A Recombinant Porcine Epidemic Diarrhea Virus with Multiple S2 Subunit Mutations from China: Isolation, Genetic Characterization, and Pathogenicity Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
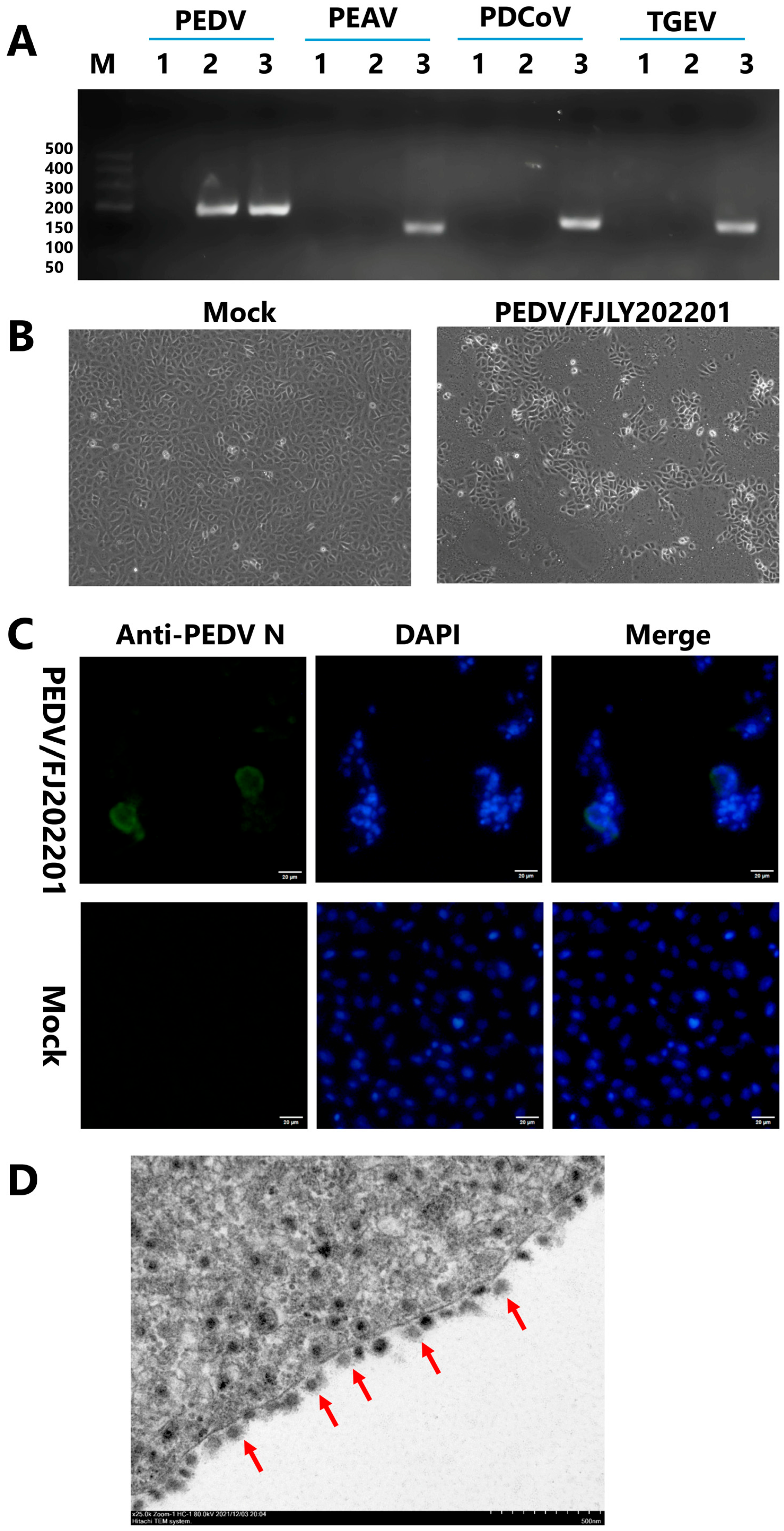
2.2. Virus Isolation and Identification
2.3. Reverse Transcription PCR (RT-PCR) Analysis and Complete Genome Sequencing
2.4. Immunofluorescence Assay (IFA)
2.5. Phylogenetic and Recombination Analysis
2.6. Animal Experiment
2.7. Histopathology and Immunohistochemistry (IHC)
2.8. Biosafety and Biocontainment Procedures
2.9. Statistical Analysis
3. Results
3.1. Isolation and Identification of the PEDV/FJLY202201 Strain
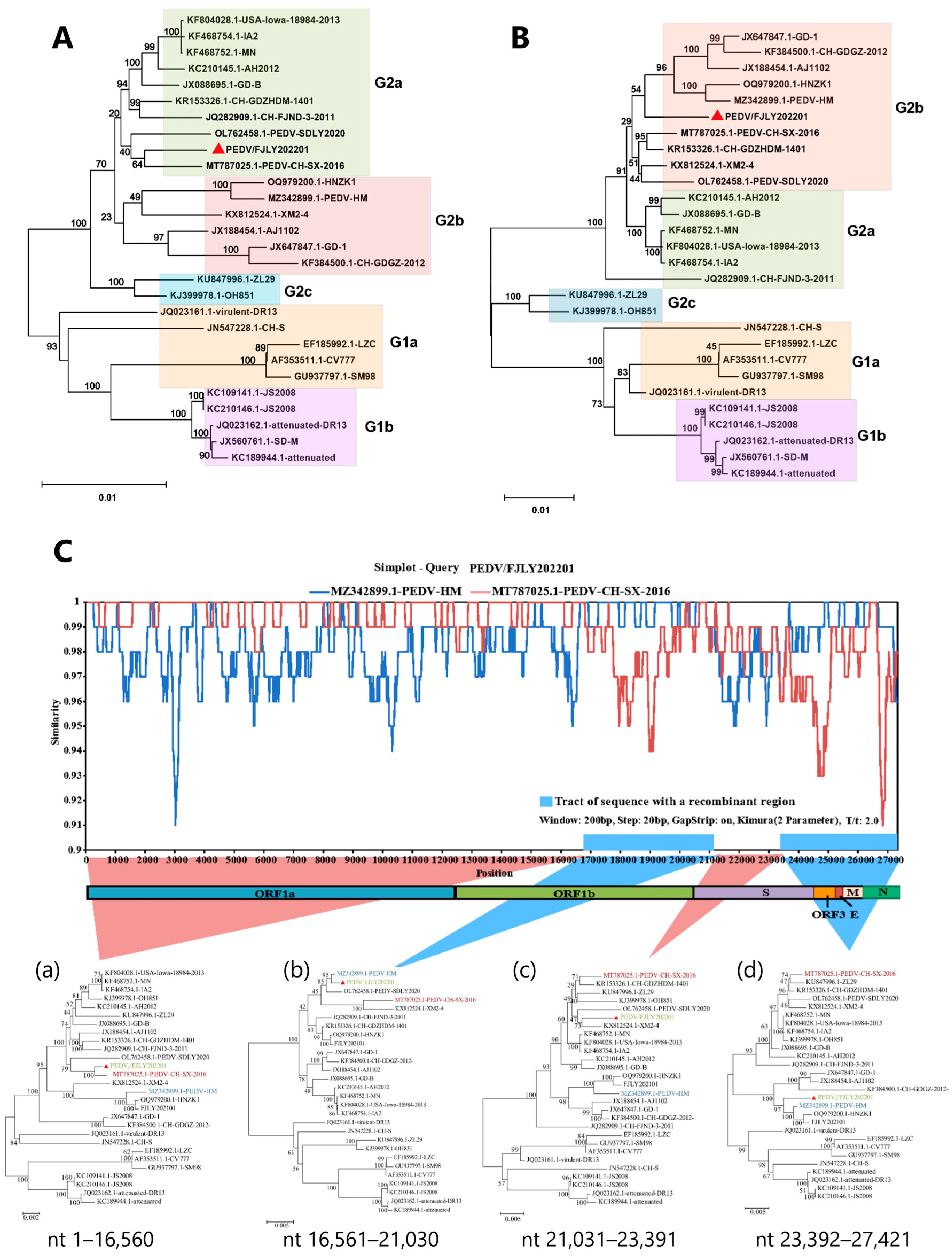
3.2. Complete Genome Sequence Analysis and Phylogenetic Characterization of PEDV/FJLY202201
3.3. Recombination Analysis
3.4. Multiple Mutations in the S2 Subunit of PEDV/FJLY202201
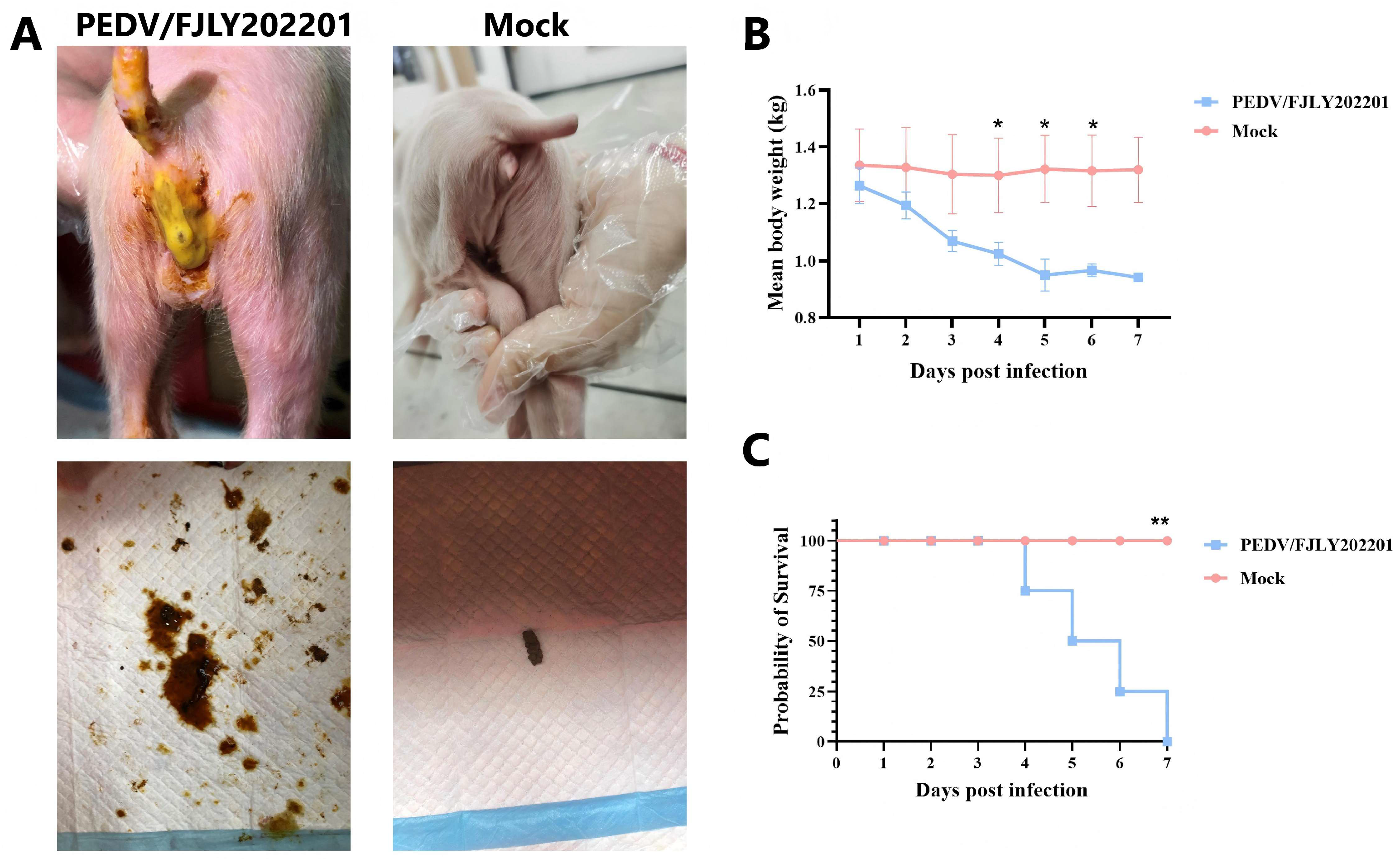
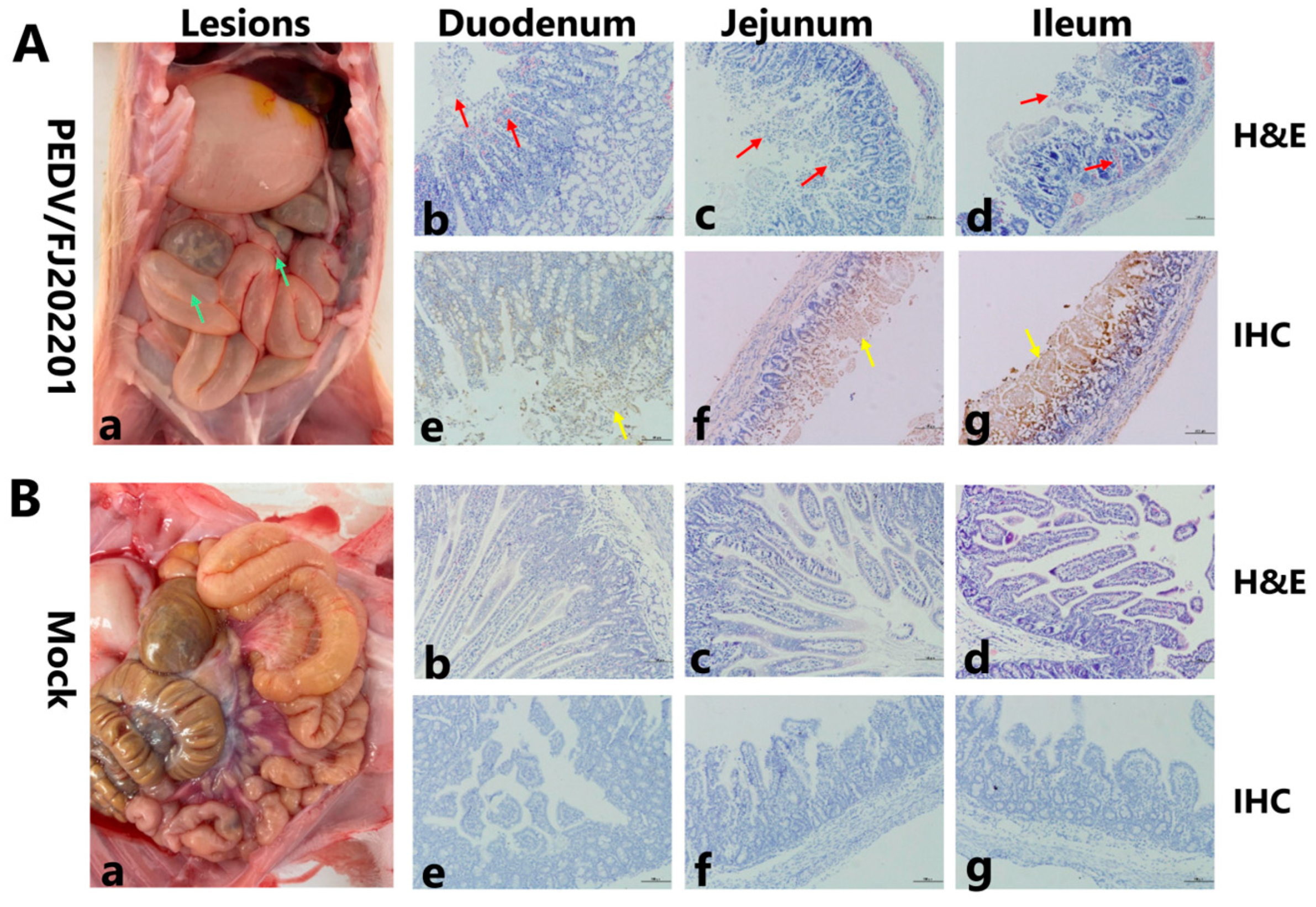
3.5. Pathogenicity of PEDV/FJLY202201 in Piglets
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BSA | Bovine serum albumin |
| BSL-2 | Biosafety level 2 |
| COE | CO-26K equivalent |
| CPE | Cytopathic effect |
| D0 | Domain 0 |
| H&E | Hematoxylin and eosin |
| HRP | Horseradish peroxidase |
| IFA | Immunofluorescence assay |
| IgG | Immunoglobulin G |
| IHC | Immunohistochemistry |
| M | Membrane |
| N | Nucleocapsid |
| ORF | Open reading frame |
| PBS | Phosphate-buffered saline |
| PDCoV | Porcine deltacoronaviruses |
| PEAV | Porcine enteric alphacoronavirus |
| PED | Porcine epidemic diarrhea |
| PEDV | Porcine epidemic diarrhea virus |
| RT | Room temperature |
| SEM | Standard error of the mean |
| S | Spike |
