.L. Oligomeric lipoprotein PelC guides Pel polysaccharide export across the outer membrane of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2017, 114, 2892–2897. [Google Scholar] [CrossRef]
- Ma, L.; Jackson, K.D.; Landry, R.M.; Parsek, M.R.; Wozniak, D.J. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 2006, 188, 8213–8221. [Google Scholar] [CrossRef]
- Dolan, S.K.; Kohlstedt, M.; Trigg, S.; Vallejo Ramirez, P.; Kaminski, C.F.; Wittmann, C.; Welch, M. Contextual flexibility in Pseudomonas aeruginosa central carbon metabolism during growth in single carbon sources. mBio 2020, 11, e02684-19. [Google Scholar] [CrossRef]
- Sauvage, S.; Gaviard, C.; Tahrioui, A.; Coquet, L.; Le, H.; Alexandre, S.; Ben Abdelkrim, A.; Bouffartigues, E.; Lesouhaitier, O.; Chevalier, S.; et al. Impact of carbon source supplementations on Pseudomonas aeruginosa physiology. J. Proteome Res. 2022, 21, 1392–1407. [Google Scholar] [CrossRef]
- Lundgren, B.R.; Shoytush, J.M.; Scheel, R.A.; Sain, S.; Sarwar, Z.; Nomura, C.T. Utilization of L-glutamate as a preferred or sole nutrient in Pseudomonas aeruginosa PAO1 depends on genes encoding for the enhancer-binding protein AauR, the sigma factor RpoN and the transporter complex AatJQMP. BMC Microbiol. 2021, 21, 83. [Google Scholar] [CrossRef]
- Mahmud, S.A.; Qureshi, M.A.; Sapkota, M.; Pellegrino, M.W. A pathogen branched-chain amino acid catabolic pathway subverts host survival by impairing energy metabolism and the mitochondrial UPR. PLoS Pathog. 2020, 16, e1008918. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.D.; Itoh, Y.; Nakada, Y.; Jiang, Y. Functional analysis and regulation of the divergent spuABCDEFGH-spuI operons for polyamine uptake and utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2002, 184, 3765–3773. [Google Scholar] [CrossRef] [PubMed]
- Holden, M.T.; Ram Chhabra, S.; de Nys, R.; Stead, P.; Bainton, N.J.; Hill, P.J.; Manefield, M.; Kumar, N.; Labatte, M.; England, D.; et al. Quorum-sensing cross talk: Isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol. Microbiol. 1999, 33, 1254–1266. [Google Scholar] [CrossRef] [PubMed]
- Imberty, A.; Wimmerová, M.; Mitchell, E.P.; Gilboa-Garber, N. Structures of the lectins from Pseudomonas aeruginosa: Insight into the molecular basis for host glycan recognition. Microbes Infect. 2004, 6, 221–228. [Google Scholar] [CrossRef]
- Kuo, S.H.; Lau, G.W. Pseudomonas aeruginosa-derived metabolites and volatile organic compounds: Impact on lung epithelial homeostasis and mucosal immune response. Front. Immunol. 2025, 16, 1553013. [Google Scholar] [CrossRef]
- Kuznetsova, M.V.; Karpunina, T.I. Cell-mediated hemolytic activity of nosocomial Pseudomonas Aeruginosa strains. Bull. Exp. Biol. Med. 2015, 159, 258–261. [Google Scholar] [CrossRef]
- Alqahtani, A.; Kopel, J.; Hamood, A. The in vivo and in vitro assessment of pyocins in treating Pseudomonas aeruginosa infections. Antibiotics 2022, 11, 1366. [Google Scholar] [CrossRef]
