1. Introduction
The pine wood nematode (
Bursaphelenchus xylophilus [Steiner & Buhrer] Nickle, PWN) represents a highly damaging plant-parasitic nematode that is the causal agent of pine wood nematode disease (PWD), or pine wilt disease [
1]. With high reproductive rates, destructive capacity, and environmental adaptability, PWN rapidly causes pine wilt, resulting in significant ecological and economic damage to forest ecosystems [
2].
PWN, originating in North America, was first identified in the United States in 1929 but caused no significant ecological damage locally. By the 1970s, transnational timber trade facilitated its cross-border transmission via wooden packaging materials, leading to its initial establishment in Japan. Its subsequent rapid spread across Asia affected China and South Korea [
3,
4]. Since the 21st century, this pathogen has invaded Europe, with confirmed outbreaks in Portugal, Spain, and the United Kingdom [
5]. Current global monitoring data indicate its presence in at least 18 countries across Asia, Europe, North America, and Africa [
6]. Notably, Asia’s extensive contiguous pine forests have rendered it the region most severely impacted by pine wilt disease [
7,
8]. Globally, PWN-susceptible tree species include
Pinus massoniana,
Pinus thunbergii,
Pinus densiflora, and other pine genus members, as well as non-pine conifers such as
Picea spp. (spruce) and
Abies spp. (fir) [
4,
9]. The cumulative area affected by PWD in China has reached millions of hectares, posing a formidable challenge in prevention and control [
10]. The alarming spread rate and severity of PWD in China have inflicted disastrous impacts on forest resources, triggering severe ecological and economic consequences [
11].
Research has revealed a complex mutualistic relationship between PWN and its associated bacteria, which enhances its pathogenicity and adaptability. Bacteria carried by PWN such as
Pseudomonas fluorescens and
Pseudomonas putida significantly increased nematode fecundity, reproduction rate, and adult growth, while the nematodes, in turn, promoted bacterial proliferation. This mutualistic symbiosis suggests long-term co-evolution, with metabolic interactions, enhancing both organisms’ adaptability [
12]. PWNs and the associated fungus,
Sporothrix sp. 1, exhibit a synergistic relationship, where their interaction enhances PWN pathogenicity and accelerates pine tree mortality. When PWN fed on
Sporothrix sp. 1, lipid metabolism gene expression and metabolite production were upregulated, leading to increased palmitoleic acid levels through both uptake and de novo synthesis, thereby enhancing nematode fecundity. Conversely, increased nematode populations promote fungus dispersal [
13]. Palmitoleic acid acts as a molecular link, enhancing the fitness and reproductive success of both species and providing insights for other investigations of biological co-evolution. The co-evolutionary relationship between bacteria and pine wood nematodes is a key factor affecting the occurrence and spread of pine wilt disease. There are significant differences in the types of endophytic bacteria associated with pine wood nematodes from different pine species, and the dynamic changes and diversity of bacterial communities have a significant impact on the pathogenicity of pine wood nematodes, especially during the host switching stage in the nematode life cycle [
14]. This dynamic symbiotic relationship reflects the synergistic effect of bacteria and nematodes in adaptive evolution. Certain symbiotic bacteria are capable of modulating the pine tree’s antioxidant system, thereby inhibiting host defense responses [
15]. The antioxidant metabolites they secrete, such as antioxidant enzymes and peptides, can lower the levels of reactive oxygen species (ROS) within the pine, consequently weakening the tree’s immune response and establishing a more suitable environment for pine wood nematode reproduction [
16,
17,
18,
19]. The expansion of gene families in the pine wood nematode genome related to the detoxification of terpenes (such as cytochrome P450 and short-chain dehydrogenase) may have partially benefited from the long-term selective pressure of bacterial metabolic products [
20,
21]. In addition, pine wood nematodes can produce cellulase (β-1,4-endoglucanase) and pectinase to digest plant tissues, which may be due to horizontal gene transfer from bacteria and fungi to the nematodes [
22,
23]. PWN-associated bacteria enhance nematode tolerance to terpenoids and phenolic compounds, facilitating survival and reproduction within pine trees that contain these defensive compounds [
24,
25]. The presence of bacteria also enhances PWN cold resistance, allowing them to maintain physiological activity and population stability under low-temperature conditions [
26]. These adaptive enhancement mechanisms provide a crucial foundation for further investigations into the PWN-bacteria interaction, furthering our understanding of this symbiotic relationship’s role and significance within the ecosystem.
Given the critical role of bacteria in PWN’s adaptability, this study aims to investigate the relations between dominant bacterial communities and PWN strains of varying virulence, and analyze bacterial-mediated gene expression in PWN using transcriptomics. This research will advance our understanding of the drivers behind PWN virulence variation and elucidate the synergistic pathogenic mechanisms of nematode-bacteria interactions.
2. Materials and Methods
2.1. PWN and Nematodes Culturing
Bursaphelenchus xylophilus (Steiner & Buhrer) Nickle isolates F (highly virulent) and Q (weakly virulent), which were originally isolated from Fushan Forestry Park and Qingdao University campus, respectively, are characterized isolates preserved in our laboratory and were utilized as they have been in our prior research for their differing levels of virulence [
27]. For subculture inoculation, the isolated nematodes were transferred to PDA plates fully colonized by
Botrytis cinerea (pre-grown at 25 °C in the dark for 7 days), which served as a food source.
