Next-Generation Nucleic Acid-Based Diagnostics for Viral Pathogens: Lessons Learned from the SARS-CoV-2 Pandemic
Abstract
1. Introduction
2. Coronavirus Epidemiology
3. Advances in Viral Nucleic Acid Detection Methods
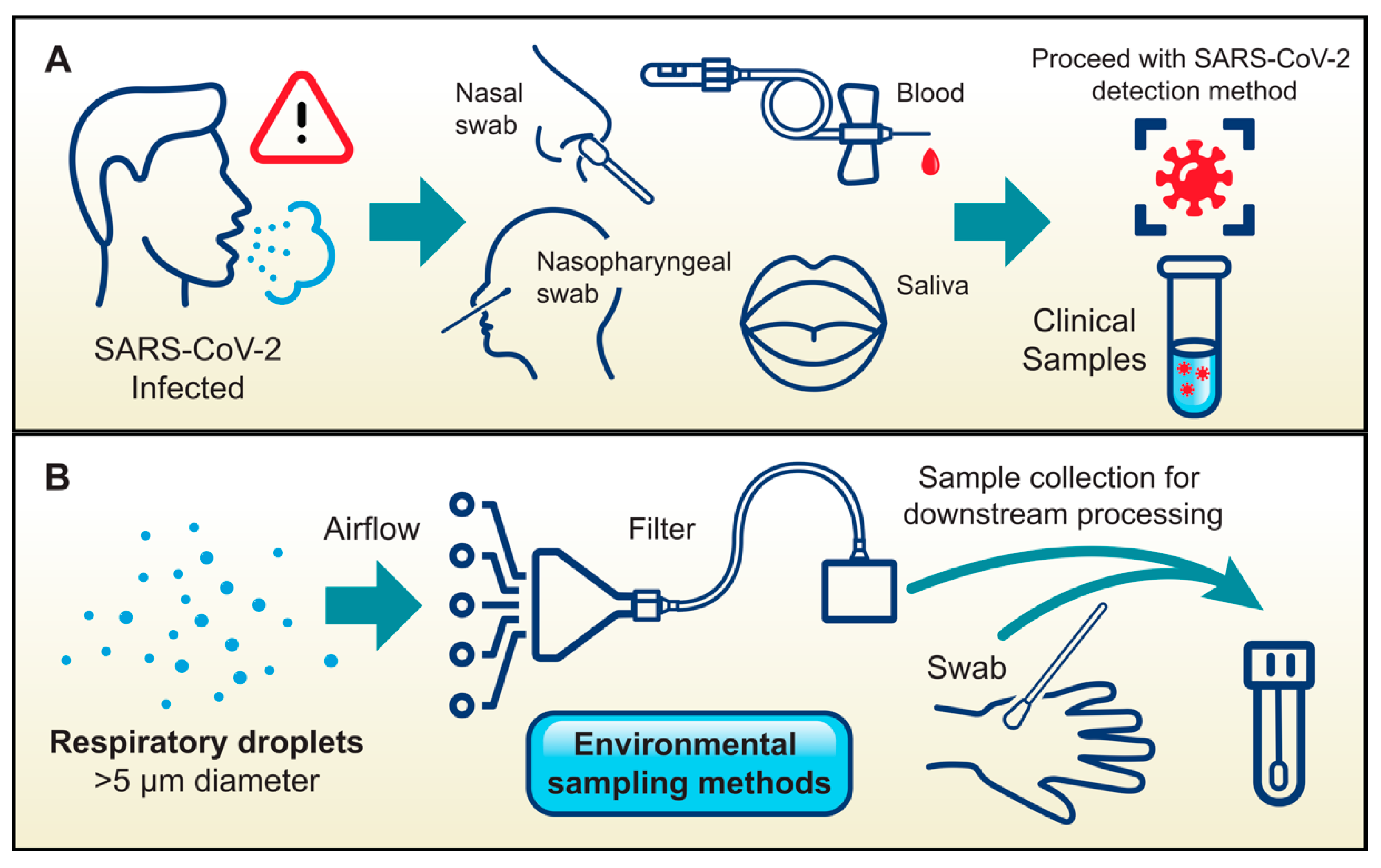
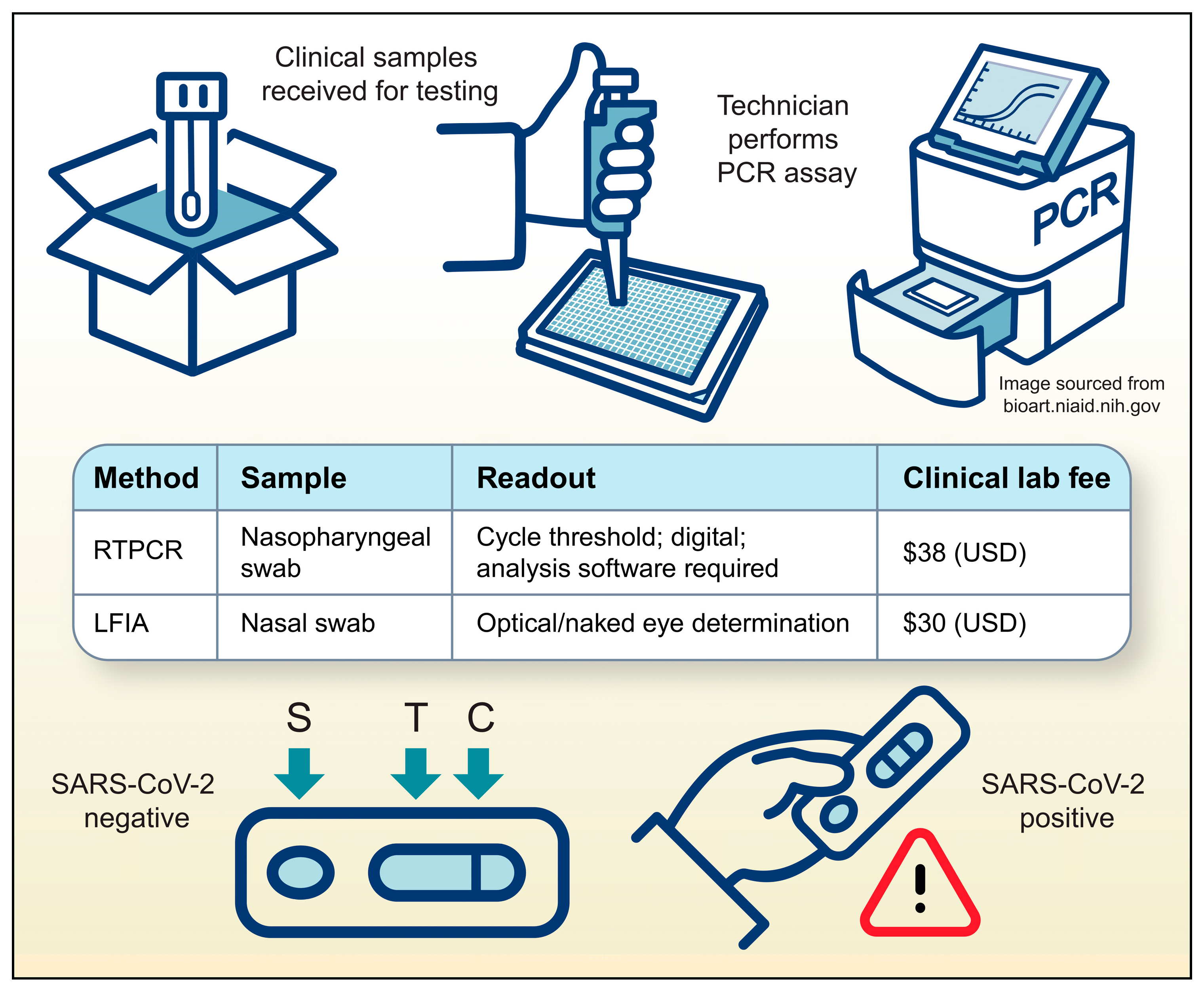
3.1. Necessity–the Mother of Invention
