Removal of Ibuprofen from Contaminated Water by Bioaugmentation with Novel Bacterial Strains Isolated from Sewage Sludge
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Bacterial Strains
2.2. Inoculum Preparation
2.3. Inhibitory Concentration of IBP for Bacterial Growth
2.4. Biodegradation Assays in Solution with Isolated IBP-Degrading Bacteria
2.5. Analytical Quantification of Ibuprofen and Its Transformation Products
2.6. Models of Biodegradation Kinetics
2.7. Statistical Analysis
3. Results and Discussion
3.1. Ibuprofen-Induced Inhibition of Bacterial Growth
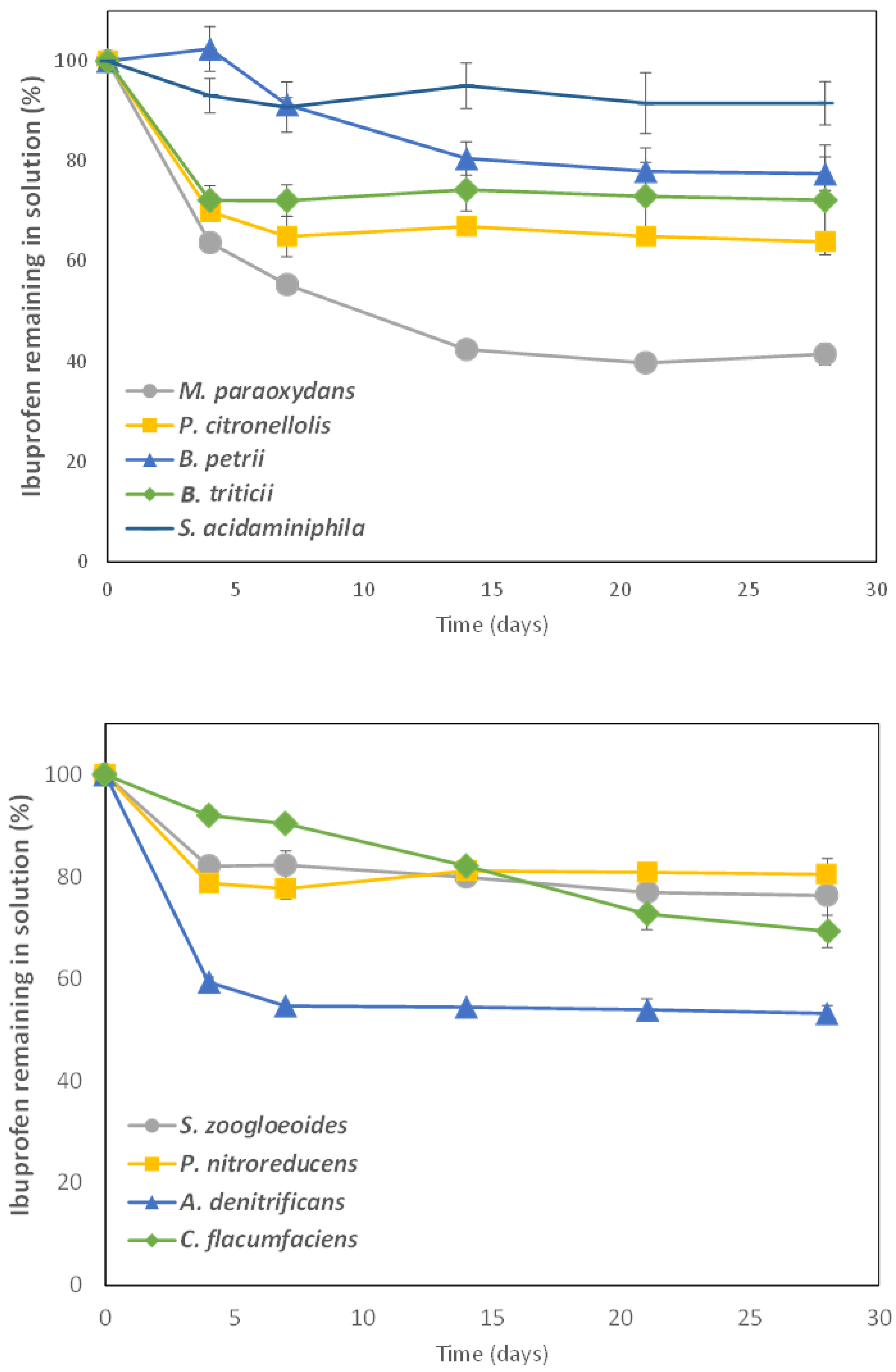
3.2. IBP Biodegradation in Aqueous Solution by Bacteria Isolated from Sewage Sludge
| Strain | Concentration (mg L−1) | Degraded Concentration (mg L−1)/Time (Days) | References |
|---|---|---|---|
| Nocardia sp. NRRL 5646 | 1000 | 1000/5 d | [59] |
| Sphingomonas sp. Ibu-2 | 500 | 500/3 d | [8,9] |
| Patulibacter sp. I11 | 1.0 0.25 | 0.28/12.5 d 0.125/12.5 d | [31] |
| Variovorax sp. Ibu-1 | 500 | 500/7 d | [60] |
| Comamonas aquatic + Bacillus sp. | 100 | 100/1.38 d | [61] |
| Bacillus thuringiensis B1(2015b) | 20 | 20/6 d | [10,29] |
| Serratia marcescens BL1 | 30 | 28/5 d | [11] |
| Pseudoxanthomonas sp. DIN-3 | 0.05 | 0.02/14 d | [64] |
| Microccocus yunnanensis | 0.2 | 0.18/0.5 d | [12,34] |
| Novosphingobium sp. Pseudomonas sp. | 0.062 0.082 | 0.062/3 d 0.082/8 d | [65] |
| Rhodococcus cerastii IEGM 1278 | 0.1 100 | 0.1/1.25 d 100/6 d | [35] |
| Sphingopyxis granuli RW412 | 456.5 | 456.5/3.1 d | [16] |
| Citrobacter freundii PYI-2 Citrobacter portucalensis YPI-2 | 8.0 8.0 | 8.0/15 d 8.0/15 d | [62] |
| Bacillus siamensis DSI-1 | 20 | 20/1 d | [66] |
| Nocardioides carbamazepine sp. Nov. | 1.5 | 1.05/49 d | [30] |
| Pseudoalteromonas sp. | 1.0 | 0.89/3 d | [63] |
| Streptomyces murinus D218 Pseudomonas alloputida M20 | 20 20 | 15.34/0.5 d 16.66/0.5 d | [67] |
| Rhizobium daejeonense IBU_18 | 1.5 | 1.36/28 d | [33] |
| Microbacterium paraoxydans | 15 | 15/5 d | [23] |
| Klebsiella pneumoniae TIBU2.1 | 5 | 5/14 d | [15] |
| Klebsiella variicola LOIBU1.1 | 5 | 3/14 d | [15] |
| Pseudomonas aeruginosa LOIBU1.2 | 5 | 3.3/14 d | [15] |
| Mycolicibacterium aubagnense HPB1.1 | 5 | 2.9/14 d | [15] |
| Labrys neptuniae CSW11 | 1.0 5.0 10 100 | 1.0/4 d 5.0/4 d 10/7 d 48.4/28 d | [17] |
| M. paraoxydans CSW08 | 10 | 6.38/14 d | This study |
| A. denitrificans CSW15 | 10 | 4.69/7 d | This study |
| P. citronellolis CSW09 | 10 | 3.61/7 d | This study |
| C. flacumfaciens CSW18 | 10 | 3.22/28 d | This study |
| B. tritici CSW06 | 10 | 2.72/7 d | This study |
| B. petrii CSW07 | 10 | 2.45/28 d | This study |
| S. zoogloeoides CSW12 | 10 | 2.40/21 d | This study |
| P. nitroreducens CSW13 | 10 | 1.98/14 d | This study |
| S. acidaminiphila CSW10 | 10 | 0.9/28 d | This study |
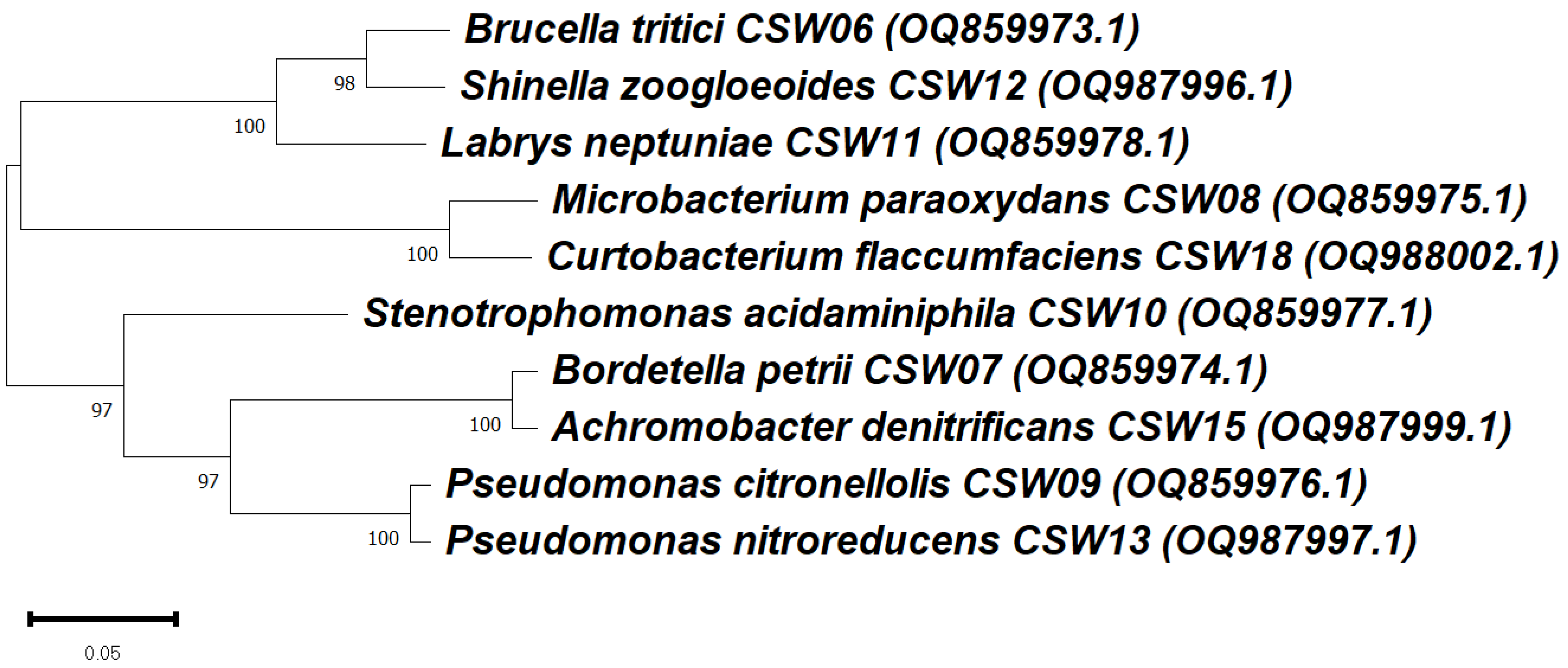
3.3. Phylogenetic Analysis of Ibuprofen-Degrading Bacterial Strains Based on 16S rRNA Gene
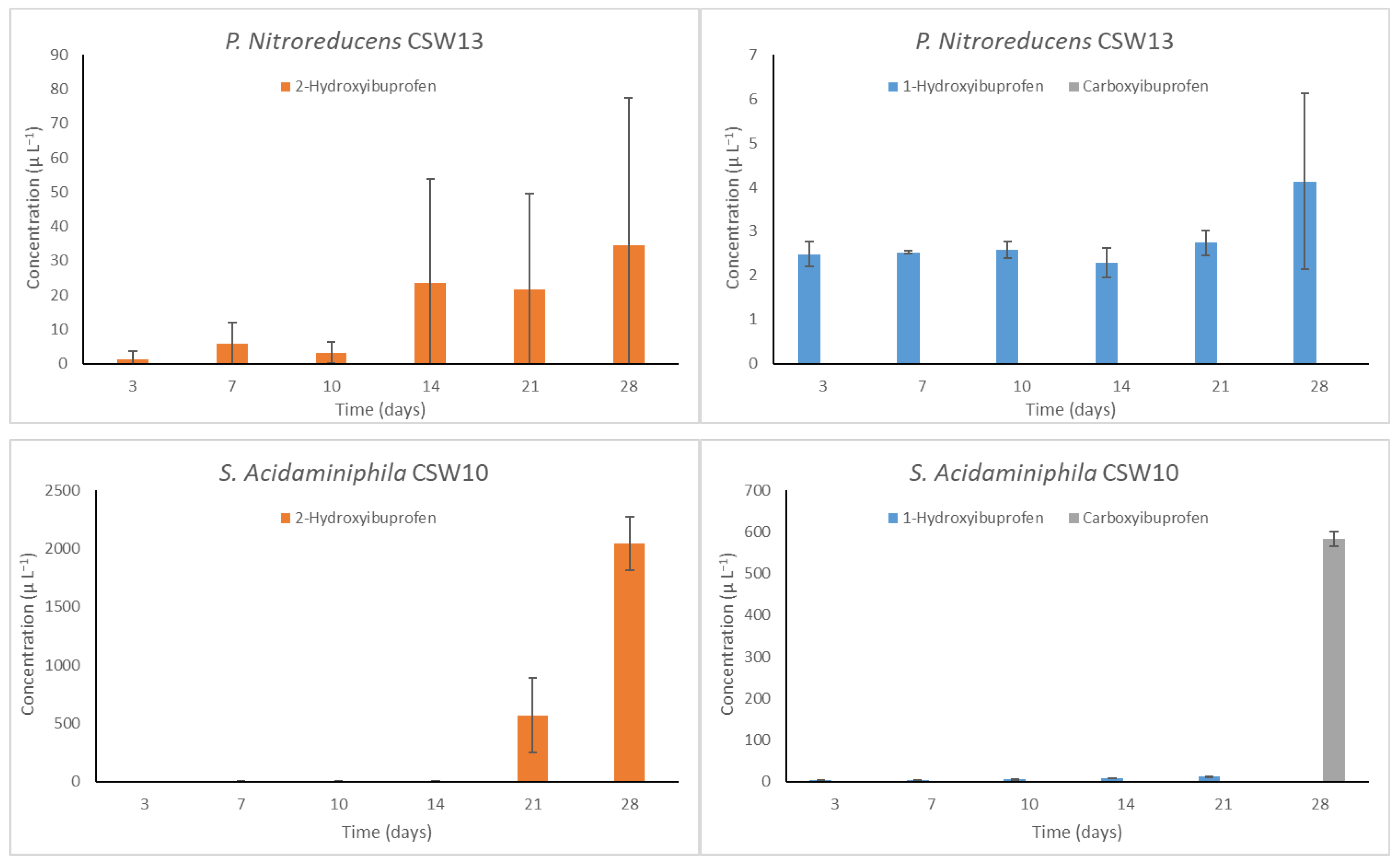
3.4. Detection of the Main Metabolites in IBP Biotransformation by Isolated Bacteria
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chopra, S.; Kumar, D. Ibuprofen as an emerging organic contaminant in environment, distribution and remediation. Heliyon 2020, 6, e04087. [Google Scholar] [CrossRef]
- Jan-Roblero, J.; Cruz-Maya, J.A. Ibuprofen: Toxicology and biodegradation of an emerging contaminant. Molecules 2023, 28, 2097. [Google Scholar] [CrossRef]
