Evaluating the Aquatic Environment as a Reservoir for Salmonella: A Comparative Analysis with Clinical Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Identification of Strains
2.2. Antimicrobial Susceptibility
2.3. Determination of Antimicrobial Resistance Genes
2.4. Analysis of PFGE and WGS
3. Results
3.1. Characteristics of Salmonella Serotypes in Streams
3.2. Antimicrobial Resistance and Molecular Characterization of Resistance Genes
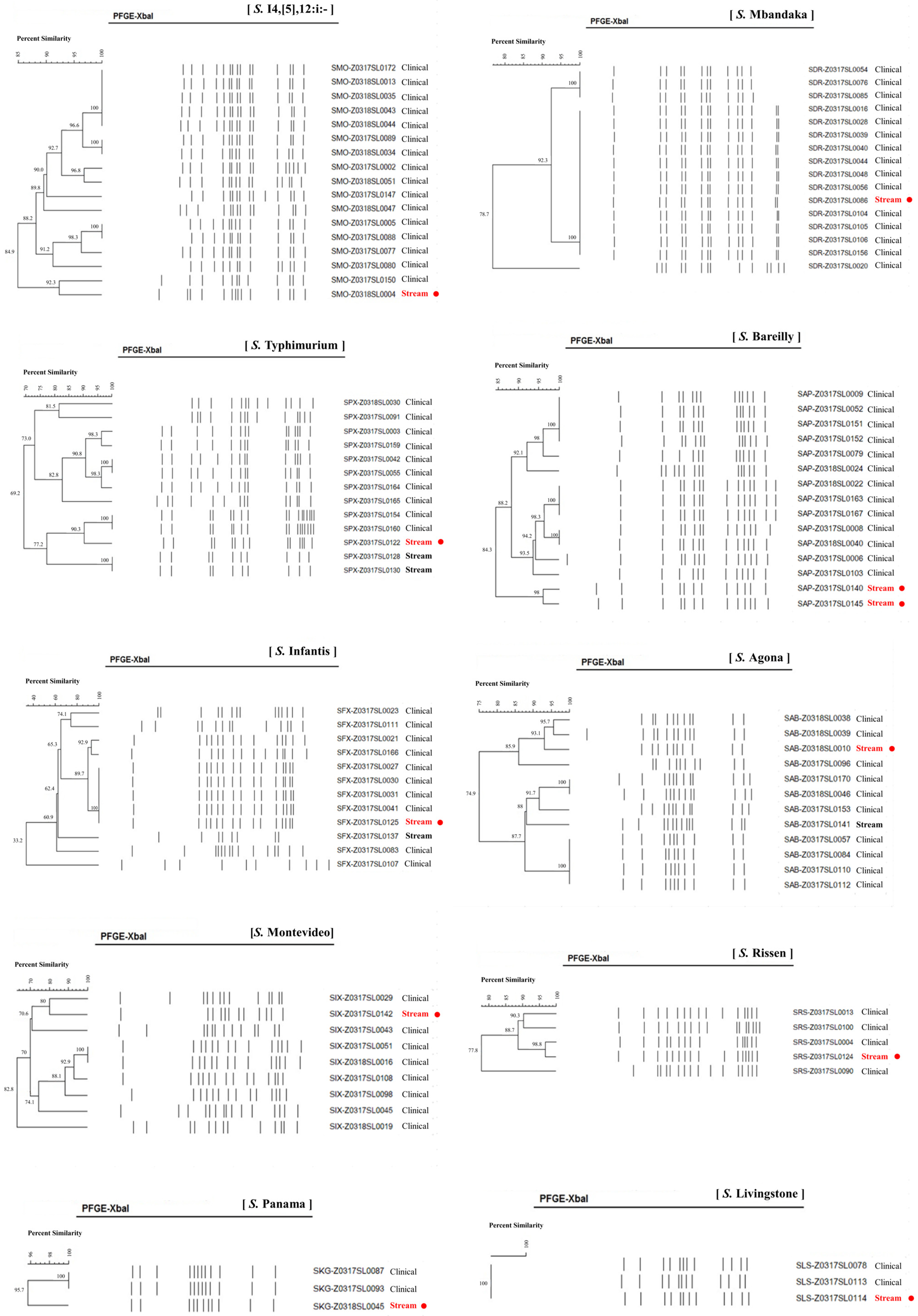
3.3. Epidemiologic Association of Salmonella from Stream and Clinical Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, S.H.; Sung, G.H.; Park, E.H.; Hwang, I.Y.; Kim, G.R.; Song, S.A.; Lee, H.K.; Uh, Y.; Kim, Y.A.; Jeong, S.H.; et al. Serotype Distribution and Antimicrobial Resistance of Salmonella Isolates in Korea between 2016 and 2017. Ann. Lab. Med. 2022, 42, 268–273. [Google Scholar] [CrossRef]
- Soyer, Y.; Moreno Switt, A.; Davis, M.A.; Maurer, J.; McDonough, P.L.; Schoonmaker-Bopp, D.J.; Dumas, N.B.; Root, T.; Warnick, L.D.; Grohn, Y.T.; et al. Salmonella enterica serotype 4,5,12:i:-, an emerging Salmonella serotype that represents multiple distinct clones. J. Clin. Microbiol. 2009, 47, 3546–3556. [Google Scholar] [CrossRef]
- Plumb, I.; Fields, P.P.; Bruce, B. Travel-Associated Infections & Diseases: Salmonellosis, Nontyphoidal. In CDC Yellow Book; Oxford University Press: Oxford, UK, 2024. [Google Scholar]
- European Centre for Disease Prevention and Control, European Food Safety Authority. Multi-country outbreak of Salmonella Mbandaka ST413 linked to consumption of chicken meat products in the EU/EEA and the UK—first update-21 March 2024. EFSA Support. Publ. 2024, 19, 7707E. [Google Scholar]
