Isolation of Ultra-Small Opitutaceae-Affiliated Verrucomicrobia from a Methane-Fed Bioreactor
Abstract
1. Introduction
2. Materials and Methods
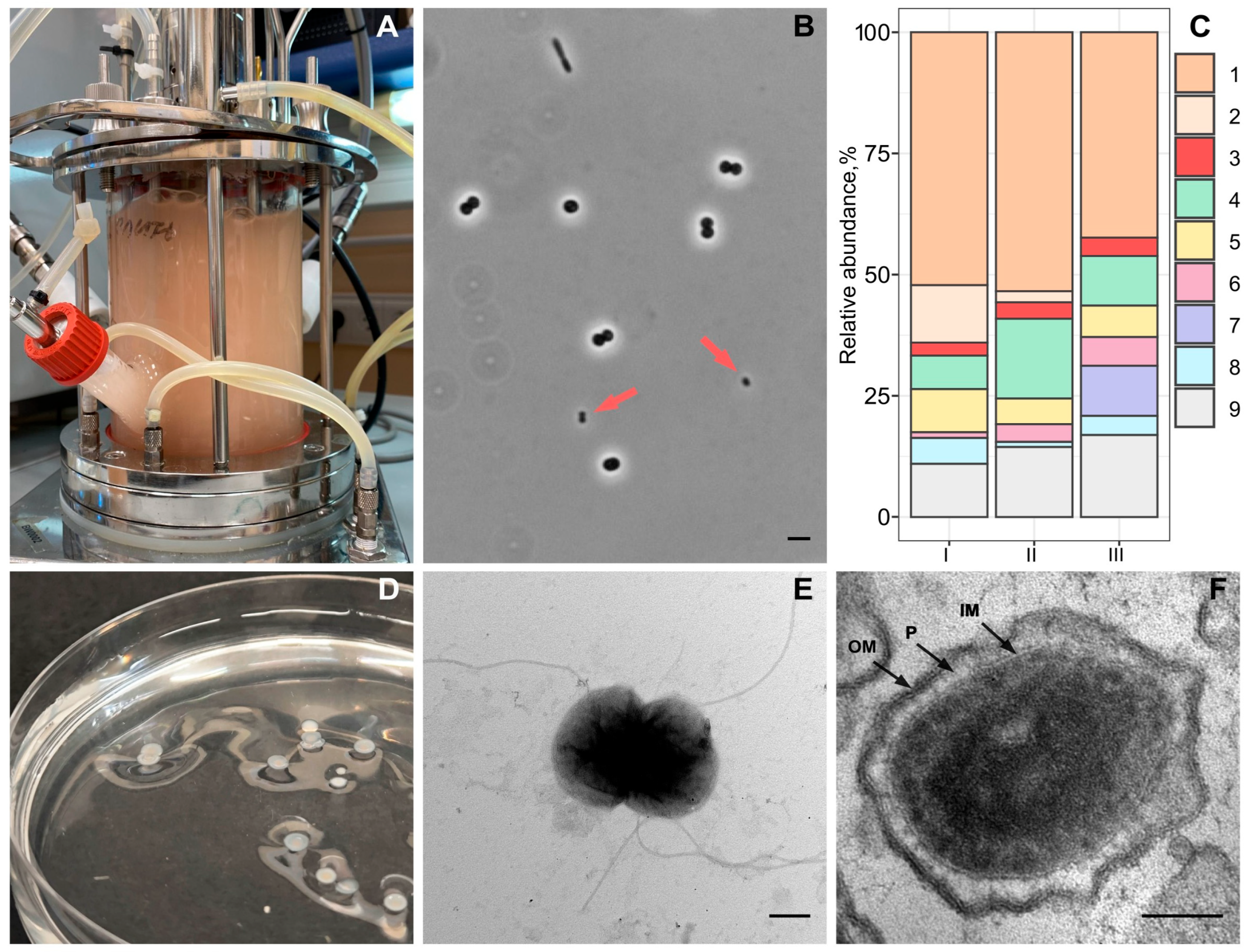
2.1. Methane-Fed Bioreactor and Methane-Oxidizing Consortium Composition
2.2. Isolation and Identification of an Ultra-Small Bacterium
2.3. Electron Microscopy
2.4. Characterization of Physiology
2.5. Co-Culture Experiments
2.6. Genome Sequencing and Annotation
2.7. Phylogenomic Analysis
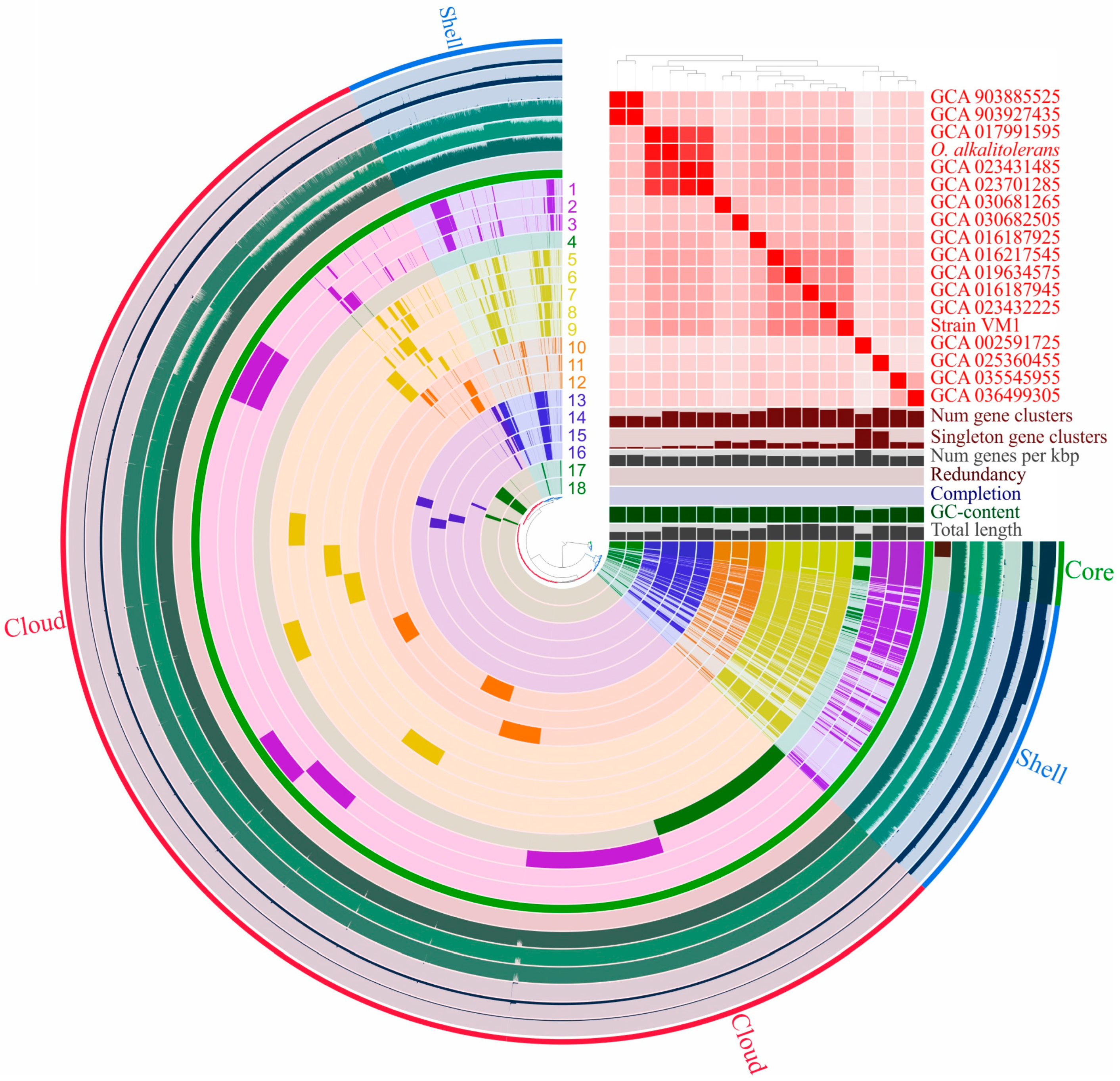
2.8. Pan-Genome Analysis
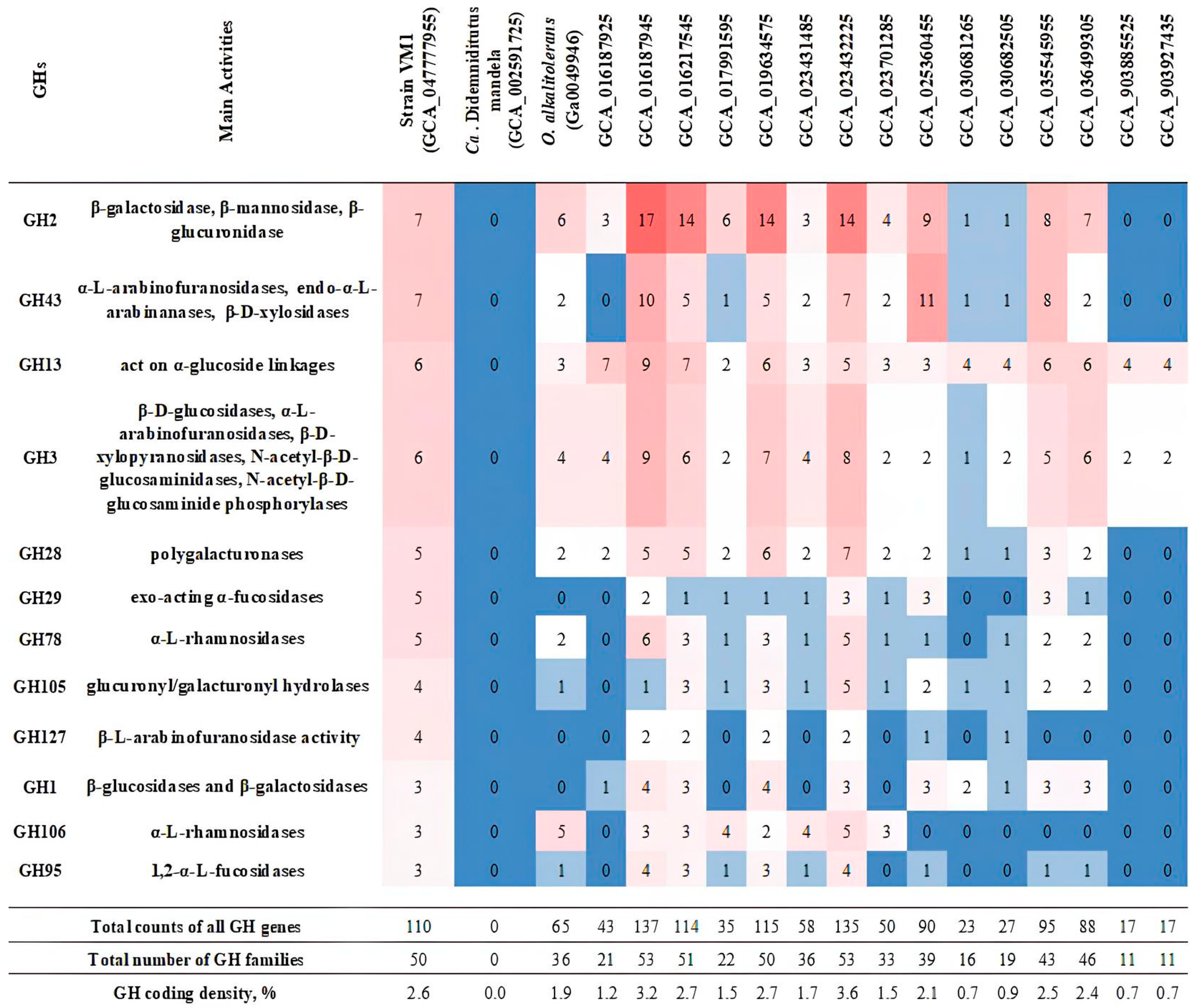
2.9. Search for the Genome-Encoded Xylan-Degrading Enzymes
3. Results
3.1. Methane-Oxidizing Consortium Composition in a Bioreactor
3.2. Isolation and Identification of Strain Vm1
3.3. Physiology and Growth Substrates
3.4. Growth in a Co-Culture with Methanotroph
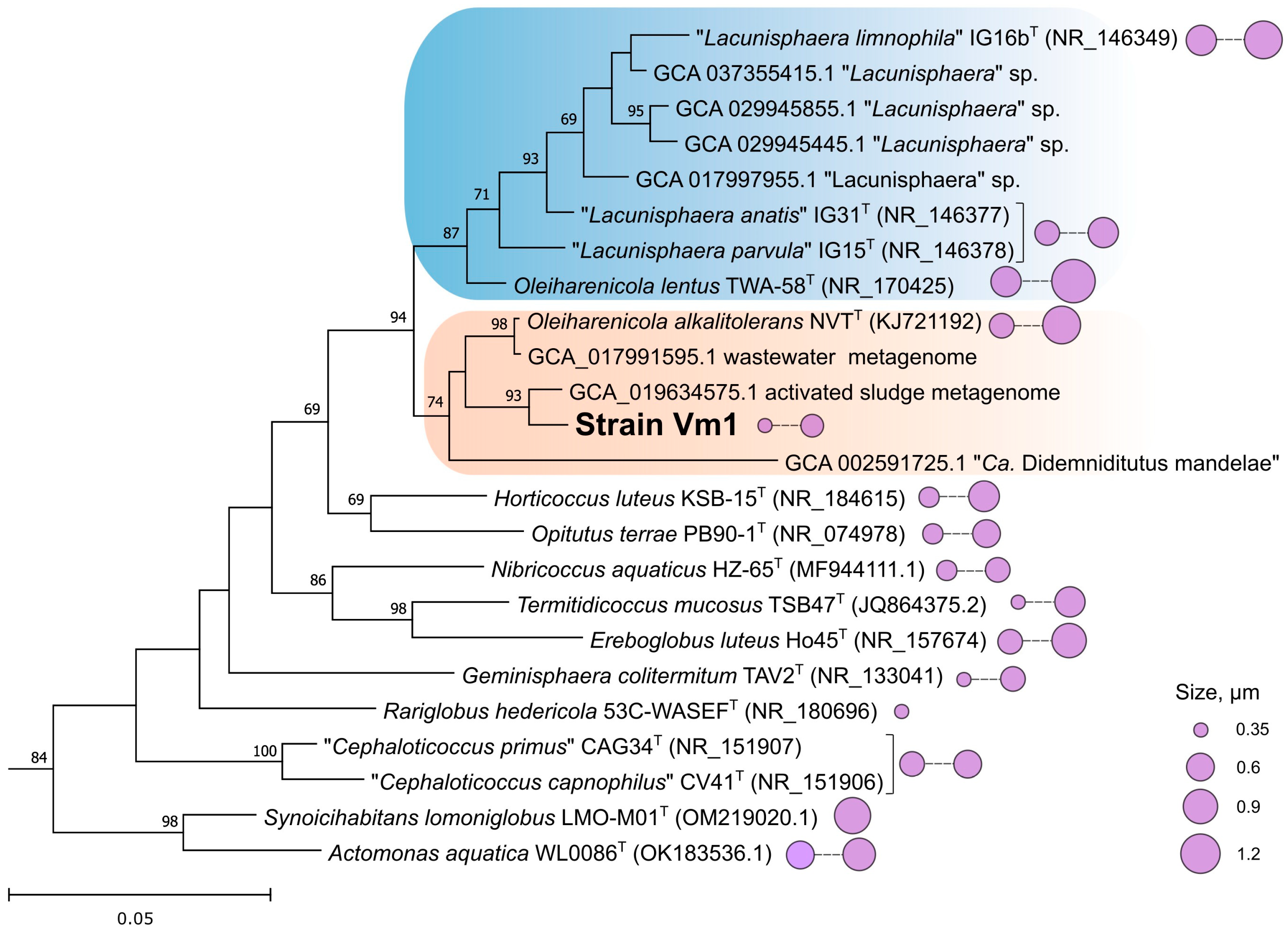
3.5. Genome Analysis and Phylogenomic Reconstructions
3.6. Pan-Genome Analysis of Oleiharenicola–Candidatus Didemniditutus Clade
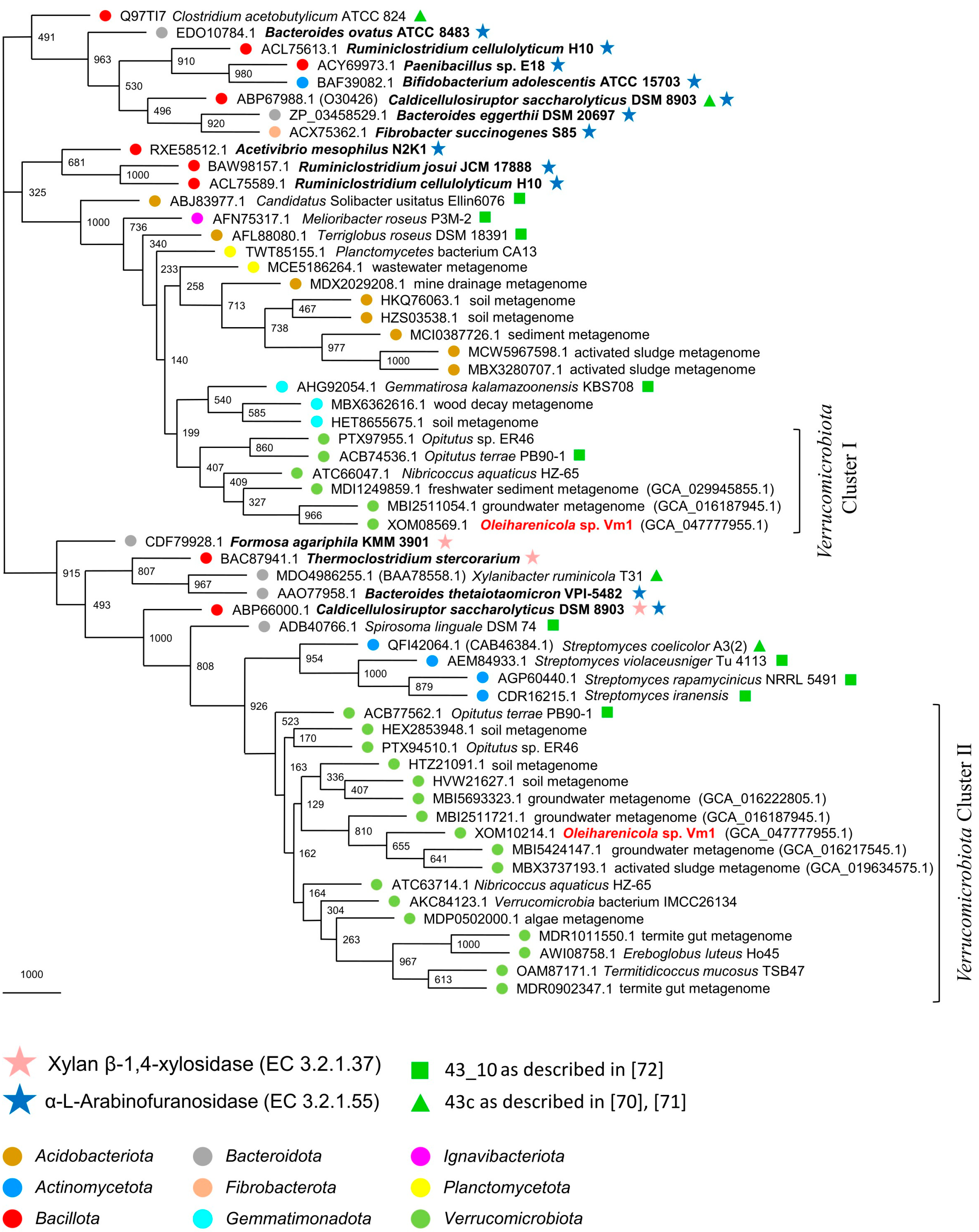
3.7. Phylogeny of Xylan-Degrading Enzymes in Strain Vm1 and Related Verrucomicrobia
4. Discussion
5. Conclusions and Future Research Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hedlund, B.P.; Gosink, J.J.; Staley, J.T. Verrucomicrobia div. nov., a new division of the bacteria containing three new species of Prosthecobacter. Antonie Leeuwenhoek 1997, 72, 29–38. [Google Scholar] [CrossRef]
- Parveen, B.; Mary, I.; Vellet, A.; Ravet, V.; Debroas, D. Temporal dynamics and phylogenetic diversity of free-living and particle-associated Verrucomicrobia communities in relation to environmental variables in a mesotrophic lake. FEMS Microbiol. Ecol. 2013, 83, 189–201. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Stevens, S.L.; Chan, L.K.; Bertilsson, S.; Glavina del Rio, T.; Tringe, S.G.; Malmstrom, R.R.; McMahon, K.D. Ecophysiology of freshwater Verrucomicrobia inferred from metagenome-assembled genomes. Msphere 2017, 2, e00277-17. [Google Scholar] [CrossRef] [PubMed]
- Chiang, E.; Schmidt, M.L.; Berry, M.A.; Biddanda, B.A.; Burtner, A.; Johengen, T.H.; Palladino, D.; Vincent, J.; Denef, V.J. Verrucomicrobia are prevalent in north-temperate freshwater lakes and display class-level preferences between lake habitats. PLoS ONE 2018, 13, e0195112. [Google Scholar]
