Ethanol-Induced Dysbiosis and Systemic Impact: A Meta-Analytical Synthesis of Human and Animal Research
Abstract
1. Introduction
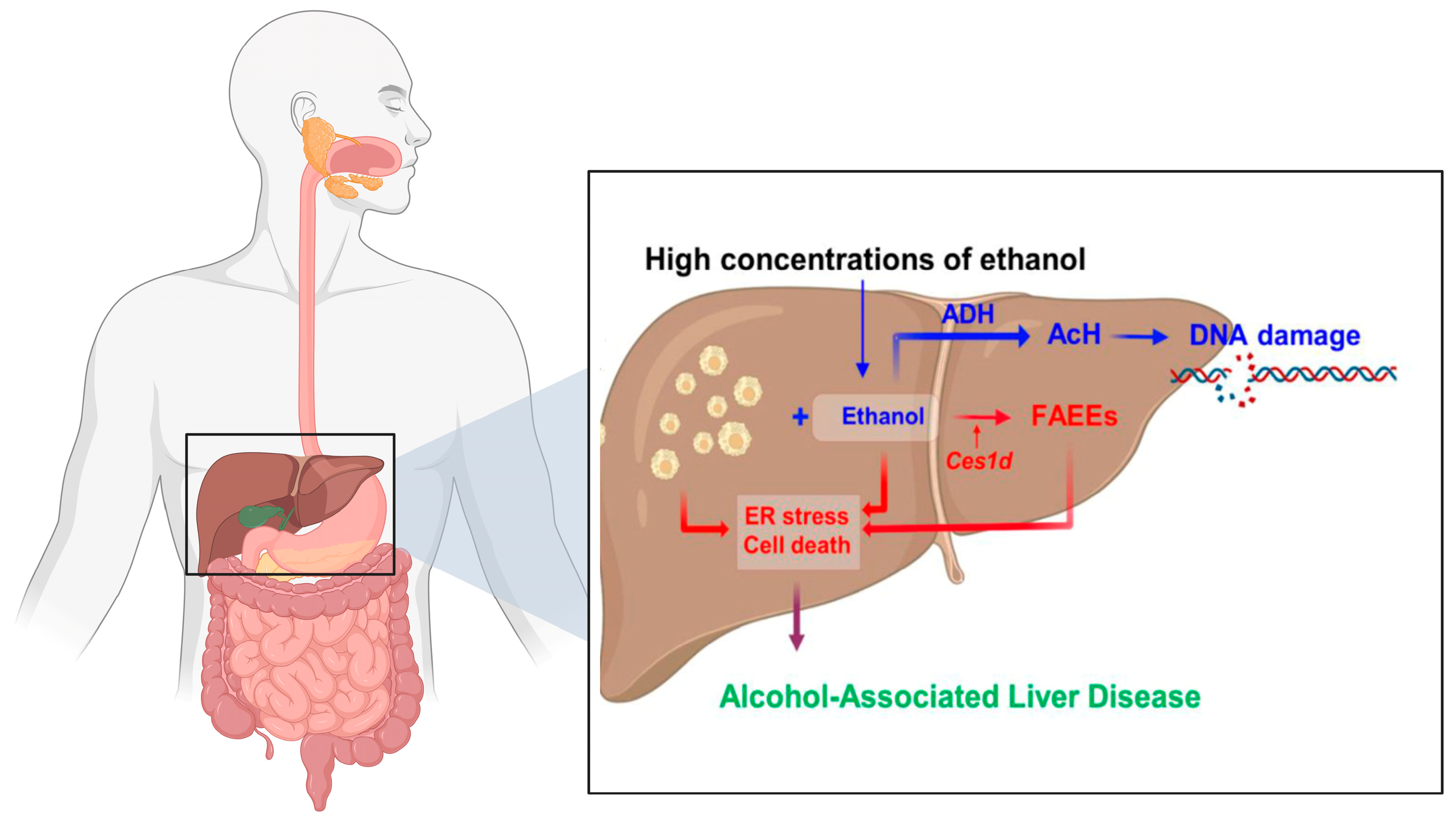
1.1. Ethanol and Its Metabolic Impact on the GI Tract
1.2. Alcohol-Induced Gut Barrier Dysfunction
1.3. Gut Microbiota Dysbiosis in Alcohol-Related Disease
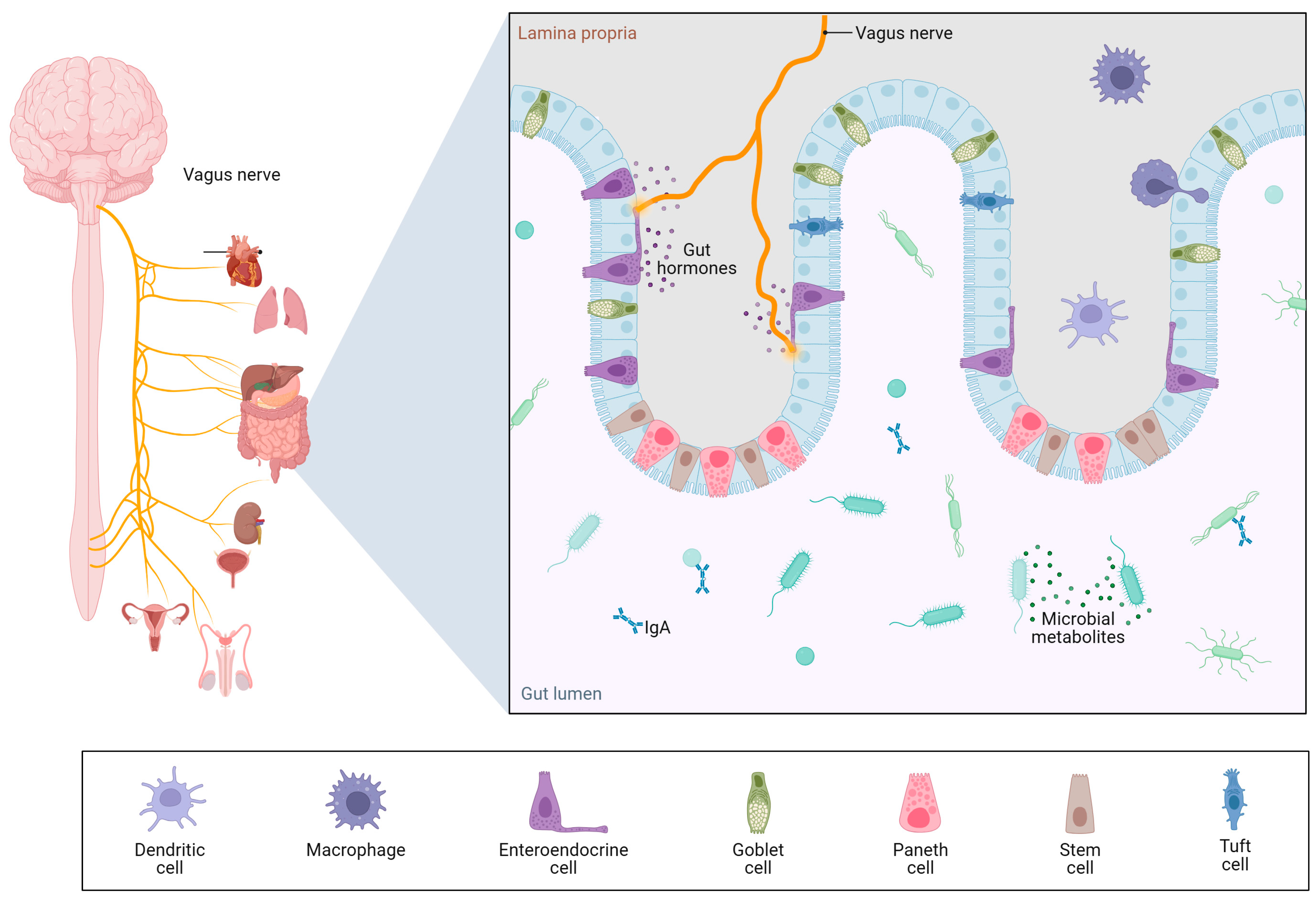
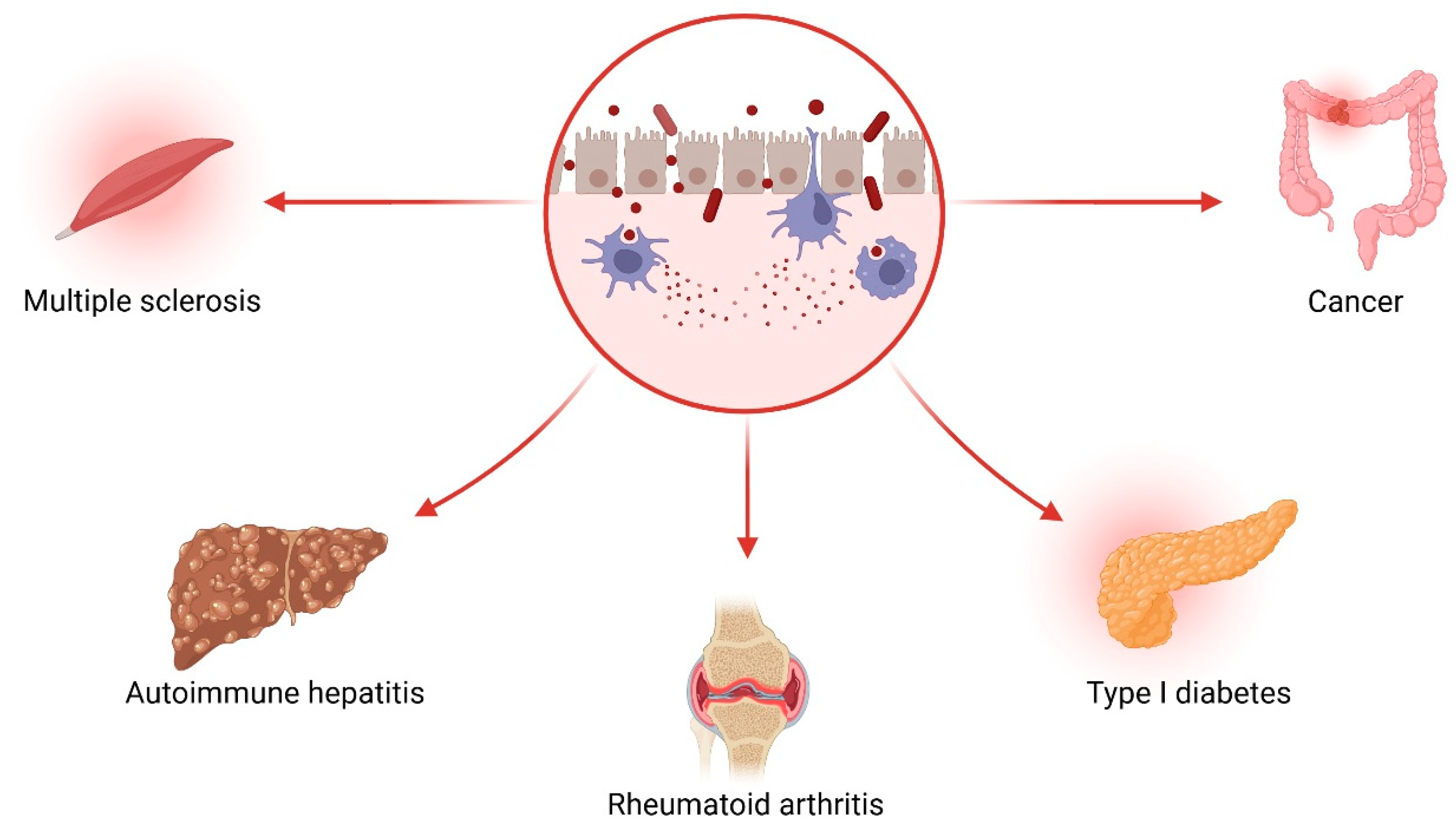
1.4. The Gut–Liver–Brain Axis: A Triangular Pathophysiological Circuit
1.5. Therapeutic Potential of Targeting the Gut Microbiota
2. Materials and Methods
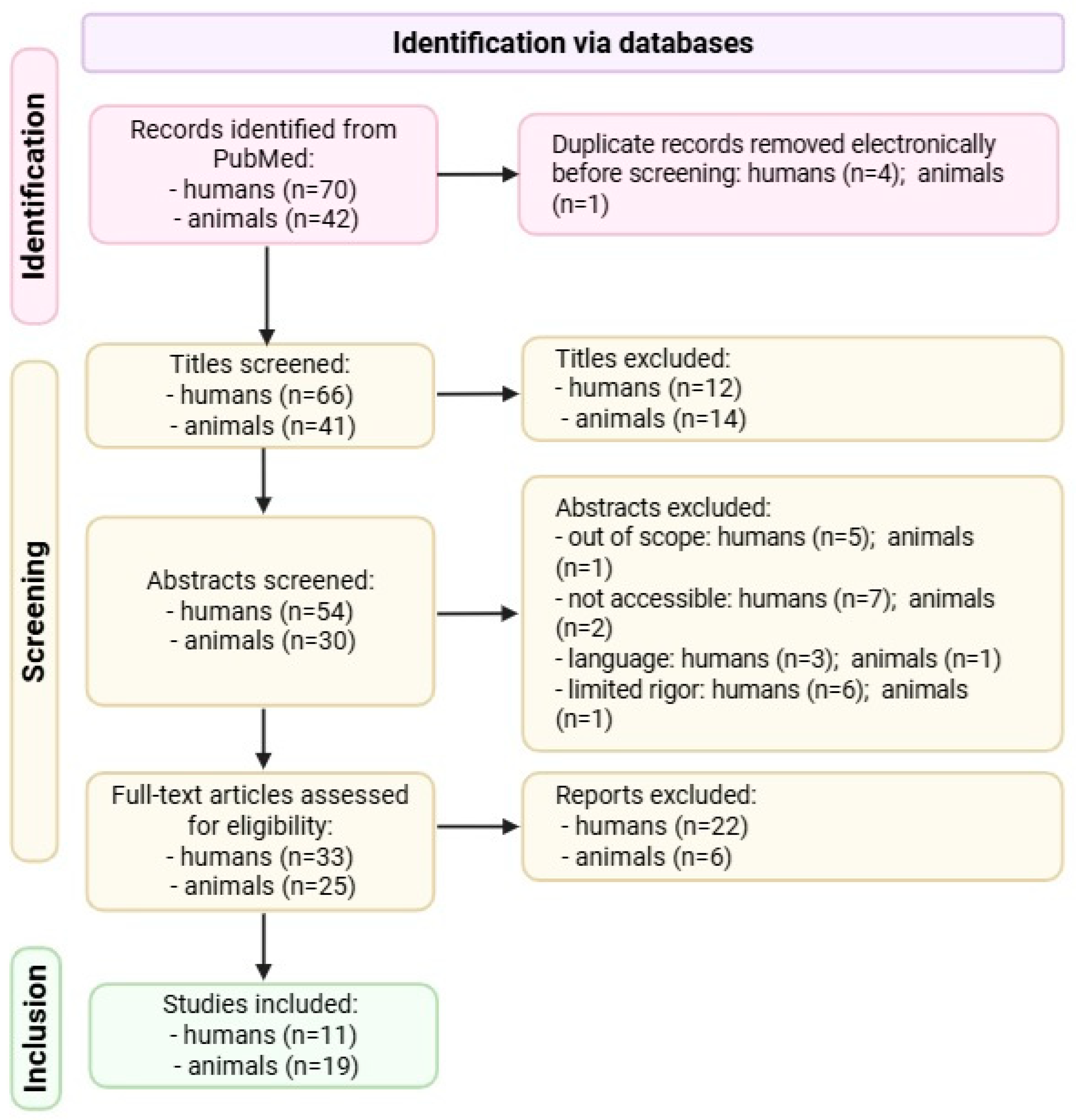
2.1. Strategy for Searching the Literature
2.2. Inclusion and Exclusion Criteria
- Human studies: eligible if they investigated individuals with chronic alcohol use or alcohol dependence, assessed gut microbiota composition using sequencing or molecular tools, and included a comparator group (e.g., healthy controls or non-drinkers).
- Animal studies: included if the animals were exposed to ethanol via drinking water, liquid diet, vapor exposure, or gavage and reported microbiota outcomes assessed via validated methods (e.g., 16S rRNA sequencing, qPCR).
- There were no geographical restrictions in study selection.
- Lack of a control group, absence of microbiota outcome data;
- Non-ethanol-related interventions (e.g., antibiotics alone, prebiotics);
- Language other than English;
- Limited methodological rigor;
- Inaccessible full text.