- WFD; EC. Directive 2000/60/EC of the European Parliament and of the Council of 23 October 2000 Establishing a Framework for Community Action in the Field of Water Policy. J 327 (L). 2000. Available online: https://www.eea.europa.eu/policy-documents/directive-2000-60-ec-of (accessed on 1 August 2025).
- EC. European Parliament Legislative Resolution of 2 July 2013 on the Proposal for a Directive of the European Parliament and of the Council Amending Directives 2000/60/EC and 2008/105/EC as Regards Priority Substances in the Field of Water Policy; European Commission: Brussels, Belgium, 2013. [Google Scholar]
- DG Env. Priority Substances Review-Next Steps. 2020. Available online: https://circabc.europa.eu/sd/a/97910629-048e-4b2b-90a2-fe520aab2adc/WGChem2020Jan(7)PrioritySubstancesReview-nextsteps.pdf (accessed on 1 August 2025).
- Nguyen, M.K.; Lin, C.; Nguyen, H.L.; Hung, N.T.Q.; La, D.D.; Nguyen, X.H.; Chang, S.W.; Chung, W.J. Occurrence, fate, and potential risk of pharmaceutical pollutants in agriculture: Challenges and environmentally friendly solutions. Sci. Total Environ. 2023, 899, 165323. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, Z.; Ma, Q.; Dai, L.; Dang, Z. Occurrence, removal and risk evaluation of ibuprofen and acetaminophen in municipal wastewater treatment plants: A critical review. Sci. Total Environ. 2023, 891, 164600. [Google Scholar] [CrossRef]
- Murdoch, R.W.; Hay, A.G. Formation of catechols via removal of acid side chains from ibuprofen and related aromatic acids. Appl. Environ. Microbiol. 2005, 71, 6121–6125. [Google Scholar] [CrossRef]
- Murdoch, R.W.; Hay, A.G. Genetic and chemical characterization of ibuprofen degradation by Sphingomonas Ibu-2. Microbiology 2013, 159, 621–632. [Google Scholar] [CrossRef]
- Marchlewicz, A.; Domaradzka, D.; Guzik, U.; Wojcieszyńska, D. Bacillus thuringiensis B1(2015b) is a Gram-Positive Bacteria Able to Degrade Naproxen and Ibuprofen. Water Air Soil Pollut. 2016, 227, 197. [Google Scholar] [CrossRef]
- Xu, B.; Xue, G.; Yang, X. Isolation and application of an ibuprofen-degrading bacterium to a biological aerated filter for the treatment of micro-polluted water. Front. Environ. Sci. Eng. 2018, 12, 15. [Google Scholar] [CrossRef]
- Sharma, K.; Kaushik, G.; Thotakura, N.; Raza, K.; Sharma, N.; Nimesh, S. Fate of ibuprofen under optimized batch biodegradation experiments using Micrococcus yunnanensis isolated from pharmaceutical sludge. Int. J. Environ. Sci. Technol. 2019, 16, 8315–8328. [Google Scholar] [CrossRef]
- Balciunas, E.M.; Kappelmeyer, U.; Harms, H.; Heipieper, H.J. Increasing ibuprofen degradation in constructed wetlands by bioaugmentation with gravel containing biofilms of an ibuprofen-degrading Sphingobium yanoikuyae. Eng. Life Sci. 2020, 20, 160–167. [Google Scholar] [CrossRef]
- Aulestia, M.; Flores, A.; Acosta-Jurado, S.; Santero, E.; Camacho, E.M. Genetic characterization of the ibuprofen-degradative pathway of Rhizorhabdus wittichii MPO218. App. Environ. Microbiol. 2022, 88, e00388-22. [Google Scholar] [CrossRef]
- Lara-Moreno, A.; Costa, M.C.; Vargas-Villagomez, A.; Carlier, J.D. New bacterial strains for ibuprofen biodegradation: Drug removal, transformation, and potential catabolic genes. Environ. Microb. Rep. 2024, 16, e13320. [Google Scholar] [CrossRef]