- Freitas, S.; Hatosy, S.; Fuhrman, J.A.; Huse, S.M.; Mark Welch, D.B.; Sogin, M.L.; Martiny, A.C. Global distribution and diversity of marine Verrucomicrobia. ISME J. 2012, 6, 1499–1505. [Google Scholar] [CrossRef]
- Sichert, A.; Corzett, C.H.; Schechter, M.S.; Unfried, F.; Markert, S.; Becher, D.; Fernandez-Guerra, A.; Liebeke, M.; Schweder, T.; Polz, M.F.; et al. Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol. 2020, 5, 1026–1039. [Google Scholar] [CrossRef] [PubMed]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef]
- Bergmann, G.T.; Bates, S.T.; Eilers, K.G.; Lauber, C.L.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol. Biochem. 2011, 43, 1450–1455. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Liu, Y.; Xie, S.; Liu, Y. Bacterioplankton communities in a high-altitude freshwater wetland. Ann. Microbiol. 2014, 64, 1405–1411. [Google Scholar] [CrossRef]
- Shen, C.; Ge, Y.; Yang, T.; Chu, H. Verrucomicrobial elevational distribution was strongly influenced by soil pH and carbon/nitrogen ratio. J. Soils Sediment. 2017, 17, 2449–2456. [Google Scholar] [CrossRef]
- Ivanova, A.A.; Dedysh, S.N. Phylogenetic diversity of Verrucomicrobiota in fens of Northern Russia. Microbiology 2023, 92 (Suppl. 1), S7–S11. [Google Scholar] [CrossRef]
- Nixon, S.L.; Daly, R.A.; Borton, M.A.; Solden, L.M.; Welch, S.A.; Cole, D.R.; Mouser, P.J.; Wilkins, M.J.; Wrighton, K.C. Genome-resolved metagenomics extends the environmental distribution of the Verrucomicrobia phylum to the deep terrestrial subsurface. Msphere 2019, 4, e00613-19. [Google Scholar] [CrossRef] [PubMed]
- Isanapong, J.; Sealy Hambright, W.; Willis, A.G.; Boonmee, A.; Callister, S.J.; Burnum, K.E.; Paša-Tolić, L.; Nicora, C.D.; Wertz, J.T.; Schmidt, T.M.; et al. Development of an ecophysiological model for Diplosphaera colotermitum TAV2, a termite hindgut Verrucomicrobium. ISME J. 2013, 7, 1803–1813. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Kuwahara, H.; Fujita, K.; Noda, S.; Kihara, K.; Yamada, A.; Ohkuma, M.; Hongoh, Y. Intranuclear verrucomicrobial symbionts and evidence of lateral gene transfer to the host protist in the termite gut. ISME J. 2014, 8, 1008–1019. [Google Scholar] [CrossRef]
- Wagner, M.; Horn, M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 2006, 17, 241–249. [Google Scholar] [CrossRef]