| SS2 | Specific site 2 |
| SS6 | Specific site 6 |
| TGEV | Transmissible gastroenteritis virus |
References
- Jung, K.; Saif, L.J.; Wang, Q. Porcine epidemic diarrhea virus (PEDV): An update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. 2020, 286, 198045. [Google Scholar] [CrossRef]
- Wood, E.N. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977, 100, 243–244. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, H.; Liu, Y.; Pan, Y.; Deng, F.; Song, Y.; Tang, X.; He, Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg Infect Dis. 2012, 18, 1350–1353. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Zeng, S.; Xiao, S.; Chen, H.; Fang, L. Complete genome sequence of porcine epidemic diarrhea virus strain AJ1102 isolated from a suckling piglet with acute diarrhea in China. J. Virol. 2012, 86, 10910–10911. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Tian, X.; Li, W.; Zhou, Q.; Wang, D.; Bi, Y.; Chen, F.; Song, Y. Isolation and characterization of a variant porcine epidemic diarrhea virus in China. Virol. J. 2012, 9, 195. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, J.; Yang, Q.; Zhuang, T.; Xiao, S.; Fang, L. Isolation, genetic characterization, and pathogenicity of the porcine epidemic diarrhea virus S-INDEL strain EJS6 in China. Anim. Dis. 2025, 5, 8. [Google Scholar] [CrossRef]
- Fu, Y.; Wang, Y.; Dai, L.; Cheng, B.; Xiao, S.; Yin, Y. Evolutionary dynamics and antigenic diversity of porcine epidemic diarrhea virus (PEDV) in China: Phylogenetic and recombination analyses based on large-scale S gene sequences. BMC Vet. Res. 2025, 21, 426. [Google Scholar] [CrossRef]