- Aguilar-Romero, I.; De la Torre-Zúñiga, J.; Quesada, J.M.; Haïdour, A.; O’Connell, G.; McAmmond, B.M.; Van Hamme, J.D.; Romero, E.; Regina-Michaela Wittich, R.M.; van Dillewijn, P. Effluent decontamination by the ibuprofen-mineralizing strain, Sphingopyxis granuli RW412: Metabolic processes. Environ. Pollut. 2021, 274, 116536. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Romero, I.; Madrid, F.; Villaverde, J.; Alonso, E.; Santos, J.L.; Morillo, E. Removal of ibuprofen in water by bioaugmentation with Labrys neptuniae CSW11 isolated from sewage sludge—Assessment of biodegradation pathway based on metabolite formation and genomic analysis. J. Xenobiot. 2025, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Romero, I.; Madrid, F.; Villaverde, J.; Morillo, E. Ibuprofen-enhanced biodegradation in solution and sewage sludge by an indigenous microbial consortium. Shift Assoc. Bact. Communities J. Hazard. Mater. 2024, 464, 132970. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Ordóñez, A.; Aguilar-Romero, I.; Villaverde, J.; Madrid, F.; Morillo, E. Isolation of novel bacterial strains Pseudomonas extremaustralis CSW01 and Stutzerimonas stutzeri CSW02 from sewage sludge for paracetamol biodegradation. Microorganisms 2023, 11, 196. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- FOCUS. Guidance Document on Estimating Persistence and Degradation Kinetics from Environmental Fate Studies on Pesticides in EU Registration. Report of the FOCUS Work Group on Degradation Kinetics, EC Document Reference Sanco/10058/2005 version 2.0; 2006; 434 p. Available online: https://esdac.jrc.ec.europa.eu/public_path/projects_data/focus/dk/docs/finalreportFOCDegKinetics.pdf (accessed on 2 July 2025).
- Okpala, O.E.; Rondevaldova, J.; Kokoska, L. Anti-inflammatory drugs as potential antimicrobial agents: A review. Front. Pharmacol. 2025, 16, 1557333. [Google Scholar] [CrossRef]
- Show, S.; Sarkar, P.; Barman, S.; Halder, G. Microbial remediation of ibuprofen contaminated water using novel isolate Microbacterium paraoxydans. Chem. Pap. 2023, 77, 517–531. [Google Scholar] [CrossRef]
- Wittich, R.M.; Haïdour, A.; Aguilar-Romero, I.; de la Torre-Zúñiga, J.; van Dillewijn, P. Biodegradation of microtoxic phenylpropanoids (phenylpropanoic acid and ibuprofen) by bacteria and the relevance for their removal from wastewater treatment plants. Genes 2023, 14, 442. [Google Scholar] [CrossRef]
- Rajapaksha, R.M.C.P. Heavy metal tolerance of culturable bacteria and fungi in a long-term cultivated tropical ultisol. Eur. J. Soil Biol. 2011, 47, 9–15. [Google Scholar] [CrossRef]
- Al-janabi, A.A.H.S. In vitro antibacterial activity of Ibuprofen and acetaminophen. J. Glob. Infect. Dis. 2010, 2, 105–108. [Google Scholar] [CrossRef]
- Lara-Moreno, A.; Aguilar-Romero, I.; Rubio-Bellido, M.; Madrid, F.; Villaverde, J.; Santos, J.L.; Alonso, E.; Morillo, E. Novel nonylphenol-degrading bacterial strains isolated from sewage sludge: Application in bioremediation of sludge. Sci. Total Environ. 2022, 847, 157647. [Google Scholar] [CrossRef]
- Mulkiewicz, E.; Wolecki, D.; Świacka, K.; Kumirska, J.; Stepnowski, P.; Caban, M. Metabolism of non-steroidal anti-inflammatory drugs by non-target wild-living organisms. Sci. Total Environ. 2021, 791, 148251. [Google Scholar] [CrossRef] [PubMed]
- Marchlewicz, A.; Guzik, U.; Smulek, W.; Wojcieszynska, D. Exploring the degradation of ibuprofen by Bacillus thuringiensis B1 (2015b): The new pathway and factors affecting degradation. Molecules 2017, 22, 1676. [Google Scholar] [CrossRef] [PubMed]
- Benedek, T.; Pápai, M.; Gharieb, K.; Bedics, A.; Táncsics, A.; Tóth, E.; Daood, H.; Maróti, G.; Wirth, R.; Menashe, O.; et al. Nocardioides carbamazepini sp. nov., an ibuprofen degrader isolated from a biofilm bacterial community enriched on carbamazepine. Syst. Appl. Microbiol. 2022, 45, 126339. [Google Scholar] [CrossRef]