- Van Niftrik, L.; Devos, D.P. Planctomycetes-Verrucomicrobia-Chlamydiae bacterial superphylum: New model organisms for evolutionary cell biology. Front. Microbiol. 2017, 8, 1458. [Google Scholar] [CrossRef]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef]
- Gómez-Gallego, C.; Pohl, S.; Salminen, S.; De Vos, W.M.; Kneifel, W. Akkermansia muciniphila: A novel functional microbe with probiotic properties. Benef. Microbes 2016, 7, 571–584. [Google Scholar] [CrossRef] [PubMed]
- de Vos, W.M. Microbe Profile: Akkermansia muciniphila: A conserved intestinal symbiont that acts as the gatekeeper of our mucosa. Microbiology 2017, 163, 646–648. [Google Scholar] [CrossRef]
- Guo, X.; Li, S.; Zhang, J.; Wu, F.; Li, X.; Wu, D.; Zhang, M.; Ou, Z.; Jie, Z.; Yan, Q.; et al. Genome sequencing of 39 Akkermansia muciniphila isolates reveals its population structure, genomic and functional diverisity, and global distribution in mammalian gut microbiotas. BMC Genom. 2017, 18, 800. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, M.; Brazel, D.M.; Swan, B.K.; Arnosti, C.; Chain, P.S.; Reitenga, K.G.; Xie, G.; Poulton, N.J.; Lluesma Gomez, M.; Masland, D.E.; et al. Capturing single cell genomes of active polysaccharide degraders: An unexpected contribution of Verrucomicrobia. PLoS ONE 2012, 7, e35314. [Google Scholar] [CrossRef]
- Herlemann, D.P.; Lundin, D.; Labrenz, M.; Jürgens, K.; Zheng, Z.; Aspeborg, H.; Andersson, A.F. Metagenomic de novo assembly of an aquatic representative of the verrucomicrobial class Spartobacteria. mBio 2013, 4, e00569-12. [Google Scholar] [CrossRef]
- Cardman, Z.; Arnosti, C.; Durbin, A.; Ziervogel, K.; Cox, C.; Steen, A.D.; Teske, A. Verrucomicrobia are candidates for polysaccharide-degrading bacterioplankton in an arctic fjord of Svalbard. Appl. Environ. Microbiol. 2014, 80, 3749–3756. [Google Scholar] [CrossRef]
- Rakitin, A.L.; Kulichevskaya, I.S.; Beletsky, A.V.; Mardanov, A.V.; Dedysh, S.N.; Ravin, N.V. Verrucomicrobia of the family Chthoniobacteraceae participate in xylan degradation in boreal peat soils. Microorganisms 2024, 12, 2271. [Google Scholar] [CrossRef]
- Op den Camp, H.J.; Islam, T.; Stott, M.B.; Harhangi, H.R.; Hynes, A.; Schouten, S.; Jetten, M.S.M.; Birkeland, N.K.; Pol, A.; Dunfield, P.F. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 2009, 1, 293–306. [Google Scholar] [CrossRef]