- Zhao, Z.; Sokhansanj, B.A.; Malhotra, C.; Zheng, K.; Rosen, G.L. Genetic grouping of SARS-CoV-2 coronavirus sequences using informative subtype markers for pandemic spread visualization. PLoS Comput. Biol. 2020, 16, e1008269. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, J.; Gu, W.; Zhao, Y.; Zhang, S.; Zuo, Y. Genetic characterization and pathogenicity analysis of three porcine epidemic diarrhea virus strains isolated from North China. Vet. Res. 2025, 56, 118. [Google Scholar] [CrossRef]
- Guo, Y.; Sui, L.; Kong, D.; Liu, D.; Gao, Y.; Jiang, Y.; Cui, W.; Li, J.; Li, Y.; Wang, L. Porcine epidemic diarrhea virus strain CH/HLJ/18 isolated in China: Characterization and phylogenetic analysis. Virol. J. 2024, 21, 28. [Google Scholar] [CrossRef]
- Li, D.; Li, Y.; Liu, Y.; Chen, Y.; Jiao, W.; Feng, H.; Wei, Q.; Wang, J.; Zhang, Y.; Zhang, G. Isolation and Identification of a Recombinant Porcine Epidemic Diarrhea Virus with a Novel Insertion in S1 Domain. Front. Microbiol. 2021, 12, 667084. [Google Scholar] [CrossRef]
- Zeng, Z.; Li, T.; Jin, X.; Peng, F.; Song, N.; Peng, G.; Ge, X. Coexistence of multiple genotypes of porcine epidemic diarrhea virus with novel mutant S genes in the Hubei Province of China in 2016. Virol Sin. 2017, 32, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Jin, Y.L.; Xing, G.; Qv, L.L.; Huang, Y.W.; Zhou, J.Y. Evidence of recombinant strains of porcine epidemic diarrhea virus, United States, 2013. Emerg Infect Dis. 2014, 20, 1735–1738. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.H.; Li, Y.Q.; Pan, Y.Q.; Guo, Y.Y.; Guo, F.; Shi, R.Z.; Xing, L. The spike glycoprotein genes of porcine epidemic diarrhea viruses isolated in China. Vet. Res. 2021, 52, 87. [Google Scholar] [CrossRef]