2.3. Screening and Data Extraction
2.4. Quality Assessment
2.5. Data Synthesis and Analysis
3. Results
3.1. Human Cohort Findings: Microbiota Diversity and Clinical Outcomes
3.1.1. Population and Study Characteristics
3.1.2. Effect Sizes for Human Studies
3.1.3. Microbiota Alterations
- Decreases: Ruminococcaceae, Lachnospiraceae, Faecalibacterium, Akkermansia, Bifidobacterium;
- Increases: Enterococcus, Streptococcus, Veillonella, Proteobacteria (especially Enterobacteriaceae) (Figure 8).
3.2. Animal Model Findings
3.2.1. Effect Sizes for Animal Model Studies
3.2.2. Intervention Outcomes
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- EE6F72A. Datadot. Available online: https://data.who.int/indicators/i/EF38E6A/EE6F72A (accessed on 12 June 2025).
- Jew, M.H.; Hsu, C.L. Alcohol, the gut microbiome, and liver disease. J. Gastroenterol. Hepatol. 2023, 38, 1205–1210. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Maccioni, L.; Fu, Y.; Horsmans, Y.; Leclercq, I.; Stärkel, P.; Kunos, G.; Gao, B. Alcohol-associated bowel disease: New insights into pathogenesis. Egastroenterology 2023, 1, e100013. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lieber, C.S. Alcohol metabolism. In Elsevier eBooks; Elsevier: Amsterdam, The Netherlands, 2004; pp. 28–32. [Google Scholar] [CrossRef]
- Scientific Image and Illustration Software|BioRender. Available online: https://www.biorender.com/ (accessed on 12 June 2025).