- Nguyen, T.H.N.; Szamosvari, D.; Böttcher, T. Synthesis of bacterial 2-alkyl-4(1H)-quinolone derivatives. Arkivoc 2021, 2021, 218–239. [Google Scholar] [CrossRef]
- Illakkiam, D.; Ponraj, P.; Shankar, M.; Muthusubramanian, S.; Rajendhran, J.; Gunasekaran, P. Identification and structure elucidation of a novel antifungal compound produced by Pseudomonas aeruginosa PGPR2 against Macrophomina phaseolina. Appl. Biochem. Biotechnol. 2013, 171, 2176–2185. [Google Scholar] [CrossRef] [PubMed]
- El-Damasy, A.K.; Kim, H.J.; Nocentini, A.; Seo, S.H.; Eldehna, W.M.; Bang, E.K.; Supuran, C.T.; Keum, G. Discovery of new 6-ureido/amidocoumarins as highly potent and selective inhibitors for the tumour-relevant carbonic anhydrases IX and XII. J. Enzym. Inhib. Med. Chem. 2023, 38, 2154603. [Google Scholar] [CrossRef] [PubMed]
- Chougoni, K.K.; Neely, V.; Ding, B.; Oduah, E.; Lam, V.T.; Hu, B.; Koblinski, J.E.; Windle, B.E.; Palit Deb, S.; Deb, S.; et al. Oncogenic mutant p53 sensitizes non-small cell lung cancer cells to proteasome inhibition via oxidative stress-dependent induction of mitochondrial apoptosis. Cancer Res. Commun. 2024, 4, 2685–2698. [Google Scholar] [CrossRef]
- García-Crespo, C.; de Ávila, A.I.; Gallego, I.; Soria, M.E.; Durán-Pastor, A.; Somovilla, P.; Martínez-González, B.; Muñoz-Flores, J.; Mínguez, P.; Salar-Vidal, L.; et al. Synergism between remdesivir and ribavirin leads to SARS-CoV-2 extinction in cell culture. Br. J. Pharmacol. 2024, 181, 2636–2654. [Google Scholar] [CrossRef]
- Liu, X.; Yang, J.; Yang, C.; Huang, X.; Han, M.; Kang, F.; Li, J. Morphine promotes the malignant biological behavior of non-small cell lung cancer cells through the MOR/Src/mTOR pathway. Cancer Cell Int. 2021, 21, 622. [Google Scholar] [CrossRef] [PubMed]
- Keil, V.C.; Funke, F.; Zeug, A.; Schild, D.; Müller, M. Ratiometric high-resolution imaging of JC-1 fluorescence reveals the subcellular heterogeneity of astrocytic mitochondria. Pflug. Arch. Eur. J. Physiol. 2011, 462, 693–708. [Google Scholar] [CrossRef]
- Martin, R.; Busch, W.; Herrmann, R.G.; Wanner, G. Efficient preparation of plant chromosomes for high-resolution scanning electron microscopy. Chromosome Res. 1994, 2, 411–415. [Google Scholar] [CrossRef]
- Abdul Rahman, M.S.; Kanakarajan, S.; Selvaraj, R.; Kamalanathan, A.; Fatima, S.; Abudawood, M.; Siddiqi, N.J.; Alanazi, H.; Sharma, B.; de Lourdes Pereira, M. Elucidation of the anticancer mechanism of durian fruit (Durio zibethinus) pulp extract in human leukemia (HL-60) cancer cells. Nutrients 2023, 15, 2417. [Google Scholar] [CrossRef]
- Agúndez, J.A.G.; García-Martín, E.; García-Lainez, G.; Miranda, M.A.; Andreu, I. Photomutagenicity of chlorpromazine and its N-demethylated metabolites assessed by NGS. Sci. Rep. 2020, 10, 6879. [Google Scholar] [CrossRef]
- Dela Ahator, S.; Sagar, S.; Zhu, M.; Wang, J.; Zhang, L.-H. Nutrient availability and phage exposure alter the Quorum-Sensing and CRISPR-cas-controlled population dynamics of Pseudomonas aeruginosa. Msystems 2022, 7, e0009222. [Google Scholar] [CrossRef]