- Silva, C.; Calva, E.; Maloy, S. One Health and Food-Borne Disease: Salmonella Transmission between Humans, Animals, and Plants. Microbiol. Spectr. 2014, 2, OH-0020-2013. [Google Scholar] [CrossRef] [PubMed]
- Denis, N.; Zhang, H.; Leroux, A.; Trudel, R.; Bietlot, H. Prevalence and trends of bacterial contamination in fresh fruits and vegetables sold at retail in Canada. Food Control 2016, 67, 225–234. [Google Scholar] [CrossRef]
- Levantesi, C.; Bonadonna, L.; Briancesco, R.; Grohmann, E.; Toze, S.; Tandoi, V. Salmonella in surface and drinking water: Occurrence and water-mediated transmission. Food Res. Int. 2012, 45, 587–602. [Google Scholar] [CrossRef]
- Callahan, M.T.; Van Kessel, J.A.; Micallef, S.A. Salmonella enterica recovery from river waters of the Maryland Eastern Shore reveals high serotype diversity and some multidrug resistance. Environ. Res. 2019, 168, 7–13. [Google Scholar] [CrossRef]
- Patchanee, P.; Molla, B.; White, N.; Line, D.E.; Gebreyes, W.A. Tracking Salmonella contamination in various watersheds and phenotypic and genotypic diversity. Foodborne Pathog. Dis. 2010, 7, 1113–1120. [Google Scholar] [CrossRef]
- Sung, G.H.; Kim, S.H.; Park, E.H.; Hwang, S.N.; Kim, J.D.; Kim, G.R.; Kim, E.Y.; Jeong, J.; Kim, S.; Shin, J.H. Association of Carbapenemase-Producing Enterobacterales Detected in Stream and Clinical Samples. Front. Microbiol. 2022, 13, 923979. [Google Scholar] [CrossRef]
- M100; Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2024.