- Chirila, S.; Hangan, T.; Gurgas, L.; Costache, M.G.; Vlad, M.A.; Nitu, B.F.; Bittar, S.M.; Craciun, A.; Condur, L.; Bjørklund, G. Pharmacy-Based Influenza Vaccination: A Study of Patient Acceptance in Romania. Risk Manag. Healthc. Policy 2024, 17, 1005–1013. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Surdu, T.-V.; Surdu, M.; Surdu, O.; Franciuc, I.; Tucmeanu, E.-R.; Tucmeanu, A.-I.; Serbanescu, L.; Tica, V.I. Microvascular Responses in the Dermis and Muscles After Balneotherapy: Results from a Prospective Pilot Histological Study. Water 2025, 17, 1830. [Google Scholar] [CrossRef]
- Madigan. Brock Biology of Microorganisms: (International Edition): With How to Write about Biology; Prentice Hall: Hoboken, NJ, USA, 2003. [Google Scholar]
- Voiosu, T.; Voiosu, A.; Danielescu, C.; Popescu, D.; Puscasu, C.; State, M.; Chiricuţă, A.; Mardare, M.; Spanu, A.; Bengus, A.; et al. Unmet needs in the diagnosis and treatment of Romanian patients with bilio-pancreatic tumors: Results of a prospective observational multicentric study. Rom. J. Intern. Med. 2021, 59, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Bansal, M.B. Role of kupffer cells in driving hepatic inflammation and fibrosis in HIV infection. Front. Immunol. 2020, 11, 1086. [Google Scholar] [CrossRef] [PubMed]
- Colella, M.; Charitos, I.A.; Ballini, A.; Cafiero, C.; Topi, S.; Palmirotta, R.; Santacroce, L. Microbiota revolution: How gut microbes regulate our lives. World J. Gastroenterol. 2023, 29, 4368–4383. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mousa, W.K.; Chehadeh, F.; Husband, S. Microbial dysbiosis in the gut drives systemic autoimmune diseases. Front. Immunol. 2022, 13, 906258. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Szychlinska, M.A.; Di Rosa, M.; Castorina, A.; Mobasheri, A.; Musumeci, G. A correlation between intestinal microbiota dysbiosis and osteoarthritis. Heliyon 2019, 5, e01134. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sosnowski, K.; Przybyłkowski, A. Ethanol-induced changes to the gut microbiome compromise the intestinal homeostasis: A review. Gut Microbes 2024, 16, 2393272. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Islam, M.M.; Mahbub, N.U.; Hong, S.; Chung, H. Gut bacteria: An etiological agent in human pathological conditions. Front. Cell. Infect. Microbiol. 2024, 14, 1291148. [Google Scholar] [CrossRef]
- Daniel-MacDougall, C. How Does Alcohol Affect the Microbiome? Available online: https://www.mdanderson.org/cancerwise/how-does-alcohol-affect-the-microbiome.h00-159696756.html (accessed on 12 June 2025).
- Sugisawa, E.; Kondo, T.; Kumagai, Y.; Kato, H.; Takayama, Y.; Isohashi, K.; Shimosegawa, E.; Takemura, N.; Hayashi, Y.; Sasaki, T.; et al. Nociceptor-derived Reg3γ prevents endotoxic death by targeting kynurenine pathway in microglia. Cell Rep. 2022, 38, 110462. [Google Scholar] [CrossRef] [PubMed]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277, Erratum in Front. Immunol. 2019, 10, 1486. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Koutromanos, I.; Legaki, E.; Gazouli, M.; Vasilopoulos, E.; Kouzoupis, A.; Tzavellas, E. Gut microbiome in alcohol use disorder: Implications for health outcomes and therapeutic strategies-a literature review. World J. Methodol. 2024, 14, 88519. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ashique, S.; Mohanto, S.; Ahmed, M.G.; Mishra, N.; Garg, A.; Chellappan, D.K.; Omara, T.; Iqbal, S.; Kahwa, I. Gut-brain axis: A cutting-edge approach to target neurological disorders and potential synbiotic application. Heliyon 2024, 10, e34092. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Santana, A.; Diaz Heijtz, R. Bacterial Peptidoglycans from Microbiota in Neurodevelopment and Behavior. Trends Mol. Med. 2020, 26, 729–743. [Google Scholar] [CrossRef] [PubMed]
- Di Vincenzo, F.; Del Gaudio, A.; Petito, V.; Lopetuso, L.R.; Scaldaferri, F. Gut microbiota, intestinal permeability, and systemic inflammation: A narrative review. Intern. Emerg. Med. 2024, 19, 275–293. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kalapatapu, N.; Skinner, S.G.; D’Addezio, E.G.; Ponna, S.; Cadenas, E.; Davies, D.L. Thiamine Deficiency and Neuroinflammation Are Important Contributors to Alcohol Use Disorder. Pathophysiology 2025, 32, 34. [Google Scholar] [CrossRef]
- Pisarello, M.J.L.; Marquez, A.; Chaia, A.P.; Babot, J.D. Targeting gut health: Probiotics as promising therapeutics in alcohol-related liver disease management. AIMS Microbiol. 2025, 11, 410–435. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Anouti, A.; Kerr, T.A.; Mitchell, M.C.; Cotter, T.G. Advances in the management of alcohol-associated liver disease. Gastroenterol. Rep. 2024, 12, goae097. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jiang, Y.; Qu, Y.; Shi, L.; Ou, M.; Du, Z.; Zhou, Z.; Zhou, H.; Zhu, H. The role of gut microbiota and metabolomic pathways in modulating the efficacy of SSRIs for major depressive disorder. Transl. Psychiatry 2024, 14, 493. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rathore, K.; Shukla, N.; Naik, S.; Sambhav, K.; Dange, K.; Bhuyan, D.; Imranul Haq, Q.M. The Bidirectional Relationship Between the Gut Microbiome and Mental Health: A Comprehensive Review. Cureus 2025, 17, e80810. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- DeMars, M.M.; Perruso, C. MeSH and text-word search strategies: Precision, recall, and their implications for library instruction. J. Med. Libr. Assoc. 2022, 110, 23–33. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- MacFarlane, A.; Russell-Rose, T.; Shokraneh, F. Search strategy formulation for systematic reviews: Issues, challenges and opportunities. Intell. Syst. Appl. 2022, 15, 200091. [Google Scholar] [CrossRef]
- PRISMA 2020 Flow Diagram—PRISMA Statement. PRISMA Statement. Available online: https://www.prisma-statement.org/prisma-2020-flow-diagram (accessed on 12 June 2025).
- Systematic Review Centre for Laboratory Animal Experimentation (SYRCLE). Available online: https://norecopa.no/3r-guide/systematic-review-centre-for-laboratory-animal-experimentation-syrcle (accessed on 12 June 2025).
- Ottawa Hospital Research Institute. Copyright 2011 Ottawa Hospital Research Institute. All Rights Reserved. Available online: https://www.ohri.ca/programs/clinical_epidemiology/oxford.asp (accessed on 12 June 2025).
- Du, Y.; Li, L.; Gong, C.; Li, T.; Xia, Y. The diversity of the intestinal microbiota in patients with alcohol use disorder and its relationship to alcohol consumption and cognition. Front. Psychiatry 2022, 13, 1054685. [Google Scholar] [CrossRef]
- Dedon, L.R.; Yuan, H.; Chi, J. Hu, H.; Arias, A.J.; Covault, J.M.; Zhou, Y. Baseline gut microbiome and metabolites are correlated with changes in alcohol consumption in participants in a randomized Zonisamide clinical trial. Sci. Rep. 2025, 15, 10486. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, R.; Deng, H.; Cui, P.; Li, C.; Yang, F.; Leong Bin Abdullah, M.F.I. Research protocol of the efficacy of probiotics for the treatment of alcohol use disorder among adult males: A comparison with placebo and acceptance and commitment therapy in a randomized controlled trial. PLoS ONE 2023, 18, e0294768. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bajaj, J.S.; Gavis, E.A.; Fagan, A.; Wade, J.B.; Thacker, L.R.; Fuchs, M.; Patel, S.; Davis, B.; Meador, J.; Puri, P.; et al. A Randomized Clinical Trial of Fecal Microbiota Transplant for Alcohol Use Disorder. Hepatology 2021, 73, 1688–1700. [Google Scholar] [CrossRef] [PubMed]