2.2. Isolation and Identification of Culturable Bacteria from Nematodes
Culturable bacteria were isolated from the bodies of PWNs by the reported method [
28], and the strains were cultured in nutrient broth (NB) liquid medium at 25 °C for 24 h. Total genomic DNA was extracted from the bacteria of
B. xylophilus using the E.Z.N.A.
® Bacterial DNA Kit (Omege Biotech, Norcross, GA, USA). The 16S rDNA was amplified by primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACCTTGTTACGACTT-3′). The PCR reaction was carried out according to the method of Yuan et al. [
29], and the PCR product sequencing was completed by Sangon Biotech Co., Ltd. (Shanghai, China). The similarity of the 16S rDNA sequence was compared with the existing sequences available in the NCBI GenBank database using a BLAST (
https://www.ncbi.nlm.nih.gov/tools/primer-blast/, accessed on 30 August 2024) search. The sequence alignment and the phylogenetic tree were constructed by MEGA11 software using the Neighbor-Joining method based on the Kimura 2-parameter (K2P) model [
30]. Bootstrap analysis was performed with 1000 replicates.
2.3. Treatment of Nematodes and Bacteria
To prepare aseptic PWN, the nematodes were extracted via Bellman funnel overnight, washed thrice with sterilized distilled water, disinfected with 9% hydrogen peroxide for 50 min, and rinsed thoroughly. Then, they were exposed to a mixture of 4% ciprofloxacin and 4% ceftriaxone sodium for 8 h before being rinsed again. Finally, PWN culturing on nutrient agar (NA) plates at 25 °C for 48 h, a bacterial staining check, and PCR analysis of 16S rDNA were used to verify the absence of bacterial contamination.
To promote stable colonization of specific bacteria associated with
B. xylophilus, the bacterial strains were first cultured in NB medium with shaking until the optical density (OD) at 600 nm reached approximately 0.8 (mid-log phase). The bacterial cells were collected by centrifugation and gently resuspended in sterile distilled water. Subsequently, the bacterial suspension was mixed with surface-sterilized and viability-confirmed nematodes and co-incubated statically at 25 °C for 12 h. After incubation, the nematodes were gently washed three times with sterile distilled water to thoroughly remove unbound free bacteria. Finally, the number of bacteria firmly attached to the nematode surface was quantified using the plate colony counting method [
31].
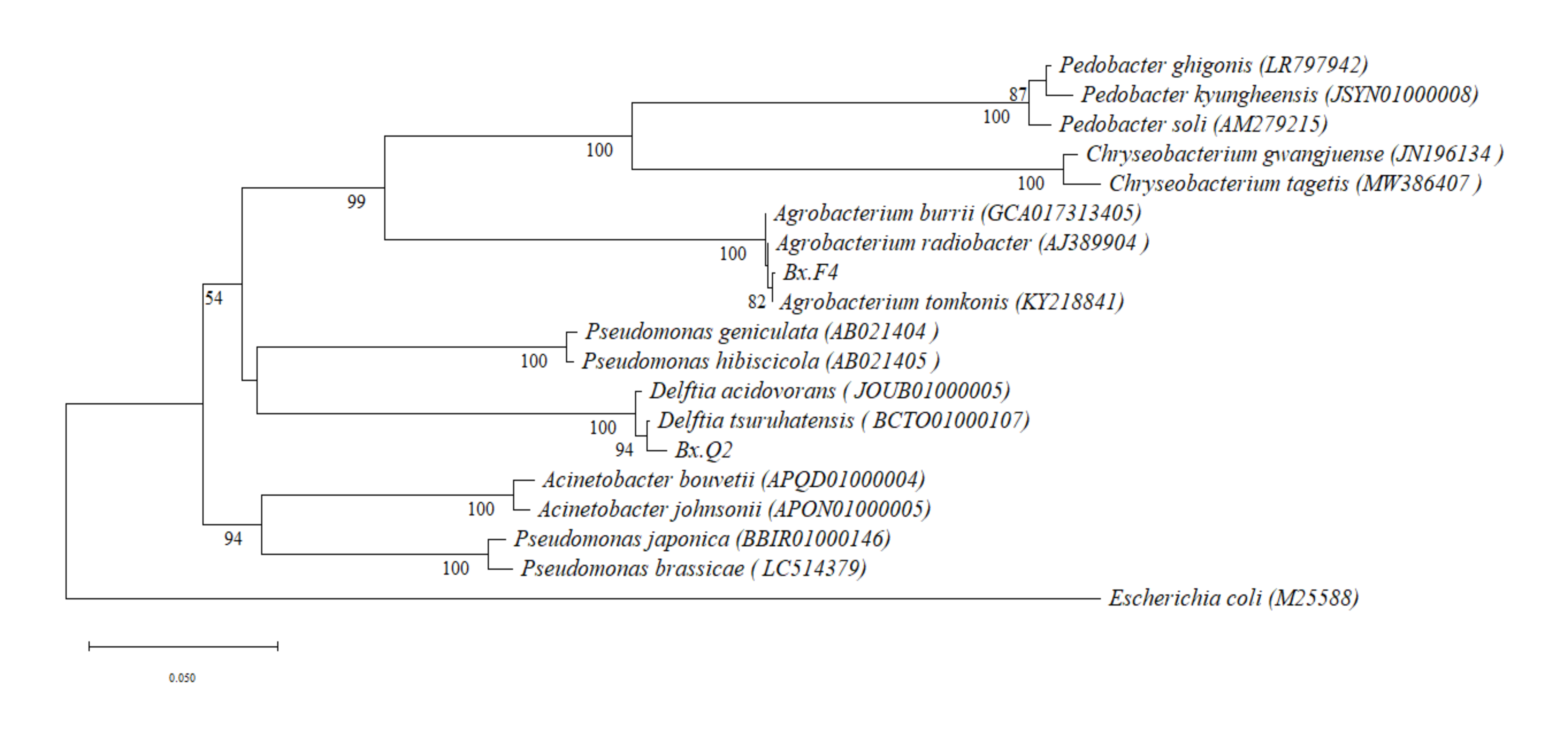
In this study,
Agrobacterium radiobacter Bx.F4, isolated from
B. xylophilus isolates F (highly virulent), and
Delftia tsuruhatensis Bx.Q2, isolated from
B. xylophilus isolates Q (weakly virulent), were used as experimental materials. Both isolated bacterial strains were identified as belonging to the dominant genera associated with
B. xylophilus based on comparative analysis with metagenomic data from prior studies [
27]. Each bacterium was co-cultured separately with aseptic PWNs. The resulting nematode samples were designated as experimental groups and named S-Bx.F4 and S-Bx.Q2, respectively.