- Schmitz, R.A.; Peeters, S.H.; Versantvoort, W.; Picone, N.; Pol, A.; Jetten, M.S.; Op den Camp, H.J. Verrucomicrobial methanotrophs: Ecophysiology of metabolically versatile acidophiles. FEMS Microbiol. Rev. 2021, 45, fuab007. [Google Scholar] [CrossRef] [PubMed]
- Lopera, J.; Miller, I.J.; McPhail, K.L.; Kwan, J.C. Increased biosynthetic gene dosage in a genome-reduced defensive bacterial symbiont. Msystems 2017, 2, e00096-17. [Google Scholar] [CrossRef]
- Sizikov, S.; Burgsdorf, I.; Handley, K.M.; Lahyani, M.; Haber, M.; Steindler, L. Characterization of sponge-associated Verrucomicrobia: Microcompartment-based sugar utilization and enhanced toxin–antitoxin modules as features of host-associated Opitutales. Environ. Microbiol. 2020, 22, 4669–4688. [Google Scholar] [CrossRef] [PubMed]
- Hedlund, B.P. Verrucomicrobia phyl. nov. In Bergey’s Manual of Systematics of Archaea and Bacteria; Trujillo, M.E., Dedysh, S., DeVos, P., Hedlund, B., Kämpfer, P., Rainey, F.A., Whitman, W.B., Eds.; Wiley: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Spring, S.; Bunk, B.; Spröer, C.; Schumann, P.; Rohde, M.; Tindall, B.J.; Klenk, H.P. Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum. ISME J. 2016, 10, 2801–2816. [Google Scholar] [CrossRef]
- Cho, J.C.; Vergin, K.L.; Morris, R.M.; Giovannoni, S.J. Lentisphaera araneosa gen. nov., sp. nov, a transparent exopolymer producing marine bacterium, and the description of a novel bacterial phylum, Lentisphaerae. Environ. Microbiol. 2004, 6, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Chin, K.J.; Liesack, W.; Janssen, P.H. Opitutus terrae gen. nov., sp. nov., to accommodate novel strains of the division ‘Verrucomicrobia’ isolated from rice paddy soil. Int. J. Syst. Evol. Microbiol. 2001, 51, 1965–1968. [Google Scholar] [CrossRef]
- Janssen, P.H.; Yates, P.S.; Grinton, B.E.; Taylor, P.M.; Sait, M. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 2002, 68, 2391–2396. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.J.; Hugenholtz, P.; Sangwan, P.; Osborne, C.A.; Janssen, P.H. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 2003, 69, 7210–7215. [Google Scholar] [CrossRef]