- Amadieu, C.; Ahmed, H.; Leclercq, S.; Koistinen, V.; Leyrolle, Q.; Stärkel, P.; Bindels, L.B.; Layé, S.; Neyrinck, A.M.; Kärkkäinen, O.; et al. Effect of inulin supplementation on fecal and blood metabolome in alcohol use disorder patients: A randomised, controlled dietary intervention. Clin. Nutr. ESPEN 2025, 66, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Philips, C.A.; Ahamed, R.; Oommen, T.T.; Nahaz, N.; Tharakan, A.; Rajesh, S.; Augustine, P. Clinical outcomes and associated bacterial and fungal microbiota changes after high dose probiotic therapy for severe alcohol-associated hepatitis: An observational study. Medicine 2024, 103, e40429. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Muthiah, M.D.; Smirnova, E.; Puri, P.; Chalasani, N.; Shah, V.H.; Kiani, C.; Taylor, S.; Mirshahi, F.; Sanyal, A.J. Development of Alcohol-Associated Hepatitis Is Associated With Specific Changes in Gut-Modified Bile Acids. Hepatol. Commun. 2022, 6, 1073–1089. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, J.; Li, C.; Duan, M.; Qu, Z.; Wang, Y.; Dong, Y.; Wu, Y.; Fang, S.; Gu, S. The Improvement Effects of Weizmannia coagulans BC99 on Liver Function and Gut Microbiota of Long-Term Alcohol Drinkers: A Randomized Double-Blind Clinical Trial. Nutrients 2025, 17, 320. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lang, S.; Fairfied, B.; Gao, B.; Duan, Y.; Zhang, X.; Fouts, D.E.; Schnabl, B. Changes in the fecal bacterial microbiota associated with disease severity in alcoholic hepatitis patients. Gut Microbes 2020, 12, 1785251. [Google Scholar] [CrossRef] [PubMed Central]
- Haas, E.A.; Saad, M.J.A.; Santos, A.; Vitulo, N.; Lemos, W.J.F.; Martins, A.M.A.; Picossi, C.R.C.; Favarato, D.; Gaspar, R.S.; Magro, D.O.; et al. A red wine intervention does not modify plasma trimethylamine N-oxide but is associated with broad shifts in the plasma metabolome and gut microbiota composition. Am. J. Clin. Nutr. 2022, 116, 1515–1529. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Han, S.H.; Suk, K.T.; Kim, D.J.; Kim, M.Y.; Baik, S.K.; Kim, Y.D.; Cheon, G.J.; Choi, D.H.; Ham, Y.L.; Shin, D.H.; et al. Effects of probiotics (cultured Lactobacillus subtilis/Streptococcus faecium) in the treatment of alcoholic hepatitis: Randomized-controlled multicenter study. Eur. J. Gastroenterol. Hepatol. 2015, 27, 1300–1306. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Ubilla, M.; Figueroa-Valdés, A.I.; Tobar, H.E.; Quintanilla, M.E.; Díaz, E.; Morales, P.; Berríos-Cárcamo, P.; Santapau, D.; Gallardo, J.; de Gregorio, C.; et al. Gut Microbiota-Derived Extracellular Vesicles Influence Alcohol Intake Preferences in Rats. J. Extracell. Vesicles 2025, 14, e70059. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xu, Z.; Wang, C.; Dong, X.; Hu, T.; Wang, L.; Zhao, W.; Zhu, S.; Li, G.; Hu, Y.; Gao, Q.; et al. Chronic alcohol exposure induced gut microbiota dysbiosis and its correlations with neuropsychic behaviors and brain BDNF/Gabra1 changes in mice. Biofactors 2019, 45, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.W.; Ge, C.; Feng, G.X.; Li, Y.; Luo, D.; Dong, J.L.; Li, H.; Wang, H.; Cui, M.; Fan, S.J. Gut microbiota modulates alcohol withdrawal-induced anxiety in mice. Toxicol. Lett. 2018, 287, 23–30. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, G.; Liu, Q.; Guo, L.; Zeng, H.; Ding, C.; Zhang, W.; Xu, D.; Wang, X.; Qiu, J.; Dong, Q.; et al. Gut Microbiota and Relevant Metabolites Analysis in Alcohol Dependent Mice. Front. Microbiol. 2018, 9, 1874. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, X.; Liang, H. Effects of Lactobacillus casei on Iron Metabolism and Intestinal Microflora in Rats Exposed to Alcohol and Iron. Turk. J. Gastroenterol. 2022, 33, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, H.; Lin, X.; Liu, J.; Feng, Y.; Bai, Y.; Liang, H.; Hu, T.; Wu, Z.; Lai, J.; et al. Gut microbiota dysbiosis induced by alcohol exposure in pubertal and adult mice. mSystems 2024, 9, e0136624. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yi, S.; Zhang, G.; Liu, M.; Yu, W.; Cheng, G.; Luo, L.; Ning, F. Citrus Honey Ameliorates Liver Disease and Restores Gut Microbiota in Alcohol-Feeding Mice. Nutrients 2023, 15, 1078. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xia, T.; Duan, W.; Zhang, Z.; Li, S.; Zhao, Y.; Geng, B.; Zheng, Y.; Yu, J.; Wang, M. Polyphenol-rich vinegar extract regulates intestinal microbiota and immunity and prevents alcohol-induced inflammation in mice. Food Res. Int. 2021, 140, 110064. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Ya-E, Z.; Ji-Dong, W.; Yu-Fan, L.; Ying, Z.; Ya-Lun, S.; Meng-Yu, M.; Rui-Ling, Z. Comparison of Microbial Diversity and Composition in Jejunum and Colon of the Alcohol-dependent Rats. J. Microbiol. Biotechnol. 2018, 28, 1883–1895. [Google Scholar] [CrossRef] [PubMed]
- Hendrikx, T.; Duan, Y.; Wang, Y.; Oh, J.H.; Alexander, L.M.; Huang, W.; Stärkel, P.; Ho, S.B.; Gao, B.; Fiehn, O.; et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 2019, 68, 1504–1515. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, P.; Miyamoto, Y.; Mazagova, M.; Lee, K.C.; Eckmann, L.; Schnabl, B. Microbiota Protects Mice Against Acute Alcohol-Induced Liver Injury. Alcohol. Clin. Exp. Res. 2015, 39, 2313–2323. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, F.; Wei, J.; Shen, M.; Ding, Y.; Lu, Y.; Ishaq, H.M.; Li, D.; Yan, D.; Wang, Q.; Zhang, R. Integrated Analyses of the Gut Microbiota, Intestinal Permeability, and Serum Metabolome Phenotype in Rats with Alcohol Withdrawal Syndrome. Appl. Environ. Microbiol. 2021, 87, e0083421. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xue, M.; Liu, Y.; Lyu, R.; Ge, N.; Liu, M.; Ma, Y.; Liang, H. Protective effect of aplysin on liver tissue and the gut microbiota in alcohol-fed rats. PLoS ONE 2017, 12, e0178684. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mittal, A.; Choudhary, N.; Kumari, A.; Yadav, K.; Maras, J.S.; Sarin, S.K.; Sharma, S. Protein supplementation differentially alters gut microbiota and associated liver injury recovery in mouse model of alcohol-related liver disease. Clin. Nutr. 2025, 46, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Thoen, R.U.; Longo, L.; Leonhardt, L.C.; Pereira, M.H.M.; Rampelotto, P.H.; Cerski, C.T.S.; Álvares-da-Silva, M.R. Alcoholic liver disease and intestinal microbiota in an experimental model: Biochemical, inflammatory, and histologic parameters. Nutrition 2023, 106, 111888. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, Y.; Gao, M.; Xue, M.; Wang, Z.; Liang, H. Nicotinamide riboside alleviates alcohol-induced depression-like behaviours in C57BL/6J mice by altering the intestinal microbiota associated with microglial activation and BDNF expression. Food Funct. 2020, 11, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yasuda, K.; Gilmore, R.A.; Westmoreland, S.V.; Platt, D.M.; Miller, G.M.; Vallender, E.J. Alcohol-induced changes in the gut microbiome and metabolome of rhesus macaques. Psychopharmacology 2019, 236, 1531–1544. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cunningham, K.C.; Smith, D.R.; Villageliú, D.N.; Ellis, C.M.; Ramer-Tait, A.E.; Price, J.D.; Wyatt, T.A.; Knoell, D.L.; Samuelson, M.M.; Molina, P.E.; et al. Human Alcohol-Microbiota Mice have Increased Susceptibility to Bacterial Pneumonia. Cells 2023, 12, 2267. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, C.; Yan, J.; Du, K.; Liu, S.; Wang, J.; Wang, Q.; Zhao, H.; Li, M.; Yan, D.; Zhang, R.; et al. Intestinal microbiome dysbiosis in alcohol-dependent patients and its effect on rat behaviors. mBio 2023, 14, e0239223. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yan, J.; Sheng, L.; Li, H. Akkermansia muciniphila: Is it the Holy Grail for ameliorating metabolic diseases? Gut Microbes 2021, 13, 1984104. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nguyen, T.L.; Vieira-Silva, S.; Liston, A.; Raes, J. How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 2015, 8, 1–16. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chancharoenthana, W.; Kamolratanakul, S.; Udompornpitak, K.; Wannigama, D.L.; Schultz, M.J.; Leelahavanichkul, A. Alcohol-induced gut permeability defect through dysbiosis and enterocytic mitochondrial interference causing pro-inflammatory macrophages in a dose dependent manner. Sci. Rep. 2025, 15, 14710. [Google Scholar] [CrossRef]