2.4. Egg-Laying Ability of Nematodes
Ten male and ten unmated female PWNs (all late J4 stage) from the aseptic PWN group (control group, CK), S-Bx.Q2, and S-Bx.F4 were randomly selected and transferred to 24-well tissue culture plates containing sterile distilled water and incubated at 25 °C. After allowing them to mate in the dark for 36 h, the number of eggs in each well was counted under a stereomicroscope (Motic SMZ-168, Fujian, China) at 80× magnification. CK, S-Bx.Q2, and S-Bx.F4 had six repeat treatments each. This data were recorded as the number of eggs laid per female nematode.
2.5. Measurement of Motility and Lifespan of Nematodes
The head thrashing frequency of B. xylophilus is a key indicator for evaluating its motility. Adult PWNs from the control group (CK), S-Bx.Q2, and S-Bx.F4 were randomly selected and transferred to 24-well tissue culture plates containing sterile distilled water for incubation at 25 °C. Within each well, 10 live adult nematodes were randomly chosen for observation, and their head-thrashing frequency per minute was recorded.
To calculate the survival rate of PWNs, the number of live nematodes in each well was counted daily until all nematodes were dead. The final recorded day was then taken as the lifespan of the nematodes. CK, S-Bx.Q2, and S-Bx.F4 had six repeat treatments each.
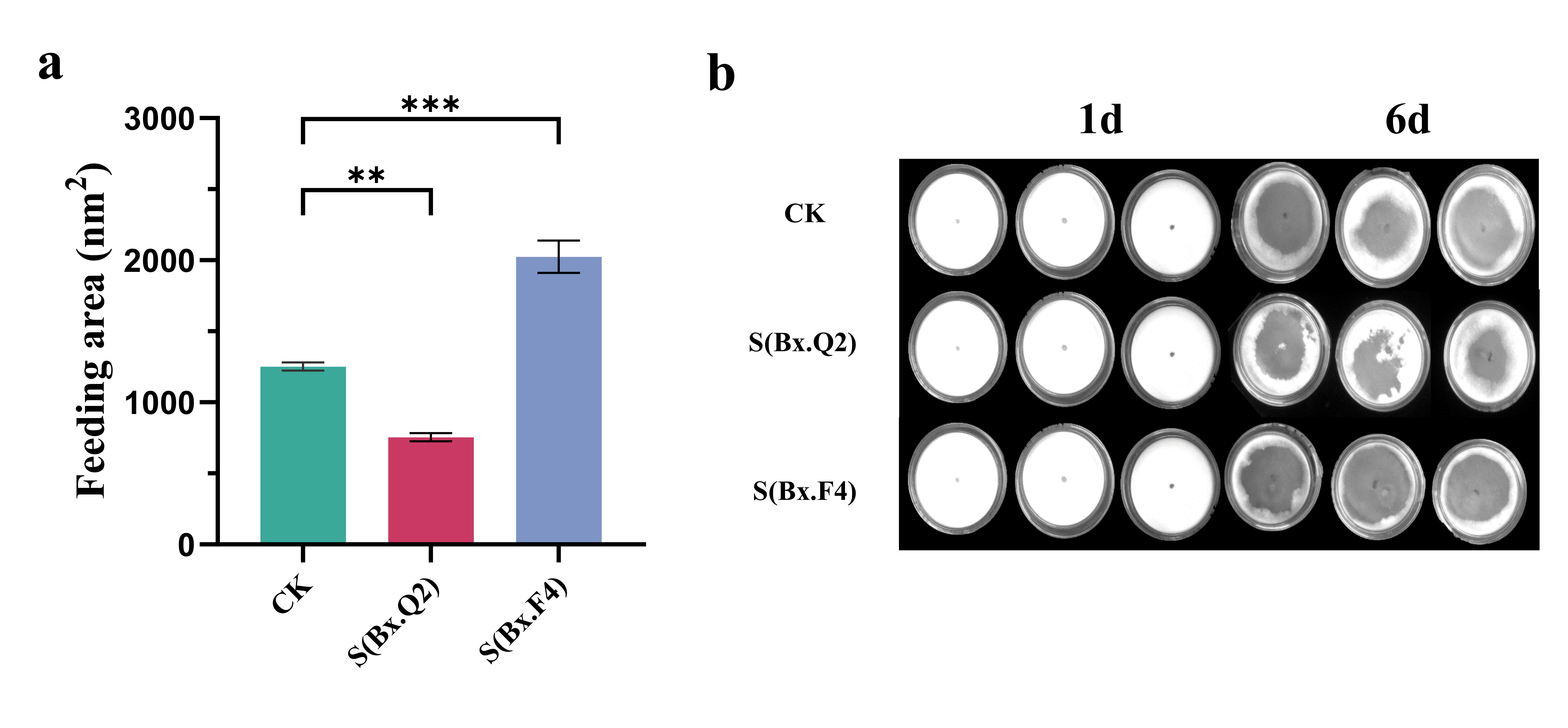
2.6. Analysis of Population Growth and Feeding Ability of Nematodes
Equal numbers of PWNs (approximately 100 per dish) from the control group (CK), S-Bx.Q2, and S-Bx.F4 were individually inoculated onto 6 cm Petri dishes overlaid with B. cinerea and cultured at 25 °C. Feeding zones were observed daily and quantified using ImageJ software version 1.8.0 (National Institutes of Health, Bethesda, MD, USA).