- Almeida, B.; Kjeldal, H.; Lolas, I.; Knudsen, A.D.; Carvalho, G.; Nielsen, K.L.; Barreto Crespo, M.T. Quantitative proteomic analysis of ibuprofen-degrading Patulibacter sp. strain I11. Biodegradation 2013, 24, 615–630. [Google Scholar] [CrossRef] [PubMed]
- Salgado, R.; Brito, D.; Noronha, J.P.; Almeida, B.; Bronze, M.R.; Oehmen, A.; Carvalho, G.; Barreto-Crespo, M.T. Metabolite identification of ibuprofen biodegradation by Patulibacter medicamentivorans under aerobic conditions. Environ. Technol. 2020, 41, 450–465. [Google Scholar] [CrossRef]
- Pápai, M.; Benedek, T.; Táncsics, A.; Bornemann, T.L.V.; Plewka, J.; Probst, A.J.; Hussein, D.; Maróti, G.; Menashe, O.; Kriszt, B. Selective enrichment, identification, and isolation of diclofenac, ibuprofen, and carbamazepine degrading bacteria from a groundwater biofilm. Environ. Sci. Pollut. Res. 2023, 30, 44518–44535. [Google Scholar] [CrossRef]
- Sharma, K.; Kaushik, G.; Thotakura, N.; Raza, K.; Sharma, N.; Nimesh, S. Enhancement effects of process optimization technique while elucidating the degradation pathways of drugs present in pharmaceutical industry wastewater using Micrococcus yunnanensis. Chemosphere 2020, 238, 124689. [Google Scholar] [CrossRef]
- Ivshina, I.B.; Tyumina, E.A.; Bazhutin, G.A.; Vikhareva, E.V. Response of Rhodococcus cerastii IEGM 1278 to toxic effects of ibuprofen. PLoS ONE 2021, 16, e0260032. [Google Scholar] [CrossRef]
- Jin, J.; Yao, J.; Liu, W.; Zhang, Q.; Liu, J. Fluoranthene degradation and binding mechanism study based on the active-site structure of ring-hydroxylatingdioxygenase in Microbacterium paraoxydans JPM1. Environ. Sci. Pollut. Res. 2017, 24, 363–371. [Google Scholar] [CrossRef]
- Singh, V.; Singh, J.; Singh, N.; Rai, S.N.; Verma, M.K.; Verma, M.; Singh, V.; Chivate, M.S.; Bilal, M.; Mishra, V. Simultaneous removal of ternary heavy metal ions by a newly isolated Microbacterium paraoxydans strain VSVM IIT(BHU) from coal washery effluent. BioMetals 2023, 36, 829–845. [Google Scholar] [CrossRef]
- Peng, W.; Lin, S.; Deng, Z.; Liang, R. Bioaugmentation removal and microbiome analysis of the synthetic estrogen 17α-ethynylestradiol from hostile conditions and environmental samples by Pseudomonas citronellolis SJTE-3. Chemosphere 2023, 317, 137893. [Google Scholar] [CrossRef]
- Reis, P.J.M.; Reis, A.C.; Ricken, B.; Kolvenbach, B.A.; Manaia, C.M.; Corvini, P.F.X.; Nunes, O.C. Biodegradation of sulfamethoxazole and other sulfonamides by Achromobacter denitrificans PR1. J. Haz. Mat. 2014, 280, 741–749. [Google Scholar] [CrossRef]
- Nguyen, P.Y.; Silva, A.F.; Reis, A.C.; Nunes, O.C.; Rodrigues, A.M.; Rodrigues, J.E.; Cardoso, V.V.; Benoliel, M.J.; Reis, M.A.M.; Oehmen, A.; et al. Bioaugmentation of membrane bioreactor with Achromobacter denitrificans strain PR1 for enhanced sulfamethoxazole removal in wastewater. Sci. Total Environ. 2019, 648, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hou, J.; Zhang, X.; Cheng, L.; Hu, W.; Zhang, Q. Biochar-assisted degradation of oxytetracycline by Achromobacter denitrificans and underlying mechanisms. Bioresour. Technol. 2023, 387, 129673. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Sun, J.; Guo, H.; Wang, C.; Fang, T.; Rogers, M.J.; He, J.; Wang, H. Anaerobic biodegradation of phenanthrene by a newly isolated nitrate-dependent Achromobacter denitrificans strain PheN1 and exploration of the biotransformation processes by metabolite and genome analyses. Environ. Microbiol. 2021, 23, 908–923. [Google Scholar] [CrossRef] [PubMed]