- Wei, L.; Pan, Y.; Guo, Y.; Zhu, Y.; Jin, H.; Gu, Y.; Li, C.; Wang, Y.; Lin, J.; Chen, Y.; et al. Symbiotic combination of Akkermansia muciniphila and inosine alleviates alcohol-induced liver injury by modulating gut dysbiosis and immune responses. Front. Microbiol. 2024, 15, 1355225. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Capurso, G.; Lahner, E. The interaction between smoking, alcohol and the gut microbiome. Best. Pract. Res. Clin. Gastroenterol. 2017, 31, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, E.; Puri, P.; Muthiah, M.D.; Daitya, K.; Brown, R.; Chalasani, N.; Liangpunsakul, S.; Shah, V.H.; Gelow, K.; Siddiqui, M.S.; et al. Fecal Microbiome Distinguishes Alcohol Consumption From Alcoholic Hepatitis But Does Not Discriminate Disease Severity. Hepatology 2020, 72, 271–286. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pu, W.; Zhang, H.; Zhang, T.; Guo, X.; Wang, X.; Tang, S. Inhibitory effects of Clostridium butyricum culture and supernatant on inflammatory colorectal cancer in mice. Front. Immunol. 2023, 14, 1004756. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nikel, K.; Stojko, M.; Smolarczyk, J.; Piegza, M. The Impact of Gut Microbiota on the Development of Anxiety Symptoms—A Narrative Review. Nutrients 2025, 17, 933. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]


| Study | Population | Age | Gender | Diagnosis | Treatment | Duration | Primary Outcomes | Microbiota Changes |
|---|---|---|---|---|---|---|---|---|
| Du et al. (2024) [33] | 32 AUD males and 35 healthy controls (HC) | 47.16 ± 9.89 (AUD participants) 48.00 ± 11.31 (controls) | Males | Severe alcohol-associated hepatitis, biopsy-confirmed | High-dose probiotic infusion, FMT, corticosteroids | 23.50 (20.00, 30.00) drinking days in the past month (days) | 90-day survival | Gut dysbiosis in AUD patients, and some specific microbiota, were considered to be related to alcohol intake and cognitive function. Compared with HCs, Megamonas, Escherichia, Coprobacillus, Clostridium, Gemella, and Rothia had increased in AUD patients. |
| Dedon et al. (2025) [34] | 32 AUD patients from UConn Health in zonisamide vs. placebo RCT, 19 HC | Mean, 51 years (23–70) | 47% female, 53% male | DSM-5 AUD | Zonisamide vs. placebo + behavioral therapy | 16 weeks | Drinking reduction; microbiome/metabolome baseline predictors | High fecal GABA and low 3-hydroxykynurenine predicted reduction in drinking. Gut markers may forecast treatment success. ↑ Veillonella (worse severity), ↓ Akkermansia; antibiotics ↓ Bacteroides; steroids ↑ Veillonella. |
| Zhang et al. (2023) [35] | 120 male AUD patients in Henan, China; 120 healthy controls in phase I | 18–65 years | 100% male | DSM-5 moderate to severe AUD | Probiotics vs. ACT vs. placebo | 24 weeks (12 weeks treatment + follow-up) | AUD symptoms, craving, depression, inflammation, ERP | Protocol paper—outcomes pending. Expected comparison of psychological and microbial effects of ACT vs. probiotics. Outcomes pending but targeting ↑ Lactobacillus, Bifidobacterium; expected ↑ Ruminococcaceae. |
| Bajaj et al. (2021) [36] | 20 cirrhotics with ≥2 HE episodes; randomized to SOC or SOC + FMT | 65 ± 6.4 years | Males | Recurrent hepatic encephalopathy on lactulose ± rifaximin | Oral FMT capsules vs. SOC | 5 months | HE recurrence, cognition, microbiota/inflammation | FMT reduced HE episodes, improved cognition, decreased IL-6 and LBP, and increased beneficial gut taxa. ↑ Lactobacillaceae, Bifidobacteriaceae; ↓ Proteobacteria (e.g., Enterobacteriaceae). |
| Amadieu et al. (2021) [37] | 21 subjects in the placebo group and 22 in the inulin group | 48.8 ± 8.8 years (placebo) group, 48.3 ± 9.8 years (inulin group) | 66.2% male | DSM-IV alcohol dependence, no liver cirrhosis | Patients were supplemented with inulin (prebiotic DF) or maltodextrin (placebo) | 17 days | Craving, gut permeability, cytokines, microbiota richness | Fecal metabolomics revealed 14 metabolites significantly modified by inulin versus placebo treatment (increased N8-acetylspermidine and decreased indole-3-butyric acid, 5-amino valeric acid betaine and bile acids). Fecal Lachnoclostridium correlated with 6 of the identified fecal metabolites, whereas plasma lipidic moieties positively correlated with fecal Ruminococcus torques and Flavonifractor. |
| Philips et al. (2022) [38] | 50 patients with a clinical diagnosis of biopsy-proven severe | 43.8 ± 9.4 years in the corticosteroid group; 48.4 ± 11.7 in the high-dose probiotic infusion (HDPI) | Male | Biopsy-proven severe alcohol-associated hepatitis | Oral prednisolone, 40 mg, once daily for 28 days | 180 days | Survival at 90 and 180 days; inflammation, microbiota | Those receiving HDPI demonstrated an increased relative abundance of Bilophila, Roseburia, Clostridium, Finegoldia, Butyricoccus, and Weissella at the end of 1 month compared to baseline. Yueomyces was the most abundant at baseline, whereas Myrothecium and Magnoliophyta increased 1 month after high probiotic infusion. |
| Muthiah et al. (2021) [39] | 20 healthy controls, 12 heavy-drinking controls, 11 MAH, 16 SAH, and 49 subjects with available stool (20 heavy-drinking controls, 8 MAH, 21 SAH) | 46.39 ± 13.63 | 60% males, 40% females | Alcohol-related hepatitis or cirrhosis, MELD ≥ 15 | Fresh donor FMT via nasojejunal infusion vs. SOC | 6 months | Survival, infection, microbiota | Citrobacter, Enterobacter, Puralibacter, Actinomycetaceae, Bifidobacteriaceae, Camobacteriaceae, Prevotellaceae, Pseudomonaceae, Propionibacteriaceae, and Veillonellaceae positively correlated with several fecal bile acids in alcohol-associated hepatitis. |
| Zhang et al. (2023) [40] | 60 long-term alcohol drinkers (30 placebo, 30 BC99 group) (18–65 years old, alcohol consumption ≥20 g/day, lasting for more than one year) | 42.87 ± 11.15 (BC99 group), 43.60 ± 11.31 (placebo group) | 99% males | DSM-5 AUD, stratified by BMI | Two groups were administered BC99 (3 g/day, 1 × 1010 CFU) or placebo (3 g/day) | 30 days and 60 days | Gut microbiota recovery by BMI | BC99 regulated the imbalance of intestinal flora, increased the beneficial bacteria abundance (Prevotella, Faecalibacterium, and Roseburia) and reduced the conditionally pathogenic bacteria abundance (Escherichia-Shigella and Klebsiella). |
| Lang et al. (2020) [41] | 73 ALD patients: 23 ARC, 19 AH, 24 compensated ALD + 18 controls | Median, 49.2 years (31.3–74.8) | 67.1% males | Alcohol-related cirrhosis, hepatitis, or compensated ALD | None (observational) | Single time point | Microbiota, neutrophil function, inflammation | Decreased relative abundances of Akkermansia while the relative abundance of Veillonella was increased; reduction in Bacteroides abundance, increase in Veillonella abundance. |
| Haas et al. (2022) [42] | 42 participants | Mean, 60 years | 100% male | Stable coronary artery disease | Red wine (250 mL/day, 3 weeks) vs. abstinence | 2 weeks wash-out period | Plasma TMAO, gut microbiota, metabolome | Red wine did not affect TMAO but shifted microbiota and metabolome. ↑ Parasutterella, Bacteroides, Prevotella; no change in TMAO-producing bacteria. |
| Han et al. (2015) [43] | 117 hospitalized AH patients: 60 probiotic, 57 placebo | Mean, 52.7 ± 11.3 years | 64% male | Mild alcoholic hepatitis, non-cirrhotic, recent drinking | L. subtilis + S. faecium vs. placebo + silymarin | 7 days (inpatient) | LPS, TNF-α, IL-1β, liver enzymes | Probiotics reduced TNF-α, LPS, and stool E. coli CFUs. Greatest benefit in cirrhotics. Additive to abstinence effects. ↓ E. coli CFU; probiotics modulated gut flora positively, especially in cirrhotics. |
| Study | Animal | Strain | Sex | Sample Size | Intervention | Dose | Exposure Duration | Key Microbiota Findings | Other Observations |