After the
B. cinerea on the plates had been consumed by the nematodes, the nematodes were collected using the Baermann funnel method [
32], and their numbers were recorded as an indicator of the population growth of the nematodes.
2.7. Pathogenicity Analysis of Nematodes
Equal numbers of PWNs (approximately 5000 per seedling) from the control group (CK), S-Bx.Q2, and S-Bx.F4 were inoculated into two-year-old
P. thunbergii seedlings, respectively, with four replicates per treatment. The inoculation method was based on previously reported methods [
33]. The disease severity index (DSI) reflected the difference in the wilting degree of indirect species among different treatments, and the disease incidence reflected the difference in wilting number among repeats of each group. The severity of infection in
P. thunbergii was divided into five levels (
Table 1). The disease grading standard and DSI were calculated according to Yu et al. [
34].
2.8. RNA Isolation, cDNA Synthesis, and RNA Sequence
Total RNA was extracted from approximately 20,000 aseptic nematodes (control group, CK), S-Bx.Q2, and S-Bx.F4, respectively, using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) and following the manufacturer’s protocol. RNA quality was evaluated by electrophoresis on RNase-free agarose gels and analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA concentration was determined using both a NanoPhotometer spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and a Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Complementary DNA libraries were prepared with the TruSeqTM RNA Sample Prep Kit (Illumina, Inc., San Diego, CA, USA), size-selected using Certified Low Range Ultra Agarose (Bio-Rad, Hercules, CA, USA), and quantified with high sensitivity using TBS380 Picogreen (Invitrogen, USA). Final sequencing was conducted on an Illumina HiSeq4000 platform by Biozeron Biotechnology Co., Ltd. (Shanghai, China).
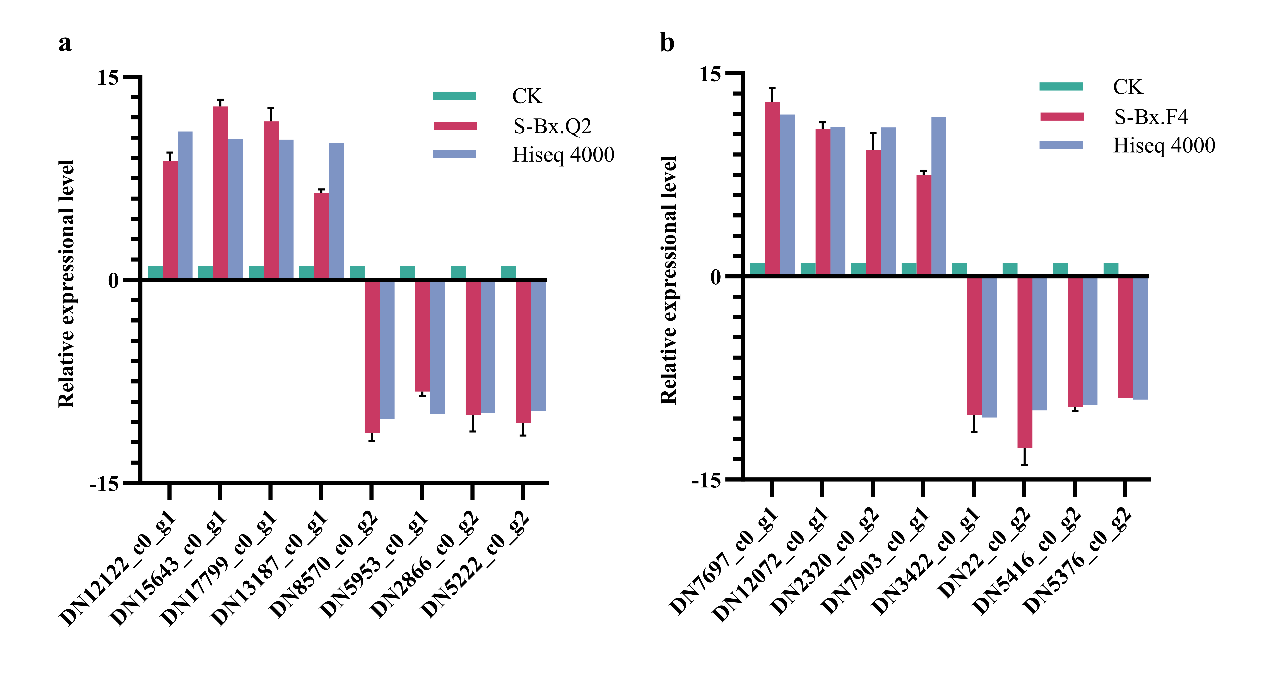
2.9. Quantitative Real-Time PCR
Total RNA was extracted using the TRIzol
® reagent, and cDNA was synthesized using the PrimeScript RT Reagent Kit (Takara Bio Inc., Kusatsu, Japan). cDNA was precisely quantified using a NanoPhotometer spectrophotometer. The qPCR was then performed on ABI7000 Fast (Thermo Fisher Scientific, USA) using the Talent qPCR PreMix (SYBR Green) Kit (TIANGEN, Beijing, China), following the manufacturer’s protocol. Primer sequences are listed in
Table 2. The actin gene (GenBank accession number: EU100952) was selected as the reference gene for qPCR. The relative expression levels of the CK group were set as 1 (
n = 6). The experiment was performed twice, with three replications for each treatment, and gene expression levels were quantified using the 2
−∆∆Ct method.