- Huang, J.; Xu, Z.; Zhou, T.; Zhang, L.H.; Xu, Z. Suppression of Pseudomonas aeruginosa type III secretion system by a novel calcium-responsive signaling pathway. iScience 2024, 27, 109690. [Google Scholar] [CrossRef] [PubMed]
- Afrdi, M.B.; Sardar, H.; Serdaroğlu, G.; Wadood Ali Shah, S.; Alsharif, K.F.; Khan, H. In silico ADMET and DFT analysis of methoxy substituted curcumin derivatives. Inorg. Chem. Commun. 2024, 168, 112943. [Google Scholar] [CrossRef]
- Valente, E.; Assis, M.C.; Alvim, I.M.; Pereira, G.M.; Plotkowski, M.C. Pseudomonas aeruginosa induces apoptosis in human endothelial cells. Microb. Pathog. 2000, 29, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.-h.; Shi, Z.-f.; Xu, H. The mitochondrial cyclophilin D/p53 complexation mediates doxorubicin-induced non-apoptotic death of A549 lung cancer cells. Mol. Cell. Biochem. 2014, 389, 17–24. [Google Scholar] [CrossRef]
- Lei, K.F.; Wu, Z.M.; Huang, C.H. Impedimetric quantification of the formation process and the chemosensitivity of cancer cell colonies suspended in 3D environment. Biosens. Bioelectron. 2015, 74, 878–885. [Google Scholar] [CrossRef]
- Moreno-Vargas, L.M.; Prada-Gracia, D. Cancer-targeting applications of cell-penetrating peptides. Int. J. Mol. Sci. 2024, 26, 2. [Google Scholar] [CrossRef]
- Taghizadeh-Hesary, F.; Akbari, H.; Bahadori, M.; Behnam, B. Targeted anti-mitochondrial therapy: The future of oncology. Genes 2022, 13, 1728. [Google Scholar] [CrossRef]
- Zorova, L.D.; Popkov, V.A.; Plotnikov, E.Y.; Silachev, D.N.; Pevzner, I.B.; Jankauskas, S.S.; Babenko, V.A.; Zorov, S.D.; Balakireva, A.V.; Juhaszova, M.; et al. Mitochondrial membrane potential. Anal. Biochem. 2018, 552, 50–59. [Google Scholar] [CrossRef]
- Sivandzade, F.; Bhalerao, A.; Cucullo, L. Analysis of the mitochondrial membrane potential using the cationic JC-1 dye as a sensitive fluorescent probe. Bio-Protocol 2019, 9, e3128. [Google Scholar] [CrossRef]
- Kciuk, M.; Gielecinska, A.; Mujwar, S.; Kolat, D.; Kaluzinska-Kolat, Z.; Celik, I.; Kontek, R. Doxorubicin-An agent with multiple mechanisms of anticancer activity. Cells 2023, 12, 659. [Google Scholar] [CrossRef]
- Szwed, M.; Kania, K.D.; Jozwiak, Z. Assessment of pro-apoptotic activity of doxorubicin-transferrin conjugate in cells derived from human solid tumors. Int. J. Biochem. Cell Biol. 2016, 70, 57–67. [Google Scholar] [CrossRef]
- Hassan, F.; Islam, S.; Mu, M.M.; Ito, H.; Koide, N.; Mori, I.; Yoshida, T.; Yokochi, T. Lipopolysaccharide prevents doxorubicin-induced apoptosis in RAW 264.7 macrophage cells by inhibiting p53 activation. Mol. Cancer Res. 2005, 3, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zheng, Z.; Cao, X.; Yang, Q.; Norton, V.; Adini, A.; Maiti, A.K.; Adini, I.; Wu, H. RIP1/RIP3/MLKL mediates myocardial function through necroptosis in experimental autoimmune myocarditis. Front. Cardiovasc. Med. 2021, 8, 696362. [Google Scholar] [CrossRef] [PubMed]
- Liebl, M.P.; Meister, S.C.; Frey, L.; Hendrich, K.; Klemmer, A.; Wohlfart, B.; Untucht, C.; Nuber, J.; Pohl, C.; Lakics, V. Robust LC3B lipidation analysis by precisely adjusting autophagic flux. Sci. Rep. 2022, 12, 79. [Google Scholar] [CrossRef]