|---|---|---|---|---|---|---|---|---|---|
| Daaz-Ubilla et al. (2025) [44] | Rat | Wistar | Males and female | 10 per group | bEVs from ethanol-naïve and ethanol-exposed UChB rats injected intraperitoneally (3 days) | 11.4 ± 1.2 g ethanol/kg/day for 120 days | 120 days exposure in donors, 3-day bEV exposure in recipients, followed by 4-day test | Exposure to blood extracellular vesicles (bEVs) derived from ethanol-exposed UChB rats resulted in significant changes in the gut–brain axis of naïve Wistar rats. Although the bEVs did not induce systemic inflammation or changes in microglial activation, they triggered microbiota-brain interactions that increased ethanol-seeking behavior. | Behavioral tests (two-bottle choice) showed increased voluntary ethanol consumption in bEV-exposed Wistar rats. This effect was abolished when the vagus nerve was surgically cut, highlighting vagal involvement. No significant IL-6, TNF-α, or CD11b elevation in liver or brain tissues. |
| Xu et al. (2018) [45] | Mouse | C57BL | Male | Varied: n = 6 for preliminary, n = 31 alcohol group, n = 16 control (main test) | Oral ethanol in drinking water with gradient concentrations | The concentration of alcohol was increased from 2%, 4%, to 6% every 3 days and reached 8% | 21 days | Microbiota profiling using 16S rRNA sequencing showed that ethanol consumption significantly ↑ Actinobacteria and Cyanobacteria phyla. At the genus level, ↑ Adlercreutzia, Allobaculum, and Turicibacter were noted and ↓ Helicobacter. | Ethanol-treated mice displayed reduced locomotor activity, higher immobility in tail suspension and forced swim tests, and decreased open-arm exploration in the elevated plus maze, suggesting anxiety- and depression-like phenotypes. These behaviors were associated with downregulation of BDNF and Gabra gene expression in both hippocampus and prefrontal cortex. |
| Xiao et al. (2018) [46] | Mouse | C57BL | Male | n = 12 per group (water group and alcohol group) | Gavage alcohol feeding: week-wise 5% to 35% ethanol; then, withdrawal | Water for mice in water group; 5%, 10%, 20%, 35% alcoholic solution force-fed into the mice’s stomach for the alcohol group | 4 weeks | Increased Erysipelotrichia, Erysipelotrichaceae, and Erysipelotrichales, whereas Lactobacillaceae, Lactobacillus, Lactobacillale, Bacilli, Bacteroides, Parabacteroides, and Alloprevotella were significantly reduced. | Alcohol withdrawal in donor mice increased immobility in forced swim and tail suspension tests, decreased sucrose preference, and increased anxiety scores. FMT alone was sufficient to transfer these behaviors. |
| Wang et al. (2018) [47] | Mouse | BALB/c | Female | n = 10 per group, 3 groups | Active vs. forced alcohol drinking (3% → 20% over 7 weeks; then, withdrawal) | 3%, 6%, 10%, 20% alcohol progressively | 8 weeks (7 weeks alcohol + 1 week withdrawal) | During active alcohol exposure, there was a marked ↑ in Firmicutes and Clostridiales, as well as specific genera like Lachnospiraceae, Alistipes, and Odoribacter. These changes persisted after withdrawal, indicating long-term dysbiosis. Concurrently, there was increased serotonin concentration in the gut. | Histological examination revealed hepatocellular degeneration and colonic epithelial damage in alcohol-exposed mice. Behavioral assessments post-withdrawal showed significant anxiety and depression. These included decreased center time in open-field tests and increased immobility. |
| Li et al. (2022) [48] | Rat | Wistar | Male | n = 20 per group, 3 groups | Gavage ethanol (8 → 12 mL/kg/day) for 12 weeks; co-exposure with dietary iron | 56% ethanol, 8–12 mL/kg/day | 12 weeks | Rats exposed to both alcohol and high dietary iron experienced significant intestinal dysbiosis characterized by ↓ Lactobacillus and ↑ Bacteroides and E. coli. Supplementation with Lactobacillus casei reversed these alterations, restoring microbial balance towards homeostasis. | Combined alcohol and iron exposure led to elevated serum ferritin, hepcidin, and increased protein expression of intestinal DMT1 and FPN1—indicators of iron overload. L. casei supplementation significantly reduced these markers. |
| Yang et al. (2024) [49] | Mouse | C57BL/6J | Male | 20 pubertal (P27–P44), 20 adult (P60–P78) | 20% ethanol in sterile water | 20% | 2 weeks (chronic exposure); fecal samples collected at 0 h, 24 h, 1 week, 2 weeks | In pubertal mice: mild gut dysbiosis; ↓ in Lactobacillus intestinalis and Limosilactobacillus reuteri; ↑ in Bifidobacterium, Butyricimonas, and Alistipes shahii; ↓ in Turicimonas muris and L. taiwanensis. In adult mice: more severe dysbiosis with ↑ Alistipes, Bacteroides; ↓ Lactobacillus, Mucispirillum schaedleri. | Pubertal mice showed less liver and intestinal injury, increased ALDH activity, decreased ADH. Adult mice had increased mucin-degrading enzymes, liver enzyme imbalance, higher oxidative stress enzymes, and more histological damage to small intestine. |
| Yi et al. (2023) [50] | Mouse | C57BL/6J | Male | 56 (7 groups, 8 mice each) | 5% ethanol in drinking water for 10 days; then, 31.5% ethanol via gavage on day 102 | 5% (daily ethanol); 31.5% (gavage ethanol) | 13 weeks + 10 days ethanol feeding + 1 ethanol gavage | Citrus honey (CH) reversed ethanol-induced gut dysbiosis: ↑ Bacteroidota; ↓ Firmicutes, Proteobacteria, Verrucomicrobiota, and Turicibacter. Improved SCFA levels (acetic, propionic, butyric, and valeric acids). | CH decreased ALT and AST levels, protected against alcohol-induced liver histopathology (reduced steatosis and inflammation). Dose-dependent effects seen with LH and HH. CH effects were superior to fructose syrup. |
| Xia et al. (2021) [51] | Mouse | ICR | Male | 8–10 mice per group (control group, model group, low- and high- dose ZAVE groups) | Oral gavage (daily) | Escalating: 2 g/kg (week 1), 4 g/kg (week 2), 6 g/kg (days 15–30) | 30 days | ZAVE ↑ Akkermansia, Lachnospiraceae, and Bacteroidetes; ↓ Firmicutes, Proteobacteria, Bilophila, and Butyricimonas. Reversed ethanol-induced dysbiosis and improved F/B ratio. | Improved gut barrier, increased IL-10, TGF-β, IgA, IL-22, Reg3b/g. Reduced ROS, LPS, TNF-α, IL-6, IL-1β, liver enzymes (ALT/AST), and histological damage. |
| Yang Fan et al. (2018) [52] | Rat | Wistar | Male | 40 total (n = 6 per group selected for sequencing) | In drinking water | Gradually from 1% to 6% (then maintained at 6%) | 30 days + withdrawal (up to 14 days) | No significant diversity/richness changes; colon: ↑ Bacteroidetes, Ruminococcaceae, Parabacteroides, Butyricimonas, ↓ Lactobacillus, Gauvreauii; Jejunum mostly unaffected. | Alcohol dependence significantly alters colonic microbiota; microbiota partially restored after withdrawal; KEGG functions ↑ in amino acid metabolism, peroxisome, polyketide sugar biosynthesis; behavioral withdrawal signs observed. |
| Hendrikx et al. (2020) [53] | Chronic–binge ethanol feeding (NIAAA model) | C57BL/6 mice and Reg3g−/− mice | Males and females | WT: n = 51; KO: n = 46 (cumulative across groups) | Lieber–DeCarli ethanol diet, followed by gavage with 5 g/kg ethanol | ~36% of total calories from ethanol (starting day 6) | 15 days total (10 days ethanol, final gavage on day 16) | Ethanol feeding reduced levels of indole-3-acetic acid (IAA), an AHR ligand; impaired IL22 and REG3G expression; increased bacterial translocation to liver. Engineered L. reuteri/IL22 restored IL22 and REG3G, reduced dysbiosis-associated liver damage. | Antibiotics restored IL22 expression and reduced damage. IAA supplementation increased IL22 and REG3G, prevented bacterial translocation. Engineered L. reuteri/IL22 strain showed therapeutic potential. No protective effect observed in Reg3g−/− mice. |