- Kim, S.H.; Park, E.H.; Hwang, I.Y.; Lee, H.; Song, S.A.; Lee, M.; Lee, S.; Kim, S.Y.; Kim, J.J.; Shin, J.H.; et al. Serotyping and Antimicrobial Susceptibility of Salmonella Isolated in Korea in 2015. Ann. Clin. Microbiol. 2019, 22, 55–60. [Google Scholar] [CrossRef]
- Gay, K.; Robicsek, A.; Strahilevitz, J.; Park, C.H.; Jacoby, G.; Barrett, T.J.; Medalla, F.; Chiller, T.M.; Hooper, D.C. Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin. Infect. Dis. 2006, 43, 297–304. [Google Scholar] [CrossRef]
- Eaves, D.J.; Randall, L.; Gray, D.T.; Buckley, A.; Woodward, M.J.; White, A.P.; Piddock, L.J. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob. Agents Chemother. 2004, 48, 4012–4015. [Google Scholar] [CrossRef]
- Li, Q.; Wang, X.; Yin, K.; Hu, Y.; Xu, H.; Xie, X.; Xu, L.; Fei, X.; Chen, X.; Jiao, X. Genetic analysis and CRISPR typing of Salmonella enterica serovar Enteritidis from different sources revealed potential transmission from poultry and pig to human. Int. J. Food Microbiol. 2018, 266, 119–125. [Google Scholar] [CrossRef]
- Baudart, J.; Lemarchand, K.; Brisabois, A.; Lebaron, P. Diversity of Salmonella strains isolated from the aquatic environment as determined by serotyping and amplification of the ribosomal DNA spacer regions. Appl. Environ. Microbiol. 2000, 66, 1544–1552. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, M.; Martinez-Urtaza, J.; Rodriguez-Alvarez, M.X.; Leon-Felix, J.; Chaidez, C. Prevalence and genetic diversity of Salmonella spp. in a river in a tropical environment in Mexico. J. Water Health 2014, 12, 874–884. [Google Scholar] [CrossRef]
- Greene, S.K.; Daly, E.R.; Talbot, E.A.; Demma, L.J.; Holzbauer, S.; Patel, N.J.; Hill, T.A.; Walderhaug, M.O.; Hoekstra, R.M.; Lynch, M.F.; et al. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol. Infect. 2008, 136, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Gorski, L.; Liang, A.S.; Walker, S.; Carychao, D.; Aviles Noriega, A.; Mandrell, R.E.; Cooley, M.B. Salmonella enterica Serovar Diversity, Distribution, and Prevalence in Public-Access Waters from a Central California Coastal Leafy Green-Growing Region from 2011 to 2016. Appl. Environ. Microbiol. 2022, 88, e0183421. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. National Enteric Disease Surveillance: Salmonella Annual Report; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2016. Available online: https://stacks.cdc.gov/view/cdc/58450 (accessed on 1 May 2025).
- Ayyagari, A.; Chander, J.; Narang, A.; Banerjee, C.K.; Panigrahi, D.; Bhakoo, O.N.; Sarkar, S. Outbreak of Salmonella worthington meningitis & septicaemia in a hospital at Chandigarh (north India). Indian J. Med. Res. 1990, 91, 15–17. [Google Scholar]