- Witzgall, F.; Depke, T.; Hoffmann, M.; Empting, M.; Broenstrup, M.; Mueller, R.; Blankenfeldt, W. The alkylquinolone repertoire of Pseudomonas aeruginosa is linked to structural flexibility of the FabH-like 2-Heptyl-3-hydroxy-4(1H)-quinolone (PQS) biosynthesis enzyme PqsBC. ChemBiochem A Eur. J. Chem. Biol. 2018, 19, 1531–1544. [Google Scholar] [CrossRef] [PubMed]
- McGrath, S.; Wade, D.S.; Pesci, E.C. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol. Lett. 2004, 230, 27–34. [Google Scholar] [CrossRef]
- Linani, A.; Benarous, K.; Bou-Salah, L.; Yousfi, M. Hispidin, Harmaline, and Harmine as potent inhibitors of bovine xanthine oxidase: Gout treatment, in vitro, ADMET prediction, and SAR studies. Bioorg. Chem. 2021, 112, 104937. [Google Scholar] [CrossRef]
- Reen, F.J.; McGlacken, G.P.; O’Gara, F. The expanding horizon of alkyl quinolone signalling and communication in polycellular interactomes. FEMS Microbiol. Lett. 2018, 365, fny076. [Google Scholar] [CrossRef]
- Liang, L.; Sproule, A.; Haltli, B.; Marchbank, D.H.; Berrué, F.; Overy, D.P.; McQuillan, K.; Lanteigne, M.; Duncan, N.; Correa, H.; et al. Discovery of a new natural product and a deactivation of a quorum sensing system by culturing a “Producer” bacterium with a heat-killed “Inducer” culture. Front. Microbiol. 2018, 9, 3351. [Google Scholar] [CrossRef]
- Depke, T.; Franke, R.; Brönstrup, M. Clustering of MS(2) spectra using unsupervised methods to aid the identification of secondary metabolites from Pseudomonas aeruginosa. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1071, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.Y.; Hoke, T.; Seravalli, J.; Switzer, B.L.; Bavitz, M.; Fliege, J.D.; Murphy, P.J.; Britigan, B.E. Pseudomonas quinolone signal induces oxidative stress and inhibits heme oxygenase-1 expression in lung epithelial Cells. Infect. Immun. 2017, 85, e00176-17. [Google Scholar] [CrossRef] [PubMed]
- Rieger, B.; Thierbach, S.; Ommer, M.; Dienhart, F.S.V.; Fetzner, S.; Busch, K.B. Pseudomonas quinolone signal molecule PQS behaves like a B Class inhibitor at the I(Q) site of mitochondrial complex I. FASEB Bioadv. 2020, 2, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Supong, K.; Thawai, C.; Supothina, S.; Auncharoen, P.; Pittayakhajonwut, P. Antimicrobial and anti-oxidant activities of quinoline alkaloids from Pseudomonas aeruginosa BCC76810. Phytochem. Lett. 2016, 17, 100–106. [Google Scholar] [CrossRef]
- Balthazar, J.D.; Soosaimanickam, M.P.; Emmanuel, C.; Krishnaraj, T.; Sheikh, A.; Alghafis, S.F.; Ibrahim, H.M. 8-Hydroxyquinoline a natural chelating agent from Streptomyces spp. inhibits A549 lung cancer cell lines via BCL2/STAT3 regulating pathways. World J. Microbiol. Biotechnol. 2022, 38, 182. [Google Scholar] [CrossRef]
- Mori, T.; Yamashita, T.; Furihata, K.; Nagai, K.; Suzuki, K.; Hayakawa, Y.; Shin-Ya, K. Burkholone, a new cytotoxic antibiotic against IGF-I dependent cells from Burkholderia sp. J. Antibiot. 2007, 60, 713–716. [Google Scholar] [CrossRef]