- Maleki Rad, M.; Moghimi, H.; Azin, E. Biodegradation of thermo-oxidative pretreated low-density polyethylene (LDPE) and polyvinyl chloride (PVC) microplastics by Achromobacter denitrificans Ebl13. Mar. Pollut. Bull. 2022, 181, 113830. [Google Scholar] [CrossRef]
- Khleifat, K.; Magharbeh, M.; Alqaraleh, M.; Al-Sarayrah, M.; Alfarrayeh, I.; Al Qaisi, Y.; Alsarayreh, A.; Alkafaween, M. Biodegradation modelling of phenol using Curtobacterium flaccumfaciens as plant-growth-promoting bacteria. Heliyon 2022, 8, e10490. [Google Scholar] [CrossRef]
- Chase, A.B.; Arevalo, P.; Polz, M.F.; Berlemont, R.; Martiny, J.B.H. Evidence for ecological flexibility in the cosmopolitan genus Curtobacterium. Front. Microbiol. 2016, 7, 1874. [Google Scholar] [CrossRef]
- Al-Saleh, E.; Hassan, A. Enhanced crude oil biodegradation in soil via biostimulation. Int. J. Phytoremediation 2016, 18, 822–831. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Zhu, L.; Wang, J.; Wang, J.; Du, Z.; Zhang, C. Influence of isolated bacterial strains on the in situ biodegradation of endosulfan and the reduction of endosulfan-contaminated soil toxicity. Ecotoxicol. Environ. Saf. 2018, 160, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.A.; Dang, H.T.C.; Koekkoek, J.; Braster, M.; Parsons, J.R.; Brouwer, A.; de Boer, T.; van Spanning, R.J.M. Species and metabolic pathways involved in bioremediation of vietnamese soil from Bien Hoa airbase contaminated with herbicides. Front. Sustain. Cities 2021, 326, 692018. [Google Scholar] [CrossRef]
- Odukkathil, G.; Vasudevan, N. Biodegradation of endosulfan isomers and its metabolite endosulfate by two biosurfactant producing bacterial strains of Bordetella petrii. J. Environ. Sci. Health B 2015, 50, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Sun, Q.; Zhao, C.; Wen, D.; Tang, X. Aerobic degradation of pyridine by a new bacterial strain Shinella zoogloeoides BC026. J. Ind. Microbiol. Biotechnol. 2009, 36, 1391–1400. [Google Scholar] [CrossRef]
- Ntougias, S.; Melidis, P.; Navrozidou, E.; Tzegkas, F. Diversity and efficiency of anthracene-degrading bacteria isolated from a denitrifying activated sludge system treating municipal wastewater. Int. Biodeterior. Biodegrad. 2015, 97, 151–158. [Google Scholar] [CrossRef]
- Palma, T.L.; Shylova, A.; Costa, M.C. Isolation and characterization of bacteria from activated sludge capable of degrading 17α-ethinylestradiol, a contaminant of high environmental concern. Microbiology 2021, 167, 001038. [Google Scholar] [CrossRef]
- Li, J.; Chen, W.J.; Zhang, W.; Zhang, Y.; Lei, Q.; Wu, S.; Huang, Y.; Mishra, S.; Bhatt, P.; Chen, S. Effects of free or immobilized bacterium Stenotrophomonas acidaminiphila Y4B on glyphosate degradation performance and indigenous microbial community structure. J. Agric. Food Chem. 2022, 70, 13945–13958. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, J.; Wu, M.; Zhou, X.; Wang, S.; Ye, H.; Xiang, W.; Zhang, Q.; Cai, T. Whole genome sequencing exploitation analysis of dibutyl phthalate by strain Stenotrophomonas acidaminiphila BDBP 071. Food Biosci. 2023, 51, 102185. [Google Scholar] [CrossRef]
- Pacholak, A.; Juzwa, W.; Zgoła-Grześkowiak, A.; Kaczorek, E. Multi-faceted analysis of bacterial transformation of nitrofurantoin. Sci. Total Environ. 2023, 874, 162422. [Google Scholar] [CrossRef] [PubMed]
- Aswathi, A.; Pandey, A.; Sukumaran, R.K. Rapid degradation of the organophosphate pesticide Chlorpyrifos by a novel strain of Pseudomonas nitroreducens AR-3. Bioresour. Technol. 2019, 292, 122025. [Google Scholar] [CrossRef]