- Plumb, I.D.; Brown, A.C.; Stokes, E.K.; Chen, J.C.; Carleton, H.; Tolar, B.; Sundararaman, P.; Saupe, A.; Payne, D.C.; Shah, H.J.; et al. Increased Multidrug-Resistant Salmonella enterica I Serotype 4,[5],12:i:- Infections Associated with Pork, United States, 2009–2018. Emerg. Infect. Dis. 2023, 29, 314–322. [Google Scholar] [CrossRef]
- Bouallegue-Godet, O.; Ben Salem, Y.; Fabre, L.; Demartin, M.; Grimont, P.A.; Mzoughi, R.; Weill, F.X. Nosocomial outbreak caused by Salmonella enterica serotype Livingstone producing CTX-M-27 extended-spectrum beta-lactamase in a neonatal unit in Sousse, Tunisia. J. Clin. Microbiol. 2005, 43, 1037–1044. [Google Scholar] [CrossRef]
- Li, Y.; Teng, L.; Xu, X.; Li, X.; Peng, X.; Zhou, X.; Du, J.; Tang, Y.; Jiang, Z.; Wang, Z.; et al. A nontyphoidal Salmonella serovar domestication accompanying enhanced niche adaptation. EMBO Mol. Med. 2022, 14, e16366. [Google Scholar] [CrossRef]
- Sjolund-Karlsson, M.; Howie, R.; Krueger, A.; Rickert, R.; Pecic, G.; Lupoli, K.; Folster, J.P.; Whichard, J.M. CTX-M-producing non-Typhi Salmonella spp. isolated from humans, United States. Emerg. Infect. Dis. 2011, 17, 97–99. [Google Scholar] [CrossRef]
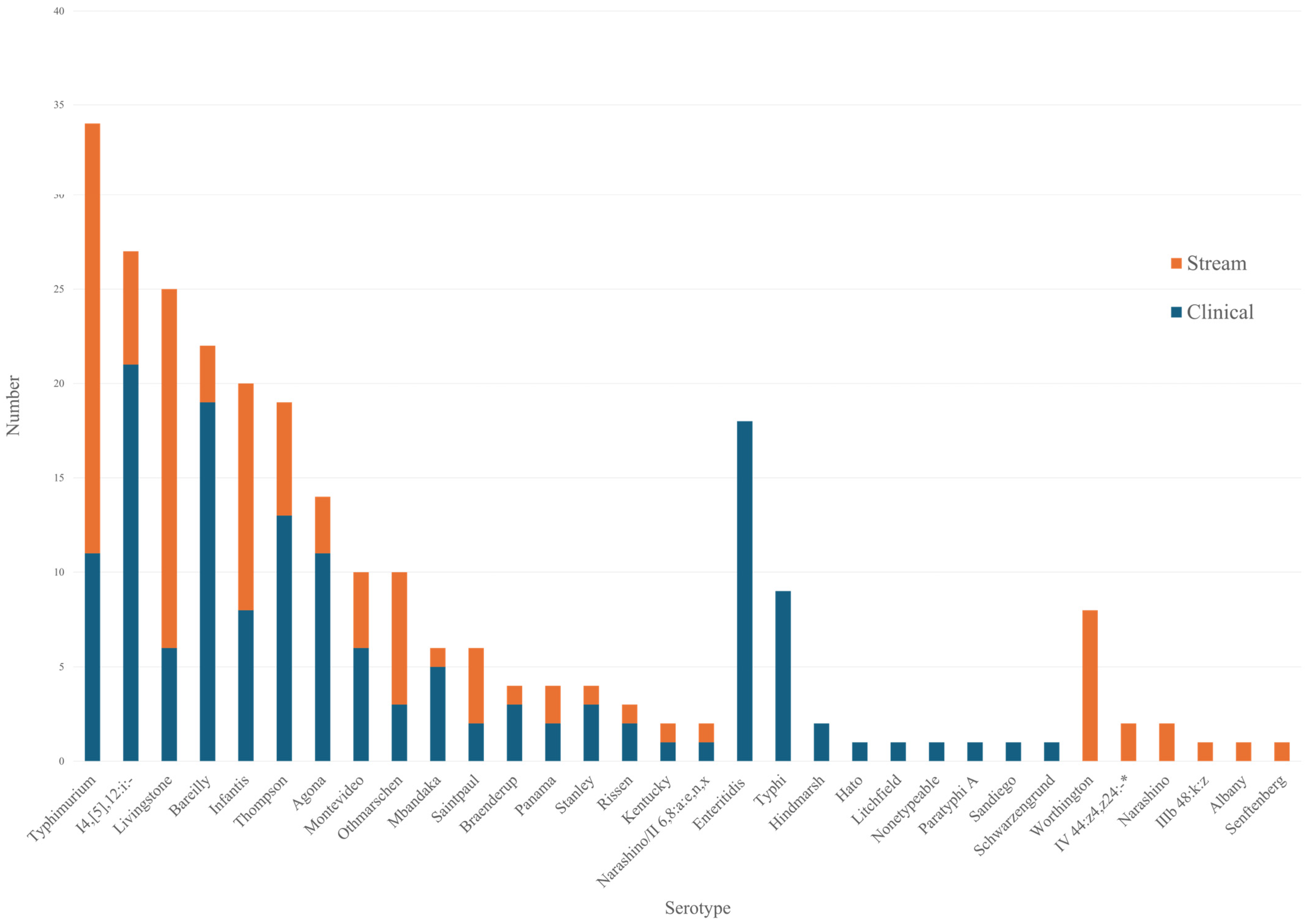
| Serogroup B (N = 37) | Serogroup C (N = 59) | Serogroup D (N = 2) | ||||||
|---|---|---|---|---|---|---|---|---|
| Serotype | No. | % | Serotype | No. | % | Serotype | No. | % |
| S. Typhimurium | 23 | 20.9% | S. Livingstone | 19 | 17.27% | S. Panama | 2 | 1.8% |
| S. I 4,[5],12:i:- | 6 | 5.5% | S. Infantis | 12 | 10.91% | Serogroup E (N = 1) | ||
| S. Saintpaul | 4 | 3.6% | S. Othmarschen | 7 | 6.36% | Serotype | No. | % |
| S. Agona | 3 | 2.7% | S. Thompson | 6 | 5.45% | S. Senftenberg | 1 | 0.9% |
| S. Stanley | 1 | 0.9% | S. Montevideo | 4 | 3.64% | Serogroup G (N = 8) | ||
| S. Bareilly | 3 | 2.73% | Serotype | No. | % | |||
| S. Narashino | 2 | 1.82% | S. Worthington | 8 | 7.3% | |||
| S. Albany | 1 | 0.91% | Serogroup V (N = 2) | |||||
| S. Braenderup | 1 | 0.91% | Serotype | No. | % | |||
| S. Kentucky | 1 | 0.91% | IV 44:z4,z24:/Christiansborg/IIIa 44:z4,z24:- | 2 | 1.8% | |||
| S. Mbandaka | 1 | 0.91% | Serogroup Y (N = 1) | |||||
| S. Rissen | 1 | 0.91% | Serotype | No. | % | |||
| S. Narashino/II 6,8:a:e,n,x | 1 | 0.91% | IIIb48:k:z | 1 | 0.9% | |||
| Antimicrobial Agents | Stream (%) | Clinical (%) | ||||
|---|---|---|---|---|---|---|
| S | I | R | S | I | R | |
| Ampicillin | 72.7 | 0 | 27.3 | 80.7 | 0 | 19.3 |
| Azithromycin | 100 | 0 | 0 | 100 | 0 | 0 |
| Chloramphenicol | 80.9 | 0 | 19.1 | 88 | 2 | 10 |