| Chen et al. (2015) [54] | Mouse | C57BL/6 | Female | GF: 7–16, Conv: 10–15 | Single oral gavage | 30% ethanol (vol/vol) | Single binge (sacrificed after 9 h) | Germ-free mice showed exacerbated liver injury, inflammation, and steatosis despite lower blood ethanol levels due to increased ethanol metabolism. No changes in microbiota after single binge in conventional mice. | Higher expression of Adh1, Aldh2, CYP2E1, Srebp-1, and increased hepatic triglycerides in GF mice. GF mice had heightened baseline hepatic inflammation and upregulated proinflammatory cytokines. |
| Yang et al. (2021) [55] | Mouse | C57BL/6 | Male | 6 mice per group | Chronic ethanol feeding plus binge (NIAAA model) | 5% (v/v) ethanol Lieber–DeCarli diet + 5 g/kg binge | 10 days Lieber–DeCarli + 1 binge (day 11) | Ethanol-fed mice showed decreased abundance of beneficial genera such as Lactobacillus and increased abundance of potentially pathogenic taxa like Escherichia-Shigella and Enterococcus. Overall microbial diversity was reduced. Probiotic Clostridium butyricum reversed dysbiosis. | Ethanol feeding elevated intestinal permeability and inflammation (TNF-α, IL-1β), while probiotic intervention restored tight junction proteins (ZO-1, occludin), reduced serum ALT/AST, and alleviated liver steatosis and oxidative stress. Clostridium butyricum modulated TLR4/NF-κB signaling. |
| Xue et al. (2017) [56] | Mouse | C57BL/6J | Male | 45 mice (15 per group—control, ethanol, aplysin, and ethanol) | Control or ethanol-containing liquid diet with varying protein sources; final dose involved binge ethanol gavage | 8 mL ethanol/kg for 2 weeks; then, 12 mL/kg for 6 weeks | Collection of liver and cecum samples for analysis | SPI and hydrolyzed SPI diets enriched beneficial gut microbes (Allobaculum, Bifidobacterium, Lactobacillus, Akkermansia) and reduced ethanol-associated increases in Helicobacter, Anaeroplasma, and Proteobacteria. Metagenomic prediction indicated enhanced bile acid metabolism and SCFA biosynthesis. | SPI diets improved liver histology: ↓ steatosis, ↓ inflammation, ↓ ALT/AST vs. casein group. Tight junction proteins were upregulated. SPI diets also modulated bile acid profiles and nuclear receptor pathways with downstream effects on lipid metabolism and hepatic inflammation. |
| Mittal et al. (2025) [57] | Mouse | C57BL/6N | Male | n = 8 per group for microbiome and biochemical studies; n = 3 per group for liver proteomic analysis due to cost constraints | Lieber–DeCarli ethanol diet to induce ALD, combined with intraperitoneal thioacetamide injections (150 mg/kg body weight, twice weekly) to enhance hepatic injury | 20–22% of total caloric intake derived from ethanol, consistent with standard ALD induction protocols | Initial 1-week acclimatization on liquid diet, followed by 8 weeks of Lieber–DeCarli + ethanol + thioacetamide. Post-alcohol abstinence phase involved 7 days of dietary intervention with standard, egg-based, or plant-based diet | Veg diet group showed significant enrichment of beneficial microbial taxa: Lachnospiraceae, Prevotellaceae, Kurthia, Christensenellaceae, Akkermansia, and Butyricicoccus. It also decreased pathogenic bacteria: Roseburia, Klebsiella, Staphylococcus, and Pseudomonas vs. egg diet group. Functional shifts included ↑NAD salvage pathway, glycolysis, TCA cycle, and urea cycle. | Vegetable protein diet significantly reduced hepatic steatosis compared to the egg diet. ALT and AST serum levels were reduced vs. egg diet. Proteomics revealed upregulation of recovery-related metabolic pathways, including fatty acid beta-oxidation, pyruvate, methionine, and cysteine metabolism. Co-expression analysis (WGCNA) showed strong correlation between veg diet and upregulated energy metabolism and antioxidant pathways. |
| Thoen et al. (2022) [58] | Wistar rats | Adults | Males | 24 (8 per group: control, ALC4, ALC8) | 10% ethanol + sunflower seed diet + binge (5 g/kg, gavage) | 10% | 4 weeks (ALC4), 8 weeks (ALC8) | ↑ Bacteroidetes, ↑ Proteobacteria, ↓ Firmicutes; correlated with liver markers (TG, ALT, AST, albumin, steatosis). | ALC4: Grade 2 micro, Grade 1 macro steatosis; ALC8: Grade 3 micro, Grade 1 macro steatosis; no fibrosis; ↑ AST, ALT, glucose; ↓ albumin, HDL-C; significant mortality in ALC8. |
| Jiang et al. (2019) [59] | Mouse | C57BL/6J | males | 21 mice (3 groups of 7); plus 18 FMT recipients (3 × 6 mice) | Oral ethanol via drinking water (4 days/week) | 15% (v/v) | 10 weeks | ↑ Akkermansia, Clostridium in alcohol group, ↓ Prevotella, Barnesiella, Alloprevotella, Alistipes. Strong correlation between inflammatory cytokines (IL-1β, IL-6, TNF-α, IL-10, TGF-β) and microbial genera. | Alcohol caused depressive-like behavior; nicotinamide riboside (NR) improved behavior and anxiety. Alcohol increased microglial activation (CD68↑); NR reduced it. |
| Zhang et al. 2019) [60] | Rhesus macaques | Macaca mulatta | Males | 12 total; alcohol drinking or control groups of adolescent alcohol (n = 6), adolescent control (n = 6), adult alcohol (n = 4), and adult control (n = 5) | Custom-designed operant drinking panel attached to one side of the cage | 4% w/v ethanol solution (0.5 g/kg; then, 1 g/kg; then, 1.5 g/kg;) | 3 months | Ethanol-exposed animals had ↑ Bacteroidetes, Firmicutes, Tenericutes, Actinobacteria, Proteobacteria, and Spirochaetes. | The effects of ethanol-exposed group were partially or wholly ameliorated following a relatively short 5-day period of abstinence, suggesting that the specific effects observed here are the direct effects of alcohol. |
| Cunningham et al. (2023) [61] | Mouse | C57BL/6 | Males and females | 9 breeding pairs in each group (AUDIT score > 8 and AUDIT score < 8, respectively) | Mice were colonized with human fecal microbiota from individuals with high and low AUDIT scores and bred to produce human alcohol-associated microbiota or human control-microbiota | Human positive fecal samples from subjects with AUDIT score of ≥8 for men and ≥5 for women | Last alcohol-containing beverage consumed within the 7 days prior to enrollment | ↑ Klebsiella pneumoniae, ↑ Streptococcus pneumoniae. | Offspring colonized with fecal microbiota from high-AUDIT adults exhibited higher mortality, pulmonary bacterial burden, and post-infection lung damage to Klebsiella pneumoniae and Streptococcus pneumoniae pneumonia. |
| Wang et al. (2023) [62] | Rat model | Antibiotics-treated conventional rats | Male | n = 8 per group; groups included control, ethanol-fed, and ethanol- + LGG-treated mice | Role of the gut microbiome on the behaviors of rats by fecal microbiota transferred orally throughout ethanol treatment | 489.42 ± 29.91 alcohol intake/day (15.82 ± 9.04 years) for humans from which fecal microbiota was collected | FMT daily for 21 days, behavioral testing for the next 6 days, alcohol preference test for the next 5 days | ↑ Lactobacillus, ↑ Akkermansia, ↑ Rikenellaceae; ↓ Enterobacteriaceae, and ↓ Bacteroides. | Alcohol dependence in rats, including increased anxiety- and depression-like behaviors, reduced exploratory and recognition memory, and higher alcohol preference. |
| Diagnosis | Participants | Percentage (%) |
|---|---|---|
| Alcohol use disorder | 289 | 41.17% |
| Alcoholic hepatitis | 203 | 28.91% |
| Alcohol-related cirrhosis | 98 | 13.96% |
| Hepatic encephalopathy (HE) | 20 | 2.85% |
| Coronary artery disease (CAD) | 42 | 5.98% |
| Other/mixed (AH + ARC or ALD) | 50 | 7.13% |
| Study | Outcome | Cohen’s d | Hedges’ g |
|---|---|---|---|
| Du et al. [33] | MoCA (AUD vs. HC) | −0.98 | −0.96 |
| MMSE (AUD vs. HC) | −1.25 | −1.22 | |
| Dedon et al. [34] | Percent drinking reduction (placebo vs. zonisamide) | 0.39 | 0.38 |
| Zhang et al. [35] | Protocol only, no outcome data available | No data available | No data available |
| Bajaj et al. [36] | ACQ-SF score at day 15 (FMT vs. placebo) | 0.29 | 0.28 |
| Amadieu et al. [37] | Metabolomics outcomes only | No data available | No data available |