- Palma, T.L.; Shylova, A.; Dias Carlier, J.; Costa, M.C. An autochthonous aerobic bacterial community and its cultivable isolates capable of degrading fluoxetine. J. Chem. Technol. Biotechnol. 2021, 96, 2813–2826. [Google Scholar] [CrossRef]
- Alamer, N.J.; Aldayel, M.F.; Khalifa, A. Isolation and characterization of Brucella spp., low-density polyethylene (LDPE) plastic degrading bacteria in Al-Ahsa Region, Saudi Arabia. Appl. Sci. 2023, 13, 4629. [Google Scholar] [CrossRef]
- Chen, Y.; Rosazza, J.P.N. Microbial transformation of ibuprofen by a Nocardia species. Appl. Environ. Microbiol. 1994, 60, 1292–1296. [Google Scholar] [CrossRef]
- Murdoch, R.W.; Hay, A.G. The biotransformation of ibuprofen to trihydroxyibuprofen in activated sludge and by Variovorax Ibu-1. Biodegradation 2015, 26, 105–113. [Google Scholar] [CrossRef]
- Fortunato, M.S.; Fuentes Abril, N.P.; Martinefski, M.; Trípodi, V.; Papalia, M.; Rádice, M.; Gutkind, G.; Gallego, A.; Korol, S.E. Aerobic degradation of ibuprofen in batch and continuous reactors by an indigenous bacterial community. Environ. Technol. 2016, 37, 2617–2626. [Google Scholar] [CrossRef]
- Chopra, S.; Kumar, D. Characteristics and growth kinetics of biomass of Citrobacter freundii strains PYI-2 and Citrobacter portucalensis strain YPI-2 during the biodegradation of Ibuprofen. Int. Microbiol. 2022, 25, 615–628. [Google Scholar] [CrossRef]
- Li, Z.; Wang, J.; Gu, C.; Guo, Y.; Wu, S. Marine bacteria-mediated abiotic-biotic coupling degradation mechanism of ibuprofen. J. Hazard. Mater. 2022, 435, 128960. [Google Scholar] [CrossRef]
- Lu, Z.; Sun, W.; Li, C.; Ao, X.; Yang, C.; Li, S. Bioremoval of non-steroidal anti-inflammatory drugs by Pseudoxanthomonas sp. DIN-3 isolated from biological activated carbon process. Water Res. 2019, 161, 459–472. [Google Scholar] [CrossRef]
- Rutere, C.; Knoop, K.; Posselt, M.; Ho, A.; Horn, M.A. Ibuprofen degradation and associated bacterial communities in hyporheic zone sediments. Microorganisms 2020, 8, 1245. [Google Scholar] [CrossRef]
- Chopra, S.; Kumar, D. Characterization and biodegradation of ibuprofen by Bacillus siamensis strain DSI-1 isolated from wastewater. Rend. Lincei. Sci. Fis. Nat. 2022, 33, 643–652. [Google Scholar] [CrossRef]
- Chen, R.; Huang, J.; Li, X.; Yang, C.; Wu, X. Functional characterization of an efficient ibuprofen-mineralizing bacterial consortium. J. Hazard. Mater. 2023, 447, 130751. [Google Scholar] [CrossRef] [PubMed]
- Aulestia, M.; Flores, A.; Mangas, E.L.; Pérez-Pulido, A.J.; Santero, E.; Camacho, E.M. Isolation and genomic characterization of the ibuprofen-degrading bacterium Sphingomonas strain MPO218. Environ. Microbiol. 2021, 23, 267–280. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Zur, J.; Pinski, A.; Marchlewicz, A.; Hupert-Kocurek, K.; Wojcieszynska, D.; Guzik, U. Organic micropollutants paracetamol and ibuprofen. Toxicity, biodegradation, and genetic background of their utilization by bacteria. Environ. Sci. Pollut. Res. 2018, 25, 21498–21524. [Google Scholar] [CrossRef]
- Grabarczyk, Ł.; Mulkiewicz, E.; Stolte, S.; Puckowski, A.; Pazda, M.; Stepnowski, P.; Białk-Bielińska, A. Ecotoxicity screening evaluation of selected pharmaceuticals and their transformation products towards various organisms. Environ. Sci. Pollut. Res. 2020, 27, 26103–26114. [Google Scholar] [CrossRef]

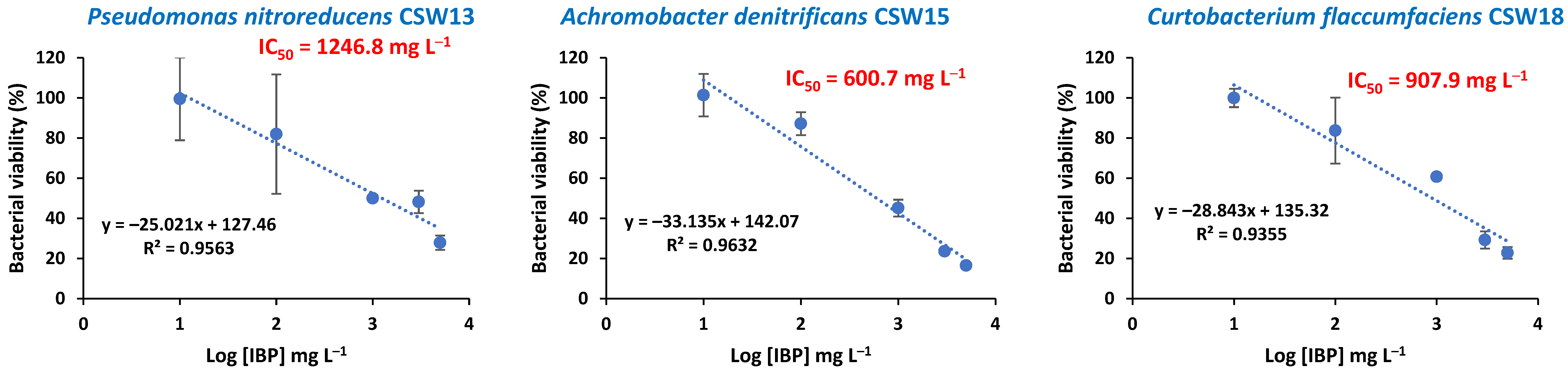

| Bacteria | IC50 (mg/L) | R2 |
|---|---|---|
| B. tritici CSW06 | 48 | 0.99 |
| B. petrii CSW07 | 1353 | 0.98 |