- Chen, B.; Dong, S.; Yu, L.; Si, F.; Li, C.; Xie, C.; Yu, R.; Li, Z. Three Amino Acid Substitutions in the Spike Protein Enable the Coronavirus Porcine Epidemic Diarrhea Virus To Infect Vero Cells. Microbiol. Spectr. 2023, 11, e0387222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-H.; Li, H.-X.; Chen, X.-M.; Zhang, L.-H.; Zhao, Y.-Y.; Luo, A.-F.; Yang, Y.-R.; Zheng, L.-L.; Chen, H.-Y. Genetic Characteristics and Pathogenicity of a Novel Porcine Epidemic Diarrhea Virus with a Naturally Occurring Truncated ORF3 Gene. Viruses 2022, 14, 487. [Google Scholar] [CrossRef]
- Mei, X.; Guo, J.; Fang, P.; Ma, J.; Li, M.; Fang, L. The Characterization and Pathogenicity of a Recombinant Porcine Epidemic Diarrhea Virus Variant ECQ1. Viruses 2023, 15, 1492. [Google Scholar] [CrossRef]
- Lee, C. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus. Virol. J. 2015, 12, 193, Erratum in Virol. J. 2016, 13, 19. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A Simple Method of Estimating Fifty Percent Endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, 12. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, e003. [Google Scholar] [CrossRef] [PubMed]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef]
- Qin, S.; Hu, C.; Yang, D.; Wu, J.; Yue, H.; Tang, C.; Zhang, B. Emergence of porcine epidemic diarrhea viruses with the novel S genes in Tibetan pigs in the Qinghai-Tibetan plateau in China. Virus Res. 2019, 270, 197652. [Google Scholar] [CrossRef]
- Sun, J.; Li, Q.; Shao, C.; Ma, Y.; He, H.; Jiang, S.; Zhou, Y.; Wu, Y.; Ba, S.; Shi, L.; et al. Isolation and characterization of Chinese porcine epidemic diarrhea virus with novel mutations and deletions in the S gene. Vet. Microbiol. 2018, 221, 81–89. [Google Scholar] [CrossRef]