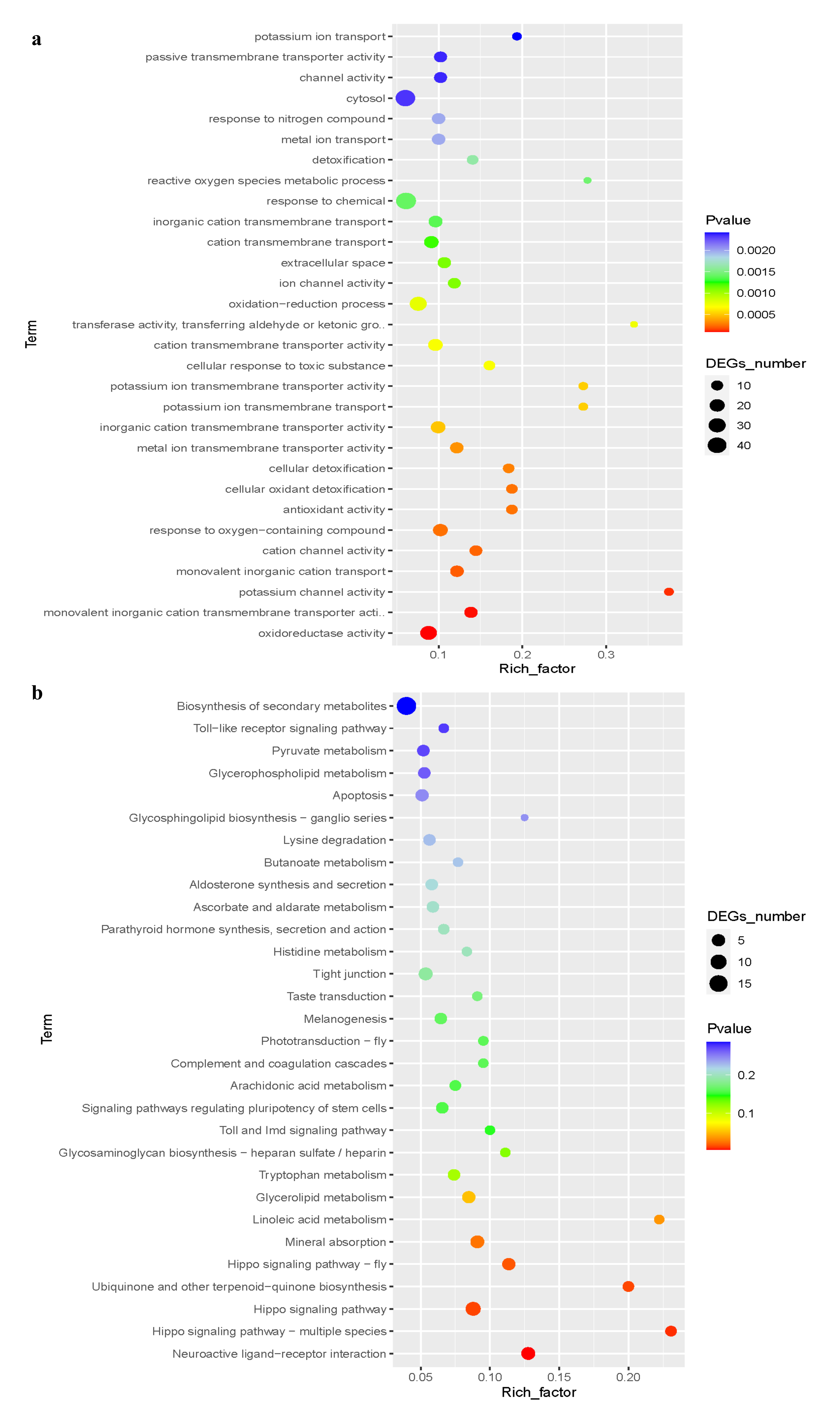
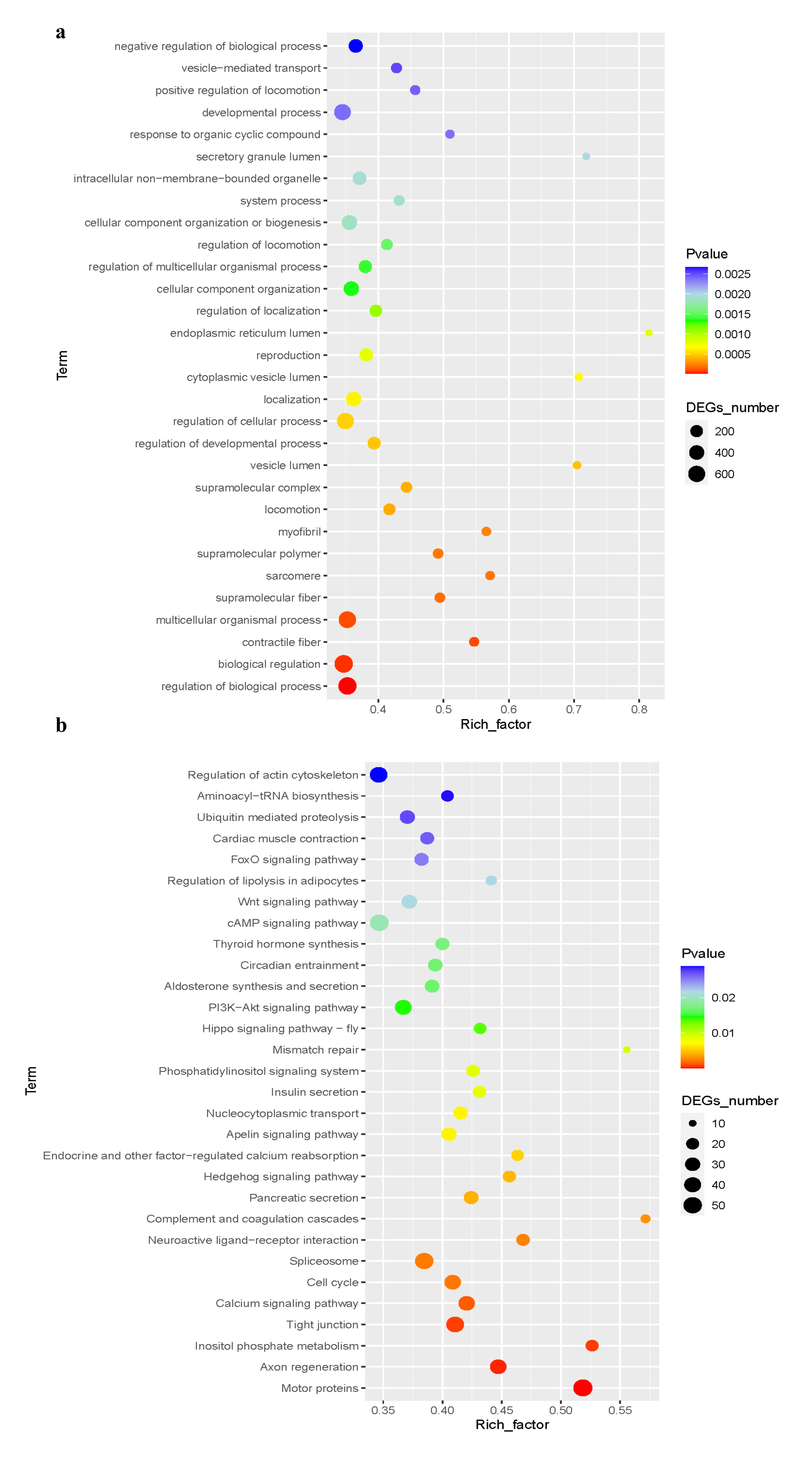
2.10. Sequencing Data Analysis
The specific parameters were as follows: adapter removal; 3′ end trimming (Q < 20); reads with >10% Ns removed; discard reads <75 bp after trimming. Goatools (
https://github.com/tanghaibao/GOatools, accessed on 5 September 2024) and KOBAS (
http://bioinfo.org/kobas, accessed on 10 September 2024) were used for differential gene expression analysis (Fisher’s exact test; FDR < 0.05, absolute fold change ≥ 2).
2.11. Statistical Analysis
All biological experiments were conducted in triplicate, with six technical replicates for each treatment, unless stated otherwise. Data are expressed as mean ± standard deviation (SD). All parameters were calculated using SPSS Statistics 17.0 (IBM, Armonk, NY, USA). The statistical analysis was performed with One-Way Analysis of Variance (* p < 0.05, ** p < 0.01, *** p < 0.001) in GraphPad Prism 10 (GraphPad Software, San Diego, CA, USA).