- Kamigiri, K.; Tokunaga, T.; Shibazaki, M.; Setiawan, B.; Rantiatmodjo, R.M.; Morioka, M.; Suzuki, K. YM-30059, a novel quinolone antibiotic produced by Arthrobacter sp. J. Antibiot. 1996, 49, 823–825. [Google Scholar] [CrossRef]
- Bultel-Poncé, V.V.; Berge, J.P.; Debitus, C.; Nicolas, J.L.; Guyot, M. Metabolites from the sponge-associated bacterium Pseudomonas species. Mar. Biotechnol. 1999, 1, 384–390. [Google Scholar] [CrossRef]
- Ding, W.Q.; Liu, B.; Vaught, J.L.; Yamauchi, H.; Lind, S.E. Anticancer activity of the antibiotic clioquinol. Cancer Res. 2005, 65, 3389–3395. [Google Scholar] [CrossRef]
- Gao, F.; Zhang, X.; Wang, T.; Xiao, J. Quinolone hybrids and their anti-cancer activities: An overview. Eur. J. Med. Chem. 2019, 165, 59–79. [Google Scholar] [CrossRef]
- Roy, S.; Bonfield, T.; Tartakoff, A.M. Non-apoptotic toxicity of Pseudomonas aeruginosa toward murine cells. PLoS ONE 2013, 8, e54245. [Google Scholar] [CrossRef] [PubMed]
- Arango, J.P.B.; Rodriguez, D.Y.M.; Cruz, S.L.; Ocampo, G.T. In silico evaluation of pharmacokinetic properties and molecular docking for the identification of potential anticancer compounds. Comput. Biol. Chem. 2025, 120, 108626. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.A.; Di, L.; Kerns, E.H. The effect of plasma protein binding on in vivo efficacy: Misconceptions in drug discovery. Nat. Rev. Drug Discov. 2010, 9, 929–939. [Google Scholar] [CrossRef] [PubMed]


| Metabolite | Function | References |
|---|---|---|
| Pyocyanin | Oxidative stress; Host damage | [30] |
| Phenazines | Redox balance; Persistence | [31] |
| Rhamnolipids | Biofilm formation; Membrane disruption | [32] |
| Pyoverdine | Iron acquisition; Virulence regulation | [33] |
| Pyochelin | Iron acquisition | [34] |
| Hydrogen cyanide | Acute virulence; Respiratory inhibition | [35] |
| Acyl-homoserine lactone | Quorum sensing | [36] |
| Quinolones | Quorum sensing; Iron acquisition; Cytotoxicity | [37] |
| Alginate | Antibiotic tolerance; Chronic infection | [38] |
| Pel polysaccharide | Surface attachment; Cell aggregation | [39] |
| Psl polysaccharide | Biofilm initiation and maintenance | [40] |
| Acetate | Energy balance; Metabolic flexibility | [41] |
| Succinate | Preferred carbon source; Growth optimization | [42] |
| Glutamate | Growth and stress tolerance | [43] |
| Branched-chain amino acids | Environmental adaptability | [44] |
| Polyamine | Stress tolerance; Biofilm stability | [45] |
| Cyclic dipeptides | Quorum sensing; interspecies interactions | [46] |
| Lectins | Host cell attachment; Biofilm stability; | [47] |
| Volatile organic compounds | Virulence | [48] |
| Hemolysin | Cell lysis; Tissue damage; Virulence | [49] |
| Pyocins | Intraspecies competition; Niche dominance | [50] |
| Cell Lines | JH62 IC50 (μM) | DOX IC50 (μM) | JH62/DOX ZIP Score |
|---|---|---|---|
| Non-cancer cell lines | |||
| MEF | 47.2 ± 4.8 | 3.9 ± 0.4 | 5.77 ± 2.17 |
| AML12 | 37.1 ± 0.7 | 5.1 ± 1.3 | 5.83 ± 0.27 |