- Li, Z.; Ma, Z.; Dong, L.; Yang, T.; Li, Y.; Jiao, D.; Han, W.; Zheng, H.; Xiao, S. Molecular Mechanism of Porcine Epidemic Diarrhea Virus Cell Tropism. mBio 2022, 13, e0373921. [Google Scholar] [CrossRef]
- Hulswit, R.J.; Haan, C.A.; Bosch, B.J. Coronavirus Spike Protein and Tropism Changes. Adv. Virus Res. 2016, 96, 29–57. [Google Scholar]
- Lin, F.; Zhang, H.; Li, L.; Yang, Y.; Zou, X.; Chen, J.; Tang, X. PEDV: Insights and Advances into Types, Function, Structure, and Receptor Recognition. Viruses 2022, 14, 1744. [Google Scholar] [CrossRef]

| Strain | Accession Number | Location | Collection Year | Genotype |
|---|---|---|---|---|
| CV777 | AF353511 | Belgium | 1978 | G1a |
| LZC | EF185992 | China | 2006 | G1a |
| SM98 | GU937797 | South Korea | 2010 | G1a |
| CH-S | JN547228 | China | 1986 | G1a |
| Virulent-DR13 | JQ023161 | South Korea | 2009 | G1a |
| Attenuated-DR13 | JQ023162 | South Korea | 2002 | G1b |
| JS2008 | KC109141 | China | 2008 | G1b |
| JS2008 | KC210146 | China | 2008 | G1b |
| SD-M | JX560761 | China | 2012 | G1b |
| PEDV | KC189944 | China | 2012 | G1b |
| OH851 | KJ399978 | USA | 2014 | G2c |
| ZL29 | KU847996 | China | 2015 | G2c |
| USA/Iowa/18984/2013 | KF804028 | USA | 2013 | G2a |
| IA2 | KF468754 | USA | 2013 | G2a |
| MN | KF468752 | USA | 2013 | G2a |
| AH2012 | KC210145 | China | 2012 | G2a |
| GD-B | JX088695 | China | 2012 | G2a |
| CH-GDZHDM-1401 | KR153326 | China | 2014 | G2b |
| CH-FJND-3-2011 | JQ282909 | China | 2011 | G2a |
| PEDV-SDLY2020 | OL762458 | China | 2020 | G2b |
| PEDV-CH-SX-2016 | MT787025 | China | 2016 | G2b |
| HNZK1 | OQ979200 | China | 2021 | G2b |