| M. paraoxydans CSW08 | 279 | 0.93 |
| P. citronellolis CSW09 | 450 | 1 |
| S. acidaminiphila CSW10 | 38 | 0.93 |
| S. zoogloeoides CSW12 | 689 | 0.99 |
| P. nitroreducens CSW13 | 1247 | 0.96 |
| A. denitrificans CSW15 | 601 | 0.96 |
| C. flaccumfaciens CSW18 | 908 | 0.94 |
| Bacterial Strain | Kinetic Model | K1 (d−1) | K2 (d−1) | tb (d) | DT50 (d) | Extent of Degradation (%) | R2 | Errscaled | Calculated χ2 * |
|---|---|---|---|---|---|---|---|---|---|
| M. paraoxydans CSW08 | HS | 0.274 | 0.021 | 1.70 | 12.5 | 63.8 | 0.967 | 0.59 | 1.925 |
| A. denitrificans CSW15 | HS | 0.434 | 0.002 | 1.34 | 44.7 | 46.9 | 0.993 | 0.05 | 0.225 |
| P. citronellolis CSW09 | HS | 0.248 | 0.003 | 1.52 | 119 | 36.1 | 0.989 | 0.14 | 0.155 |
| C. flacumfaciens CSW18 | SFO | 0.013 | - | - | 51.6 | 32.2 | 0.986 | 1.01 | 0.120 |
| B. tritici CSW06 | HS | 0.226 | 0.000 | 1.40 | 426 | 27.2 | 0.987 | 0.29 | 0.113 |
| B. petrii CSW07 | SFO | 0.011 | - | - | 61.2 | 24.5 | 0.857 | 3.08 | 1.029 |
| S. zoogloeoides CSW12 | HS | 0.134 | 0.003 | 1.38 | 150 | 24.0 | 0.995 | 0.05 | 0.020 |
| P. nitroreducens CSW13 | HS | 0.132 | 0.000 | 1.36 | 907 | 19.8 | 0.974 | 0.25 | 0.292 |
| S. acidaminiphila CSW10 | HS | 0.083 | 0.001 | 0.83 | 669 | 9.02 | 0.805 | 0.37 | 0.143 |
| Strain | Phylum/Class/Order/Family | Reference |
|---|---|---|
| Sphingomonas sp. Ibu-2 | Pseudomonadota/Alphaproteobacteria/Sphingomonadales/Sphingomonadaceae | [8,9] |
| Sphingobium yanoikuyae | Pseudomonadota/Alphaproteobacteria/Sphingomonadales/Sphingomonadaceae | [13] |
| Novosphingobium | Pseudomonadota/Alphaproteobacteria/Sphingomonadales/Sphingomonadaceae | [65] |
| Rhizorhabdus wittichii MPO218 | Pseudomonadota/Alphaproteobacteria/Sphingomonadales/Sphingomonadaceae | [14] |
| Sphingopyxis granuli TFA | Pseudomonadota/Alphaproteobacteria/Sphingomonadales/Sphingomonadaceae | [68] |
| Sphingopyxis granuli RW412 | Pseudomonadota/Alphaproteobacteria/Sphingomonadales/Sphingomonadaceae | [16] |
| Rhizobium daejeonense IBU_18 | Pseudomonadota/Alphaproteobacteria/Hyphomicrobiales/Rhizobiaceae | [33] |
| Shinella zoogloeoides CSW12 | Pseudomonadota/Alphaproteobacteria/Hyphomicrobiales/Rhizobiaceae | This study |
| Labrys neptuniae CSW11 | Pseudomonadota/Alphaproteobacteria/Hyphomicrobiales/Xanthobacteraceae | [17] |
| Brucella tritici CSW06 | Pseudomonadota/Alphaproteobacteria/Rhizobiales/Brucellaceae | This study |
| Variovorax sp. Ibu-1 | Pseudomonadota/Betaproteobacteria/Burkholderiales/Comamonadaceae | [60] |
| Comamonas aquatic (+ Bacillus sp.) | Pseudomonadota/Betaproteobacteria/Burkholderiales/Comamonadaceae | [61] |
| Bordetella petrii CSW07 | Pseudomonadota/Betaproteobacteria/Burkholderiales/Alcaligenaceae | This study |
| Achromobacter denitrificans CSW15 | Pseudomonadota/Betaproteobacteria/Burkholderiales/Alcaligenaceae | This study |
| Serratia marcescens BL1 | Pseudomonadota/Gammaproteobacteria/Enterobacterales/Yersiniaceae | [11] |
| Pseudoxanthomonas sp. DIN-3 | Pseudomonadota/Gammaproteobacteria/Xanthomonadales/Xanthomonadaceae | [64] |
| Stenotrophomonas acidaminiphila CSW10 | Pseudomonadota/Gammaproteobacteria/Xanthomonadales/Xanthomonadaceae | This study |
| Pseudoalteromonas sp. | Pseudomonadota/Gammaproteobacteria/Alteromonadales/Pseudoalteromonadaceae | [63] |
| Pseudomonas citronellolis RW422 + RW423+ RW424 | Pseudomonadota/Gammaproteobacteria/Pseudomonadales/Pseudomonadaceae | [24] |
| Pseudomonas alloputida M20 | Pseudomonadota/Gammaproteobacteria/Pseudomonadales/Pseudomonadaceae | [67] |