| WI-38 | 44.1 ± 0.4 | 1.9 ± 0.3 | 4.91 ± 0.67 |
| THLE-2 | 27.3 ± 4.9 | 1.2 ± 0.1 | 12.06 ± 1.79 |
| Cancer cell lines | |||
| HEPG2 | 17.2 ± 0.6 | 0.8 ± 0.1 | 8.31 ± 3.96 |
| HEP3B | 10.8 ± 0.4 | 3.3 ± 0.2 | 14.61 ± 1.18 |
| BxPC3 | 15.0 ± 0.4 | 1.9 ± 0.1 | 13.79 ± 1.08 |
| U87 | 10.0 ± 0.2 | 1.4 ± 1.0 | 13.85 ± 0.32 |
| DU145 | 13.8 ± 3.3 | 1.9 ± 0.2 | 20.33 ± 6.66 |
| A549 | 14.8 ± 0.1 | 2.1 ± 0.5 | 19.66 ± 5.28 |
| MCF7 | 13.9 ± 1.3 | 1.2 ± 0.5 | 5.31 ± 3.23 |
| HeLa | 13.6 ± 2.7 | 0.7 ± 0.7 | 11.72 ± 3.18 |
| HCT116 | 8.6 ± 0.1 | 3.9 ± 0.7 | 12.19 ± 2.30 |
| Properties | JH62 |
|---|---|
| Physicochemical properties | |
| Molecular Weight | 325.24 |
| No. heavy atoms | 24 |
| No. aromatic heavy atom | 10 |
| No. rotatable bonds | 11 |
| No. H-bond donors | 1 |
| No. H-bond acceptors | 1 |
| Molar refractivity | 106.74 |
| TPSA | 32.86 Å2 |
| Medicinal chemistry | |
| PAINS | 0 |
| Brenk | 1 |
| Synthetic accessibility | 3.14 |
| Lipinski rule | Accepted |
| Golden triangle rule | Accepted |
| Absorption | |
| Intestinal permeability | −5.004 ● |
| MDCK permeability | −4.703 ● |
| PAMPA | 0.003 ● |
| Human intestinal absorption | 0.065 ● |
| Oral bioavailability | 0.122 ● |
| Distribution | |
| BBB | 0.0005 ● |
| VDss | 0.5690 ● |
| PPB | 99.0% ● |
| Metabolism | |
| CYP1A2 inhibitor | Inhibitor |
| CYP2C9 inhibitor | Inhibitor |
| CYP2D6 inhibitor | Inhibitor |
| CYP3A4 inhibitor | Inhibitor |
| CYP2B6 inhibitor | Inhibitor |
| Excretion | |
| Plasma clearance | 4.937 ● |
| The half-life | 0.352 |
| Toxicity | |
| Drug-induced liver injury | 0.179 ● |
| AMES Toxicity | 0.132 ● |
| Rat oral acute toxicity | 0.181 ● |
| Carcinogenicity | 0.186 ● |
| Eye corrosion | 0.014 ● |
| Drug-induced Neurotoxicity | 0.258 ● |
| Genotoxicity | 0.014 ● |
| RPMI-8226 Immunotoxicity | 0.132 ● |
| Ototoxicity | 0.342 ● |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Chen, Q.; Wang, J.; Wu, X.; Xiong, L.; Zhang, L.; Cui, Z. A Novel Quinolone JH62 (E-2-(Tridec-4-en-1-yl)-quinolin-4(1H)-one) from Pseudomonas aeruginosa Exhibits Potent Anticancer Activity. Microorganisms 2026, 14, 78. https://doi.org/10.3390/microorganisms14010078
Chen Q, Wang J, Wu X, Xiong L, Zhang L, Cui Z. A Novel Quinolone JH62 (E-2-(Tridec-4-en-1-yl)-quinolin-4(1H)-one) from Pseudomonas aeruginosa Exhibits Potent Anticancer Activity. Microorganisms. 2026; 14(1):78. https://doi.org/10.3390/microorganisms14010078
Chicago/Turabian StyleChen, Qunyi, Jianhe Wang, Xiaoyan Wu, Lantu Xiong, Lianhui Zhang, and Zining Cui. 2026. "A Novel Quinolone JH62 (E-2-(Tridec-4-en-1-yl)-quinolin-4(1H)-one) from Pseudomonas aeruginosa Exhibits Potent Anticancer Activity" Microorganisms 14, no. 1: 78. https://doi.org/10.3390/microorganisms14010078
APA StyleChen, Q., Wang, J., Wu, X., Xiong, L., Zhang, L., & Cui, Z. (2026). A Novel Quinolone JH62 (E-2-(Tridec-4-en-1-yl)-quinolin-4(1H)-one) from Pseudomonas aeruginosa Exhibits Potent Anticancer Activity. Microorganisms, 14(1), 78. https://doi.org/10.3390/microorganisms14010078