| PEDV-HM | MZ342899 | China | 2017 | G2b |
| XM2-4 | KX812524 | China | 2016 | G2b |
| AJ1102 | JX188454 | China | 2011 | G2b |
| GD-1 | JX647847 | China | 2011 | G2b |
| CH-GDGZ-2012 | KF384500 | China | 2012 | G2b |
| Virus | Primer Name | Sequence (5′ to 3′) | Product Size (bp) |
|---|---|---|---|
| PEDV | PEDV-JD180-F | CCTGAAACAGACGCGCTTCT | 180 |
| PEDV-JD180-R | CTTGGCGACTGTGACGAAATT | ||
| PEAV | PEAV-JD120-F | CATGCCAGTCCAGGCCTCAA | 120 |
| PEAV-JD120-R | CACGCTTCCATTCAGGTTTGT | ||
| TGEV | TGEV-JD135-F | GGCCAACGTAAAGAGCTTCCT | 135 |
| TGEV-JD135-F | CCAAGCGTGGTTGGTTTGTT | ||
| PDCoV | PDCoV-JD135-F | TGGGTACATGGAGGTGCATTC | 143 |
| PDCoV-JD135-R | CCATATCCTGTGGCGGATTT |
| Recombination Events | Major Parent | Minor Parent | p Value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Beginning | Ending | RDP | GENECONV | Bootscan | MaxChi | Chimaera | SiScan | 3Seq | ||
| 16,560 | 21,030 | PEDV-HM | PEDV-CH-SX-2016 | 1.762 × 10−8 | 3.749 × 10−21 | - | 9.337 × 10−2 | - | - | 1.279 × 10−10 |
| 23,391 | 27,421 | PEDV-HM | PEDV-CH-SX-2016 | 1.249 × 10−30 | 3.749 × 10−21 | - | 2.357 × 10−12 | 3.844 × 10−18 | 6.306 × 10−23 | 9.264 × 10−18 |
| Parameter | PEDV/FJLY202201-Challenged Group | Control Group |
|---|---|---|
| Clinical signs | Diarrhea, vomiting, anorexia, bloody stool, and significant weight loss. | No clinical signs observed throughout the study. |
| Gross lesions | Thin and transparent intestinal walls, gas distension in the intestinal lumen, intestinal bleeding, and congestion/bleeding of mesenteric lymph nodes. | No significant gross lesions were observed |
| Histopathology | Severe atrophy and shortening of intestinal villi, and inflammatory cell infiltration. | Well-preserved tissue structure with no obvious lesions. |