- Sangwan, P.; Kovac, S.; Davis, K.E.; Sait, M.; Janssen, P.H. Detection and cultivation of soil Verrucomicrobia. Appl. Environ. Microbiol. 2005, 71, 8402–8410. [Google Scholar] [CrossRef]
- Davis, K.E.; Joseph, S.J.; Janssen, P.H. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 2005, 71, 826–834. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.E.R.; Sangwan, P.; Janssen, P.H. Acidobacteria, Rubrobacteridae and Chloroflexi are abundant among very slow-growing and mini-colony-forming bacteria. Environ. Microbiol. 2011, 13, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Tamaki, H.; Tanaka, K.; Tozawa, E.; Matsuzawa, H.; Toyama, T. “Duckweed-microbe co-cultivation method” for isolating a wide variety of microbes including taxonomically novel microbes. Microbes Environ. 2018, 33, 402–406. [Google Scholar] [CrossRef]
- Tanaka, Y.; Tozawa, E.; Iwashita, T.; Morishita, Y.; Tamaki, H.; Toyama, T.; Morikawa, M.; Kamagata, Y.; Moriet, K. Successful Isolation of Diverse Verrucomicrobiota Strains through the Duckweed-Microbes Co-cultivation Method. Microbes Environ. 2024, 39, ME24019. [Google Scholar] [CrossRef]
- Oshkin, I.Y.; Belova, S.E.; Khokhlachev, N.S.; Semenova, V.A.; Chervyakova, O.P.; Chernushkin, D.V.; Tikhonova, E.N.; Mardanov, A.V.; Ravin, N.V.; Popov, V.O.; et al. Molecular analysis of the microbial community developing in continuous culture of Methylococcus sp. Concept-8 on natural gas. Microbiology 2020, 89, 551–559. [Google Scholar] [CrossRef]
- Saltykova, V.A.; Danilova, O.V.; Oshkin, I.Y.; Belova, S.E.; Suzina, N.E.; Pimenov, N.V.; Dedysh, S.N. Methyloraptor flagellatus gen. nov., sp. nov., novel Ancalomicrobiaceae-affiliated facultatively methylotrophic bacteria that feed on methanotrophs of the genus Methylococcus. Syst. Appl. Microbiol. 2025, 48, 126565. [Google Scholar] [CrossRef]
- Frey, B.; Rime, T.; Phillips, M.; Stierli, B.; Hajdas, I.; Widmer, F.; Hartmann, M. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 2016, 92, fiw018. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Knight, R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, G.J. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. Peer J. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Beletsky, A.V.; Ivanova, A.A.; Kulichevskaya, I.S.; Suzina, N.E.; Philippov, D.A.; Rakitin, A.L.; Mardanov, A.V.; Ravin, N.V. Wide distribution of Phycisphaera-like planctomycetes from WD2101 soil group in peatlands and genome analysis of the first cultivated representative. Environ. Microbiol. 2021, 23, 1510–1526. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001, 56, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Eren, A.M.; Esen, Ö.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T. O Anvi’o: An advanced analysis and visualization platform for ’omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. dbCAN3: Automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef]
- Rochman, F.F.; Kim, J.J.; Rijpstra, W.I.C.; Sinninghe Damsté, J.S.; Schumann, P.; Verbeke, T.J.; Dunfield, P.F. Oleiharenicola alkalitolerans gen. nov., sp. nov., a new member of the phylum Verrucomicrobia isolated from an oilsands tailings pond. Int. J. Syst. Evol. Microbiol. 2018, 68, 1078–1084. [Google Scholar] [CrossRef]
- Rast, P.; Glockner, I.; Boedeker, C.; Jeske, O.; Wiegand, S.; Reinhardt, R.; Schumann, P.; Rohde, M.; Spring, S.; Glockner, F.O.; et al. Three Novel Species with Peptidoglycan Cell Walls form the New Genus Lacunisphaera gen. nov. in the Family Opitutaceae of the Verrucomicrobial Subdivision 4. Front. Microbiol. 2017, 8, 202. [Google Scholar] [CrossRef]
- Chen, W.M.; Chen, T.Y.; Yang, C.C.; Sheu, S.Y. Oleiharenicola lentus sp. nov., isolated from irrigation water. Int. J. Syst. Evol. Microbiol. 2020, 70, 3440–3448. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Peyraud, R.; Kiefer, P.; Christen, P.; Delmotte, N.; Massou, S.; Portais, J.-C.; Vorholt, J.A. The ethylmalonyl-CoA pathway is used in place of the glyoxylate cycle by Methylobacterium extorquens AM1 during growth on acetate. J. Biol. Chem. 2012, 287, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.M.; Stülke, J. Malate metaboli in Bacillus subtilis: Distinct roles for three classes of malate-oxidizing enzymes. FEMS Microbiol. Lett. 2013, 339, 17–22. [Google Scholar] [CrossRef]
- Cabello-Yeves, P.J.; Ghai, R.; Mehrshad, M.; Picazo, A.; Camacho, A.; Rodriguez-Valera, F. Reconstruction of diverse verrucomicrobial genomes from metagenome datasets of freshwater reservoirs. Front. Microbiol. 2017, 8, 2131. [Google Scholar] [CrossRef]