| Cefotaxime | 99.1 | 0 | 0.9 | 97.2 | 0.4 | 2.4 |
| Ciprofloxacin | 73.6 | 26.4 | 0 | 77.1 | 20.5 | 2.4 |
| Trimethoprime-sulfamethoxazole | 72.7 | 0 | 27.3 | 93.2 | 0.8 | 6 |
| Tetracycline | 79.1 | 0 | 20.9 | 82.3 | 0 | 17.7 |
| Imipenem | 100 | 0 | 0 | 100 | 0 | 0 |
| Serotype | gyrA | parC | No. |
|---|---|---|---|
| Othmarschen | D87G | T57S | 1 |
| I 4,[5],12:i:- | D87N | No mutation | 6 |
| Agona | No mutation | T57S | 1 |
| Senftenberg | No mutation | T57S | 1 |
| Albany | S83F | T57S | 1 |
| Total | 10 |
| Serotype | Origin | Genome Size (bp) | Contigs | Percent G + C | No. Proteins | MLST | cgMLST | % Allele Matches |
|---|---|---|---|---|---|---|---|---|
| S. Mbandaka | Stream | 4,659,350 | 9 | 52.22 | 4332 | 413 | 110,743 | 91.91 |
| Patient | 4,809,463 | 7 | 52.06 | 4502 | 413 | 123,743 | 91.84 | |
| Patient | 4,648,559 | 12 | 52.22 | 4333 | 413 | 110,743 | 91.97 | |
| Patient | 4,648,996 | 13 | 52.22 | 4333 | 413 | 110,743 | 91.94 | |
| Patient | 4,648,490 | 12 | 52.22 | 4332 | 413 | 110,743 | 91.94 | |
| Patient | 4,655,192 | 10 | 52.22 | 4334 | 413 | 110,743 | 91.91 | |
| S. Livingstone | Stream | 4,702,613 | 12 | 52.18 | 4375 | 543 | 73,367 | 91.64 |
| Patient | 4,707,635 | 11 | 52.16 | 4369 | 543 | 73,367 | 91.54 | |
| Patient | 4,692,629 | 9 | 52.16 | 4359 | 543 | 73,367 | 91.57 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.H.; Sung, G.-H.; Park, E.H.; Hwang, S.N.; Kim, E.-Y.; You, E.; Lee, J.Y.; Kim, G.R.; Jeong, J.; Kim, S.; et al. Evaluating the Aquatic Environment as a Reservoir for Salmonella: A Comparative Analysis with Clinical Strains. Microorganisms 2025, 13, 2072. https://doi.org/10.3390/microorganisms13092072
Kim SH, Sung G-H, Park EH, Hwang SN, Kim E-Y, You E, Lee JY, Kim GR, Jeong J, Kim S, et al. Evaluating the Aquatic Environment as a Reservoir for Salmonella: A Comparative Analysis with Clinical Strains. Microorganisms. 2025; 13(9):2072. https://doi.org/10.3390/microorganisms13092072
Chicago/Turabian StyleKim, Si Hyun, Gyung-Hye Sung, Eun Hee Park, Suk Nam Hwang, Eun-Young Kim, Eunkyoung You, Ja Young Lee, Gyu Ri Kim, Joseph Jeong, Sunjoo Kim, and et al. 2025. "Evaluating the Aquatic Environment as a Reservoir for Salmonella: A Comparative Analysis with Clinical Strains" Microorganisms 13, no. 9: 2072. https://doi.org/10.3390/microorganisms13092072
APA StyleKim, S. H., Sung, G.-H., Park, E. H., Hwang, S. N., Kim, E.-Y., You, E., Lee, J. Y., Kim, G. R., Jeong, J., Kim, S., & Shin, J. H. (2025). Evaluating the Aquatic Environment as a Reservoir for Salmonella: A Comparative Analysis with Clinical Strains. Microorganisms, 13(9), 2072. https://doi.org/10.3390/microorganisms13092072