| IHC staining | PEDV antigens (brown signals) were detected in the cytoplasm of atrophied villous epithelial cells. | No viral antigens were detected. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Yan, N.; Xu, J.; Li, Y.; Fan, S.; Qiu, S.; Huang, L.; Xiao, X.; Liao, Y.; Lin, W.; Dong, B.; et al. A Recombinant Porcine Epidemic Diarrhea Virus with Multiple S2 Subunit Mutations from China: Isolation, Genetic Characterization, and Pathogenicity Analysis. Microorganisms 2026, 14, 242. https://doi.org/10.3390/microorganisms14010242
Yan N, Xu J, Li Y, Fan S, Qiu S, Huang L, Xiao X, Liao Y, Lin W, Dong B, et al. A Recombinant Porcine Epidemic Diarrhea Virus with Multiple S2 Subunit Mutations from China: Isolation, Genetic Characterization, and Pathogenicity Analysis. Microorganisms. 2026; 14(1):242. https://doi.org/10.3390/microorganisms14010242
Chicago/Turabian StyleYan, Nana, Jingru Xu, Yuqi Li, Sisi Fan, Shuqi Qiu, Linjie Huang, Xiaoziyi Xiao, Yuting Liao, Weiye Lin, Bo Dong, and et al. 2026. "A Recombinant Porcine Epidemic Diarrhea Virus with Multiple S2 Subunit Mutations from China: Isolation, Genetic Characterization, and Pathogenicity Analysis" Microorganisms 14, no. 1: 242. https://doi.org/10.3390/microorganisms14010242
APA StyleYan, N., Xu, J., Li, Y., Fan, S., Qiu, S., Huang, L., Xiao, X., Liao, Y., Lin, W., Dong, B., Dai, A., & Fan, K. (2026). A Recombinant Porcine Epidemic Diarrhea Virus with Multiple S2 Subunit Mutations from China: Isolation, Genetic Characterization, and Pathogenicity Analysis. Microorganisms, 14(1), 242. https://doi.org/10.3390/microorganisms14010242