4. Discussion
Bursaphelenchus xylophilus, as a plant-parasitic nematode, harbors a diverse bacterial community on its surface, which is actively involved in its growth, reproduction, environmental adaptation, and infection of host pine trees [
35]. The composition of these associated bacterial communities exhibits significant geographical heterogeneity [
36]. Notably, significant differences in pathogenicity exist among different
B. xylophilus isolates, which may be partly attributable to variations in the structure and function of their associated bacterial communities. Comparative studies of the bacterial communities associated with high- and low-virulence isolates have revealed that highly virulent nematodes often possess higher bacterial diversity and demonstrate enhanced capabilities in utilizing carbohydrates and carboxylic acids [
14,
37]. The presence of specific bacterial groups, such as
Stenotrophomonas and
Pseudomonas, is closely correlated with nematode virulence [
37]. These findings indicate that the associated bacterial community serves as a critical regulator that shapes the pathogenicity of
B. xylophilus through modulating its key physiological functions.
Certain dominant associated bacteria enhance the physiological activity and pathogenicity of
B. xylophilus by providing nutritional support, improving environmental adaptation, or directly assisting in host invasion. Key to disease progression is the nematode’s reproductive capacity and its migration rate within the xylem, both of which jointly determine population expansion in the host and the development of pine wilt disease. Studies have shown that when co-cultured with
B. xylophilus,
P. fluorescens and
P. putida can significantly increase egg production and reproduction rates, while obviously enhancing the nematode’s motility, specifically manifested as increased frequency of head swinging and body bending [
38].
Furthermore, pine trees produce reactive oxygen species (ROS) as a defense response upon infection, and various associated bacteria can assist the nematode in coping with such oxidative stress.
Serratia spp. possesses intrinsic antioxidant capacity, which not only significantly improves the survival of
B. xylophilus under oxidative stress, but also upregulates the expression of nematode antioxidant enzyme genes (e.g., Bxy-ctl-1 and Bxy-ctl-2), thereby enhancing its ability to counteract host defenses and promoting successful colonization and survival during early infection [
39]. On the other hand, certain associated or environmental bacteria can suppress the survival and reproduction of pine wood nematodes by producing inhibitory metabolites, highlighting their promise as potential biocontrol agents [
40,
41,
42].
This study further revealed that the dominant associated bacteria isolated from highly pathogenic PWN significantly enhanced their motility, egg-laying capacity, population growth, lifespan, feeding ability, and pathogenicity. In contrast, when the dominant bacteria isolated from low-pathogenic PWN were introduced into highly pathogenic PWN, these parameters were markedly suppressed. These findings suggest that dominant associated bacteria may play a crucial regulatory role in the physiology and pathogenicity of PWN, providing a new theoretical foundation for the prevention and control of PWD. We propose the following hypothesis: in PWD, bacteria may function as conditional pathogens. Within the microbial community of healthy pine trees, these bacteria are generally harmless or even beneficial to the host. However, when they co-infect alongside pine wood nematodes, the bacteria may modulate the adaptability of the nematodes to the host by secreting specific metabolites or expressing their own detoxification-related genes, thereby influencing the occurrence and progression of PWD.
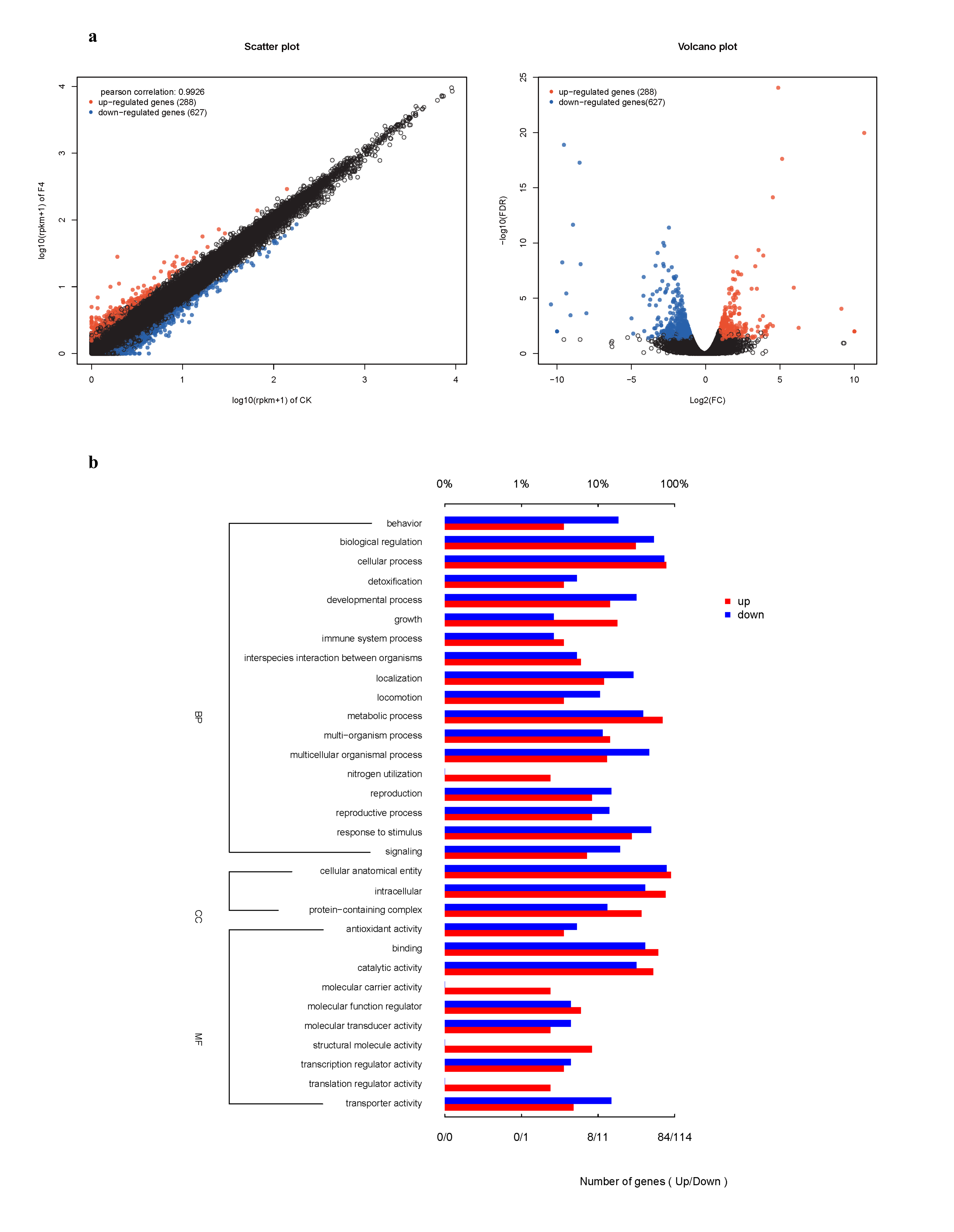
Our results showed that RNA sequencing revealed a significant impact on the gene expression of
B. xylophilus treated with
A. radiobacter Bx.F4 and
D. tsuruhatensis Bx.Q2. according to RNA sequencing results, the analysis of differentially expressed genes between the S-Bx.F4 treatment group and the aseptic PWNs (CK) group (
Table 7) revealed significant upregulation of several genes. Among them, the expression of MKK 7 was increased. As an upstream regulatory hub of the JNK signaling pathway, elevated MKK7 expression can enhance nematode environmental adaptability, homeostasis maintenance, and lifespan [