- Khmelenina, V.N.; Gayazov, R.R.; Suzina, N.E.; Doronina, N.V.; Mshensky, Y.N.; Trotsenko, Y.A. The synthesis of polysaccharides by Methylococcus capsulatus under various conditions of cultivation. Mikrobiologiya 1992, 61, 404–410. [Google Scholar]
- Erbilgin, O.; McDonald, K.L.; Kerfeld, C.A. Characterization of a planctomycetal organelle: A novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl. Environ. Microbiol. 2014, 80, 2193–2205. [Google Scholar] [CrossRef]
- McDonald, A.G.; Boyce, S.; Tipton, K.F. ExplorEnz: The primary source of the IUBMB enzyme list. Nucleic Acids Res. 2009, 37, D593–D597. [Google Scholar] [CrossRef] [PubMed]
- Naumoff, D.G. β-Fructosidase superfamily: Homology with some α-L-arabinases and β-D xylosidases. Prot. Struct. Funct. Genet. 2001, 42, 66–76. [Google Scholar] [CrossRef]
- Pons, T.; Naumoff, D.G.; Martínez-Fleites, C.; Hernández, L. Three acidic residues are at the active site of a β-propeller architecture in glycoside hydrolase families 32, 43, 62, and 68. Prot. Struct. Funct. Bioinform. 2004, 54, 424–432. [Google Scholar] [CrossRef]
- Mewis, K.; Lenfant, N.; Lombard, V.; Henrissat, B. Dividing the large glycoside hydrolase family 43 into subfamilies: A motivation for detailed enzyme characterization. Appl. Environ. Microbiol. 2016, 82, 1686–1692. [Google Scholar] [CrossRef]
- Schut, F.; Prins, R.A.; Gottschal, J.C. Oligotrophy and pelagic marine bacteria: Facts and fiction. Aquat. Microb. Ecol. 1997, 12, 177–202. [Google Scholar] [CrossRef]
- Duda, V.I.; Suzina, N.E.; Polivtseva, V.N.; Boronin, A.M. Ultramicrobacteria: Formation of the concept and contribution of ultramicrobacteria to biology. Microbiology 2002, 81, 379–390. [Google Scholar] [CrossRef]
- Janssen, P.H.; Schuhmann, A.; Mörschel, E.; Rainey, F.A. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl. Environ. Microbiol. 1997, 63, 1382–1388. [Google Scholar] [CrossRef] [PubMed]
- Pitt, A.; Schmidt, J.; Koll, U.; Hahn, M.W. Rariglobus hedericola gen. nov., sp. nov., belonging to the Verrucomicrobia, isolated from a temperate freshwater habitat. Int. J. Syst. Evol. Microbiol. 2020, 70, 1830–1836. [Google Scholar] [CrossRef]
- Velimirov, B. Nanobacteria, ultramicrobacteria and starvation forms: A search for the smallest metabolizing bacterium. Microbes Environ. 2001, 16, 67–77. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. Candidatus list no. 3. Lists of names of prokaryotic Candidatus taxa. Int. J. Syst. Evol. Microbiol. 2022, 72, 5186. [Google Scholar] [CrossRef]