| Philips et al. [38] | 90-day survival (FMT vs. HDPI) | 0.69 (large) | |
| Muthiah et al. [39] | Metabolomics/microbiome data only | No data available | No data available |
| Zhang et al. [40] | AST at 60 days (BC99 vs. placebo) | 0.66 | 0.65 |
| γ-GT at 60 days (BC99 vs. placebo) | 0.87 | 0.86 | |
| Lang et al. [41] | Observational microbiome outcomes | No data available | No data available |
| Haas et al. [42] | Crossover metabolomics data only | No data available | No data available |
| Han et al. [43] | Change in serum LPS (probiotics vs. placebo, day 7) | 0.18 | 0.18 |
| Treatment Type | Number of Studies | Microbiota Outcome |
|---|---|---|
| FMT | 4 | ↑ Diversity, ↓ Proteobacteria |
| Probiotics | 2 | ↑ Lactobacillus, ↓ E. coli |
| Abstinence | 2 | Partial recovery in SCFA-producers |
| Pharmacologic (Zonisamide [34], Pentoxifylline [38]) | 2 | Predictive microbial/metabolite markers |
| Observational | 1 | ↓ Ruminococcaceae, ↑ craving and zonulin |
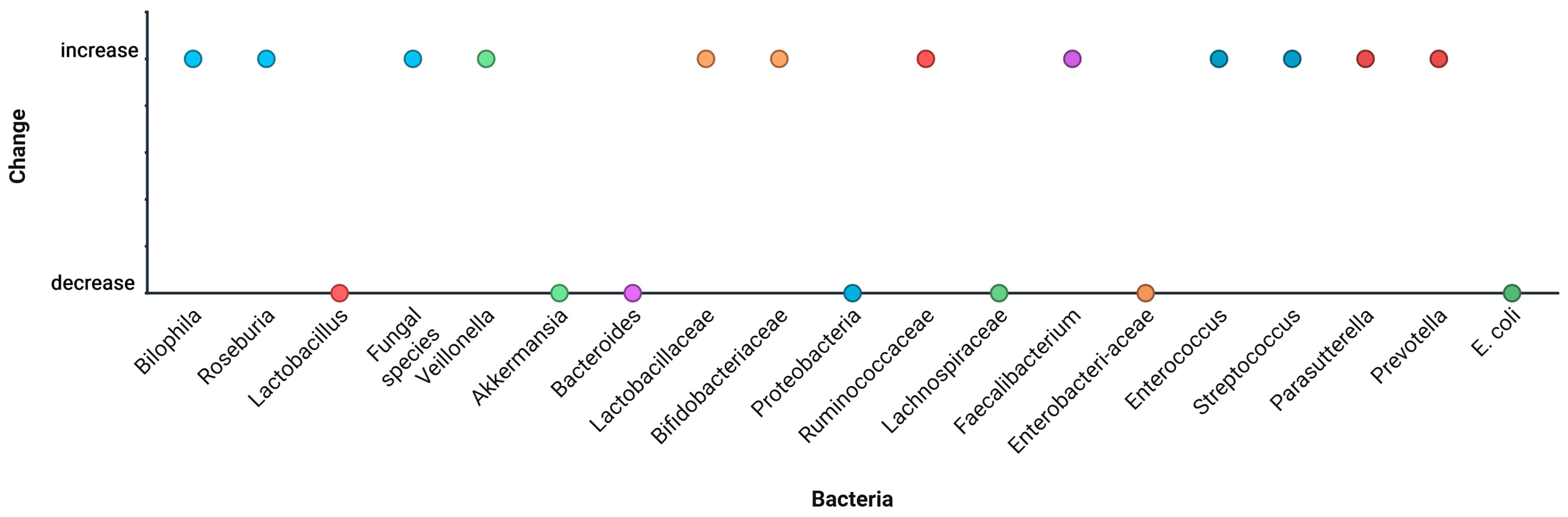
| Bacteria | Change | Study |
|---|---|---|
| Megamonas | Increase | Du et al. [33] |
| Escherichia | Increase | Du et al. [33] |
| Coprobacillus | Increase | Du et al. [33] |
| Lactobacillus | Decrease | Du et al. [33]; Philips et al. [38]; Bajaj et al. [36] |
| Fungal species | Increase | Du et al. [33] |
| Veillonella | Increase | Dedon et al. [34] |
| Akkermansia | Decrease | Dedon et al. [34] |
| Bacteroides | Decrease | Dedon et al. [34]; Haas et al. [42] |
| Lactobacillaceae | Increase | Bajaj et al. [36] |
| Bifidobacteriaceae | Increase | Bajaj et al. [36] |
| Proteobacteria | Decrease | Bajaj et al. [36]; Philips et al. [38] |
| Ruminococcaceae | Increase | Amadieu et al. [37]; Muthiah et al. [39]; Zhang et al. [35] |
| Lachnospiraceae | Decrease | Amadieu et al. [37]; Lang et al. [41] |
| Faecalibacterium | Increase | Philips et al. [38]; Zhang et al. [35] |
| Enterobacteriaceae | Decrease | Muthiah et al. [39] |
| Enterococcus | Increase | Lang et al. [41] |
| Streptococcus | Increase | Lang et al. [41] |
| Parasutterella | Increase | Haas et al. [42] |
| Prevotella | Increase | Haas et al. [42] |
| E. coli | Decrease | Han et al. [43] |
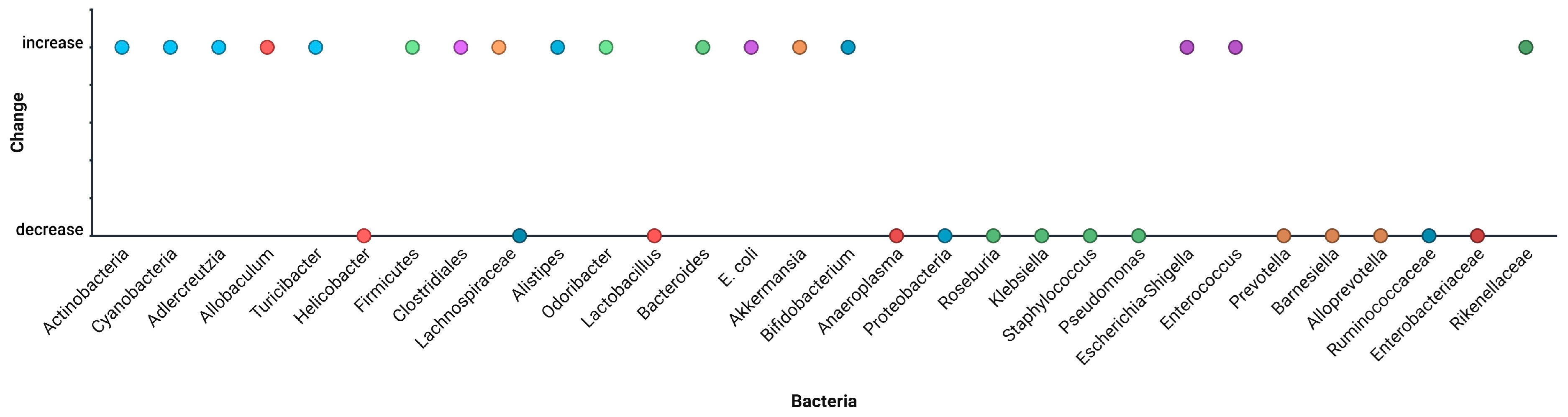
| Bacteria | Change | Study |
|---|---|---|
| Actinobacteria | Increase | Xu et al. [45] |
| Cyanobacteria | Increase | Xu et al. [45] |
| Adlercreutzia | Increase | Xu et al. [45] |
| Allobaculum | Increase | Xu et al. [45]; Xue et al. [56] |
| Turicibacter | Increase | Xu et al. [45] |
| Helicobacter | Decrease | Xu et al. [45]; Xue et al. [56] |
| Firmicutes | Increase | Wang et al. [47] |
| Clostridiales | Increase | Wang et al. [47]; |
| Lachnospiraceae | Increase | Wang et al. [47]; Mittal et al. [57] |
| Alistipes | Increase | Wang et al. [47]; Jiang et al. [59] |
| Odoribacter | Increase | Wang et al. [47] |
| Lactobacillus | Decrease | Li et al. [48]; Yang et al. [49] |
| Bacteroides | Increase | Li et al. [48]; Wang et al. [62] |
| E. coli | Increase | Li et al. [48] |
| Akkermansia | Increase | Mittal et al. [57]; Jiang et al. [59] |
| Bifidobacterium | Increase | Xue et al. [56]; |
| Anaeroplasma | Decrease | Xue et al. [56] |
| Proteobacteria | Decrease | Xue et al. [56]; |
| Roseburia | Decrease | Mittal et al. [57] |
| Klebsiella | Decrease | Mittal et al. [57]; Cunningham et al. [61] |
| Staphylococcus | Decrease | Mittal et al. [57] |
| Pseudomonas | Decrease | Mittal et al. [57] |
| Escherichia-Shigella | Increase | Yang et al. [49]; Zhang et al. [60] |
| Enterococcus | Increase | Yang et al. [49]; Zhang et al. [60] |
| Prevotella | Decrease | Jiang et al. [59] |
| Barnesiella | Decrease | Jiang et al. [59] |
| Alloprevotella | Decrease | Jiang et al. [59] |
| Ruminococcaceae | Decrease | Zhang et al. [60] |
| Lachnospiraceae | Decrease | Zhang et al. [60] |
| Enterobacteriaceae | Decrease | Wang et al. [62] |
| Rikenellaceae | Increase | Wang et al. [62] |
| Streptococcus | Increase | Cunningham et al. [61] |
| Humans | Animals | ||
|---|---|---|---|
| Akkermansia | Decrease [34] | Akkermansia | Increase [57,59,61] |
| Bacteroides | Decrease [34,42] | Bacteroides | Increase [48,62] |
| E. coli | Decrease [43] | E. coli | Increase [48] |
| Lachnospiraceae | Decrease [37,41] | Lachnospiraceae | Increase [60] |
| Prevotella | Increase [42] | Prevotella | Decrease [59] |
| Roseburia | Increase [33] | Roseburia | Decrease [57] |
| Study | Outcome | Cohen’s d | Hedges’ g |
|---|---|---|---|
| Daaz-Ubilla et al. [44] | Ethanol intake (g/kg/day) | 5.31 | 4.79 |
| Ethanol intake (g/kg/day) | 5.01 | 4.52 | |
| Xu et al. [45] | OFT—time in inner zone | 0.69 | 0.68 |
| EPM—time in open arms | 0.93 | 0.90 | |
| Xiao et al. [46] | FST—immobility time | 1.84 | 1.71 |
| TST—immobility time | 2.04 | 1.90 | |
| Wang et al. [47] | Light–dark test—time in dark | 1.19 | 1.10 |
| Open field—distance traveled | 5.10 | 4.69 | |
| Open field—time in center | 3.85 | 3.54 | |
| Li et al. [48] | Serum ferritin (ng/mL) | 1.10 | 1.01 |
| Serum hepcidin (ng/mL) | 1.47 | 1.35 | |
| Yang et al. [49] | ALT (U/L)—pubertal mice | 1.57 | 1.46 |
| ALT (U/L)—adult mice | 2.21 | 2.06 | |
| AST (U/L)—pubertal mice | 1.33 | 1.24 | |
| AST (U/L)—adult mice | 1.50 | 1.39 | |
| Yi et al. [50] | ALT (U/L) | 1.28 | 1.19 |
| AST (U/L) | 1.40 | 1.30 | |
| Xia et al. [51] | ALT (U/L) | 4.04 | 3.76 |
| AST (U/L) | 3.67 | 3.41 | |
| Yang Fan et al. [52] | Withdrawal severity score | 2.34 | 2.18 |
| Hendrikx et al. [53] | Plasma ALT (U/L) | 1.07 | 1.00 |
| Chen et al. [54] | Plasma ALT (U/L) | 1.86 | 1.73 |
| Yang et al. [55] | D-lactate (μmol/L) | 1.77 | 1.65 |
| DAO (ng/mL) | 1.70 | 1.58 | |
| LPS (EU/L) | 3.43 | 3.19 | |
| Xue et al. [56] | ALT (U/L) | 0.67 | 0.62 |
| AST (U/L) | 0.76 | 0.71 | |
| Mittal et al. [57] | ALT (U/L) | 2.45 | 2.28 |
| AST (U/L) | 1.49 | 1.38 | |
| Thoen et al. [58] | ALT (U/L) | 5.38 | 5.00 |
| AST (U/L) | 13.47 | 12.53 | |
| Jiang et al. [59] | SPT (%) | 3.31 | 3.08 |
| FST (s) | 2.30 | 2.14 | |
| Zhang et al. [60] | Chao1 index | 2.75 | 2.56 |
| Shannon index | 1.94 | 1.80 | |
| Cunningham et al. [61] | Bacterial burden (log CFU/lung)—K. pneumoniae | 1.23 | 1.14 |
| Bacterial burden (log CFU/lung)—S. pneumoniae | 1.64 | 1.53 | |
| Wang et al. [62] | OFT—time in center | 2.21 | 2.05 |
| EPM—time in open arms | 1.81 | 1.68 | |
| FST—immobility time | 1.80 | 1.67 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexandrescu, L.; Tofolean, I.T.; Tofolean, D.E.; Nicoara, A.D.; Twakor, A.N.; Rusu, E.; Preotesoiu, I.; Dumitru, E.; Dumitru, A.; Tocia, C.; et al. Ethanol-Induced Dysbiosis and Systemic Impact: A Meta-Analytical Synthesis of Human and Animal Research. Microorganisms 2025, 13, 2000. https://doi.org/10.3390/microorganisms13092000
Alexandrescu L, Tofolean IT, Tofolean DE, Nicoara AD, Twakor AN, Rusu E, Preotesoiu I, Dumitru E, Dumitru A, Tocia C, et al. Ethanol-Induced Dysbiosis and Systemic Impact: A Meta-Analytical Synthesis of Human and Animal Research. Microorganisms. 2025; 13(9):2000. https://doi.org/10.3390/microorganisms13092000
Chicago/Turabian StyleAlexandrescu, Luana, Ionut Tiberiu Tofolean, Doina Ecaterina Tofolean, Alina Doina Nicoara, Andreea Nelson Twakor, Elena Rusu, Ionela Preotesoiu, Eugen Dumitru, Andrei Dumitru, Cristina Tocia, and et al. 2025. "Ethanol-Induced Dysbiosis and Systemic Impact: A Meta-Analytical Synthesis of Human and Animal Research" Microorganisms 13, no. 9: 2000. https://doi.org/10.3390/microorganisms13092000
APA StyleAlexandrescu, L., Tofolean, I. T., Tofolean, D. E., Nicoara, A. D., Twakor, A. N., Rusu, E., Preotesoiu, I., Dumitru, E., Dumitru, A., Tocia, C., Herlo, A., Alexandrescu, D. M., Popescu, I., & Cimpineanu, B. (2025). Ethanol-Induced Dysbiosis and Systemic Impact: A Meta-Analytical Synthesis of Human and Animal Research. Microorganisms, 13(9), 2000. https://doi.org/10.3390/microorganisms13092000