43,
44,
45,
46], providing a possible molecular explanation for the improved survival rate of nematodes treated with
A. radiobacter Bx.F4.
Simultaneously, the expression of Axin, a core negative regulator of the Wnt/β-catenin signaling pathway, was also significantly increased. This pathway is involved in regulating embryonic development, cell fate determination, and organ formation, which may help explain the increased egg production and population growth observed after
D. tsuruhatensis Bx.F4 treatment [
47,
48]. Furthermore, the expression of F-actin, a core cytoskeletal protein, was elevated. Its dynamic reorganization directly affects body wall muscle contraction patterns and pharyngeal pumping efficiency, potentially contributing to the enhanced locomotor ability and feeding range of the nematodes [
49,
50,
51,
52]. The expression of HSP60, a key molecular chaperone maintaining mitochondrial protein homeostasis, was also upregulated. By modulating pathways such as mitochondrial stress response, insulin/IGF-1 signaling, and apoptosis, HSP60 may further influence nematode development, locomotor ability, and lifespan [
53,
54,
55].
In the comparison of differentially expressed genes between the S-Bx.Q2 treatment group and the aseptic PWNs (CK) group (
Table 8), several genes showed significant downregulation. The expression of DAF-16, a core transcription factor in the DAF-16/FOXO signaling pathway and longevity-related pathways, was reduced by approximately 10.26-fold, while its cofactor SIR-2.1 was downregulated by 1.67-fold. This change may impair nematode lifespan and resistance to environmental stress and pathogens, potentially explaining the decreased survival rate and population growth after
D. tsuruhatensis Bx.Q2 treatment [
56,
57,
58,
59]. Additionally, reduced expression of 14-3-3 zeta, a key adaptor in intracellular signaling, may further affect lifespan, stress resistance, and embryonic development, consistent with the observed decreases in survival rate, egg production, and population growth [
60]. Moreover, decreased expression of cystatin-like protein, a cysteine protease inhibitor, may disrupt basic protein metabolism and growth development [
61,
62]. The downregulation of vitellogenin-1, a key yolk protein precursor [
63], may impair nutrient transport from the intestine to the gonads, thereby compromising embryonic development and reproductive capacity. Together, these gene expression changes provide a molecular basis for the reduced survival rate, egg production, and population growth observed in nematodes treated with
D. tsuruhatensis Bx.Q2.
In summary, the results revealed that their individual inoculation led to significant and even opposite trends in key physiological indicators of the nematodes. Transcriptomic analysis further demonstrated that the two bacterial strains differentially regulated the expression of multiple key signaling pathways and related functional genes in the nematodes. This indicates that their mechanisms of action are not limited to a single functional process; rather, they may systematically activate or inhibit interconnected gene expression networks in the nematodes through bacterial metabolites or other effector molecules, thereby enhancing or diminishing the overall health and fitness of the nematodes.
At the transcriptomic level, this study reveals that microbial symbionts can significantly influence host phenotypes by modulating the overall architecture of the host gene expression network, providing molecular evidence for the role of bacteria in the host adaptation of pine wood nematodes. To further elucidate the underlying mechanisms, follow-up studies could employ RNA interference technology for functional validation of key genes, combined with metabolomics analysis of metabolites produced by the associated bacteria. This approach would facilitate the screening of strains and metabolites with inhibitory activity for development as potential biocontrol agents. Subsequent field evaluations of their efficacy against pine wilt disease could offer a practical strategy for green control. Through this framework, the study aims to systematically clarify how these bacteria regulate host gene expression and their potential roles in the onset and progression of PWD.