| Pseudomonas aeruginosa LOIBU1.2 | Pseudomonadota/Gammaproteobacteria/Pseudomonadales/Pseudomonadaceae | [15] |
| Pseudomonas citronellolis CSW09 | Pseudomonadota/Gammaproteobacteria/Pseudomonadales/Pseudomonadaceae | This study |
| Pseudomonas nitroreducens CSW13 | Pseudomonadota/Gammaproteobacteria/Pseudomonadales/Pseudomonadaceae | This study |
| Citrobacter freundii PYI-2 | Pseudomonadota/Gammaproteobacteria/Enterobacterales/Enterobacteriaceae | [62] |
| Citrobacter portucalensis YPI-2 | Pseudomonadota/Gammaproteobacteria/Enterobacterales/Enterobacteriaceae | [62] |
| Klebsiella pneumoniae TIBU2.1 | Pseudomonadota/Gammaproteobacteria/Enterobacterales/Enterobacteriaceae | [15] |
| Klebsiella variicola c | Pseudomonadota/Gammaproteobacteria/Enterobacterales/Enterobacteriaceae | [15] |
| Patulibacter sp. I11 | Actinomycetota/Thermoleophilia/Solirubrobacterales/Patulibacteraceae | [31] |
| Microccocus yunnanensis | Actinomycetota/Actinomycetes/Micrococcales/Micrococcaceae | [12,34] |
| Patulibacter medicamentivorans | Actinomycetota/Thermoleophilia/Solirubrobacterales/Patulibacteraceae | [32] |
| Rhodococcus cerastii IEGM 1278 | Actinomycetota/Actinomycetia/Mycobacteriales/Nocardiaceae | [35] |
| Nocardioides carbamazepine sp. | Actinomycetota/Actinomycetia/Propionibacteriales/Nocardioidaceae | [30] |
| Streptomyces murinus D218 | Actinomycetota/Streptomycetales/Streptomycineae/Streptomycetaceae | [67] |
| Mycolicibacterium aubagnense HPB1.1 | Actinomycetota/Actinomycetes/Mycobacteriales/Mycobacteriaceae | [15] |
| Nocardia sp. NRRL 5646 | Actinomycetota/Actinomycetes/Mycobacteriales/Nocardiaceae | [59] |
| Microbacterium paraoxydans | Actinomycetota/Actinomycetes/Micrococcales/Microbacteriaceae | [23] |
| Microbacterium paraoxydans CSW08 | Actinomycetota/Actinomycetes/Micrococcales/Microbacteriaceae | This study |
| Curtobacterium flaccumfaciens CSW18 | Actinomycetota/Actinomycetes/Micrococcales/Microbacteriaceae | This study |
| Bacillus sp. (+Comamonas aquatic) | Bacillota/Bacilli/Bacillales/Bacillaceae | [61] |
| Bacillus thuringiensis B1(2015b) | Bacillota/Bacilli/Bacillales/Bacillaceae | [10,29] |
| Bacillus siamensis DSI-1 | Bacillota/Bacilli/Bacillales/Bacillaceae | [66] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguilar-Romero, I.; Lara-Moreno, A.; Madrid, F.; Villaverde, J.; Alonso, E.; Santos, J.L.; Morillo, E. Removal of Ibuprofen from Contaminated Water by Bioaugmentation with Novel Bacterial Strains Isolated from Sewage Sludge. Microorganisms 2025, 13, 1927. https://doi.org/10.3390/microorganisms13081927
Aguilar-Romero I, Lara-Moreno A, Madrid F, Villaverde J, Alonso E, Santos JL, Morillo E. Removal of Ibuprofen from Contaminated Water by Bioaugmentation with Novel Bacterial Strains Isolated from Sewage Sludge. Microorganisms. 2025; 13(8):1927. https://doi.org/10.3390/microorganisms13081927
Chicago/Turabian StyleAguilar-Romero, Inés, Alba Lara-Moreno, Fernando Madrid, Jaime Villaverde, Esteban Alonso, Juan Luis Santos, and Esmeralda Morillo. 2025. "Removal of Ibuprofen from Contaminated Water by Bioaugmentation with Novel Bacterial Strains Isolated from Sewage Sludge" Microorganisms 13, no. 8: 1927. https://doi.org/10.3390/microorganisms13081927
APA StyleAguilar-Romero, I., Lara-Moreno, A., Madrid, F., Villaverde, J., Alonso, E., Santos, J. L., & Morillo, E. (2025). Removal of Ibuprofen from Contaminated Water by Bioaugmentation with Novel Bacterial Strains Isolated from Sewage Sludge. Microorganisms, 13(8), 1927. https://doi.org/10.3390/microorganisms13081927