- Chakravorty, D.; Khan, M.F.; Patra, S. Multifactorial level of extremostability of proteins: Can they be exploited for protein engineering? Extremophiles 2017, 21, 419–444. [Google Scholar] [CrossRef]
- Saidi-Mehrabad, A.; He, Z.; Tamas, I.; Sharp, C.E.; Brady, A.L.; Rochman, F.F.; Bodrossy, L.; Abell, G.C.; Penner, T.; Dong, X.; et al. Methanotrophic bacteria in oilsands tailing ponds of northern Alberta. ISME J. 2013, 7, 908–921. [Google Scholar] [CrossRef]
- Shi, L.D.; Lv, P.L.; McIlroy, S.J.; Wang, Z.; Dong, X.L.; Kouris, A.; Lai, C.-Y.; Tyson, G.W.; Strous, M.; Zhao, H.P. Methane-dependent selenate reduction by a bacterial consortium. ISME J. 2021, 15, 3683–3692. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Tao, Y.; Mao, Z.; Gu, Q.; Han, Y.; Hu, B.; Wang, H.; Lai, A.; Xing, P.; Wu, Q.L. Iron oxides act as an alternative electron acceptor for aerobic methanotrophs in anoxic lake sediments. Water Res. 2023, 234, 119833. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danilova, O.V.; Salova, V.D.; Oshkin, I.Y.; Naumoff, D.G.; Ivanova, A.A.; Suzina, N.E.; Dedysh, S.N. Isolation of Ultra-Small Opitutaceae-Affiliated Verrucomicrobia from a Methane-Fed Bioreactor. Microorganisms 2025, 13, 1922. https://doi.org/10.3390/microorganisms13081922
Danilova OV, Salova VD, Oshkin IY, Naumoff DG, Ivanova AA, Suzina NE, Dedysh SN. Isolation of Ultra-Small Opitutaceae-Affiliated Verrucomicrobia from a Methane-Fed Bioreactor. Microorganisms. 2025; 13(8):1922. https://doi.org/10.3390/microorganisms13081922
Chicago/Turabian StyleDanilova, Olga V., Varvara D. Salova, Igor Y. Oshkin, Daniil G. Naumoff, Anastasia A. Ivanova, Natalia E. Suzina, and Svetlana N. Dedysh. 2025. "Isolation of Ultra-Small Opitutaceae-Affiliated Verrucomicrobia from a Methane-Fed Bioreactor" Microorganisms 13, no. 8: 1922. https://doi.org/10.3390/microorganisms13081922
APA StyleDanilova, O. V., Salova, V. D., Oshkin, I. Y., Naumoff, D. G., Ivanova, A. A., Suzina, N. E., & Dedysh, S. N. (2025). Isolation of Ultra-Small Opitutaceae-Affiliated Verrucomicrobia from a Methane-Fed Bioreactor. Microorganisms, 13(8), 1922. https://doi.org/10.3390/microorganisms13081922