3.2. Isothermal Amplification Methods (IAM)
3.2.1. Loop-Mediated Isothermal Amplification (LAMP)
3.2.2. Recombinase Polymerase Amplification (RPA)
3.2.3. Nicking Endonuclease Amplification Reaction (NEAR)
3.2.4. Rolling Circle Amplification (RCA)
3.2.5. Nucleic Acid Sequence-Based Amplification (NASBA)
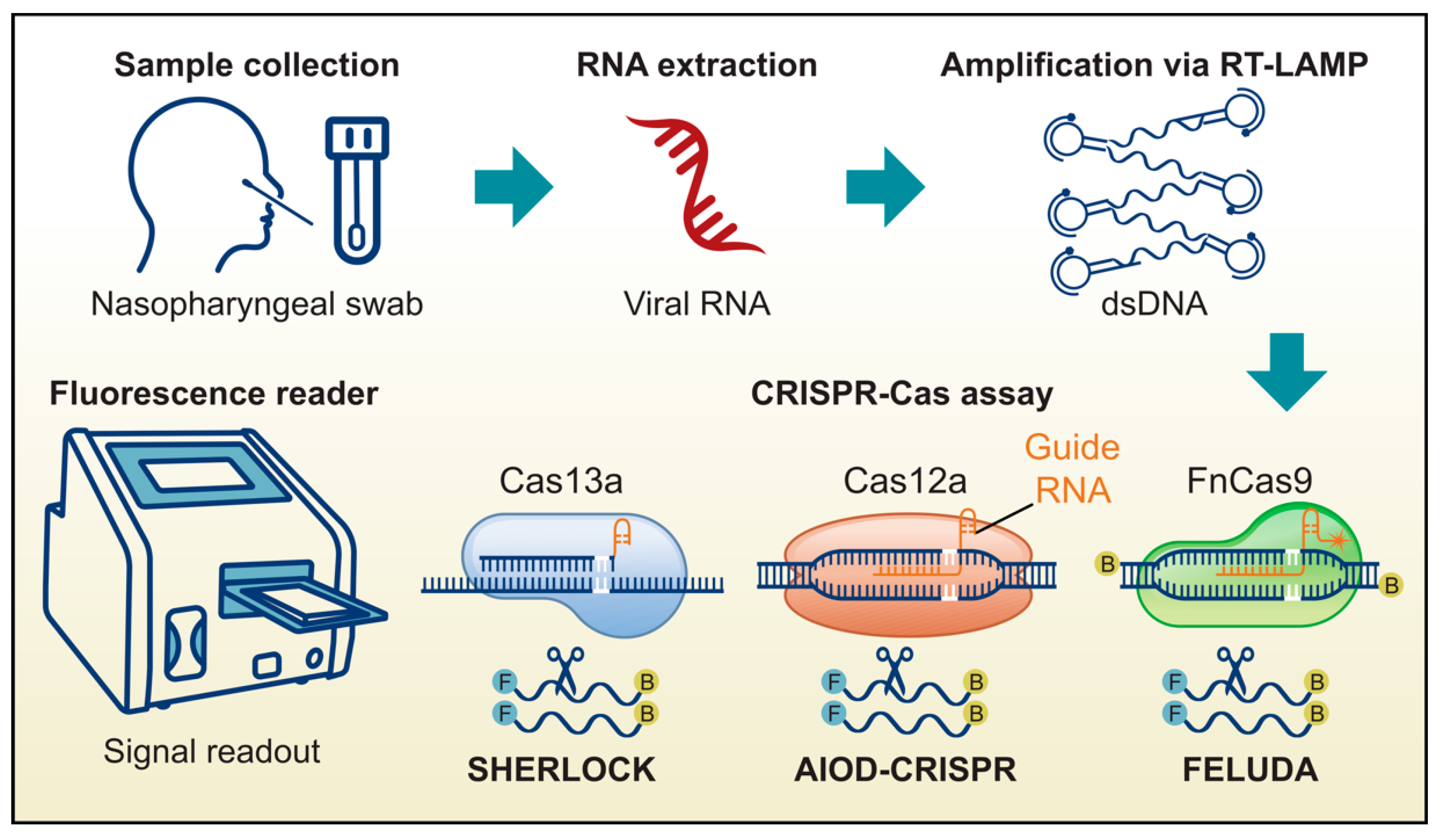
3.3. Target Sensing Methods (TSM)
3.3.1. Specific High Sensitivity Enzymatic Reporter Unlocking (SHERLOCK)
3.3.2. DNA Endonuclease Targeted CRISPR Trans Reporter (DETECTR) and All-In-One Dual CRISPR-Cas12a (AIOD-CRISPR)
3.3.3. FnCAS9 Editor Linked Uniform Detection Assay (FELUDA)
3.3.4. Aptamer-Based Biosensors (Aptasensors)
4. Discussion
4.1. Complexities of Implementation
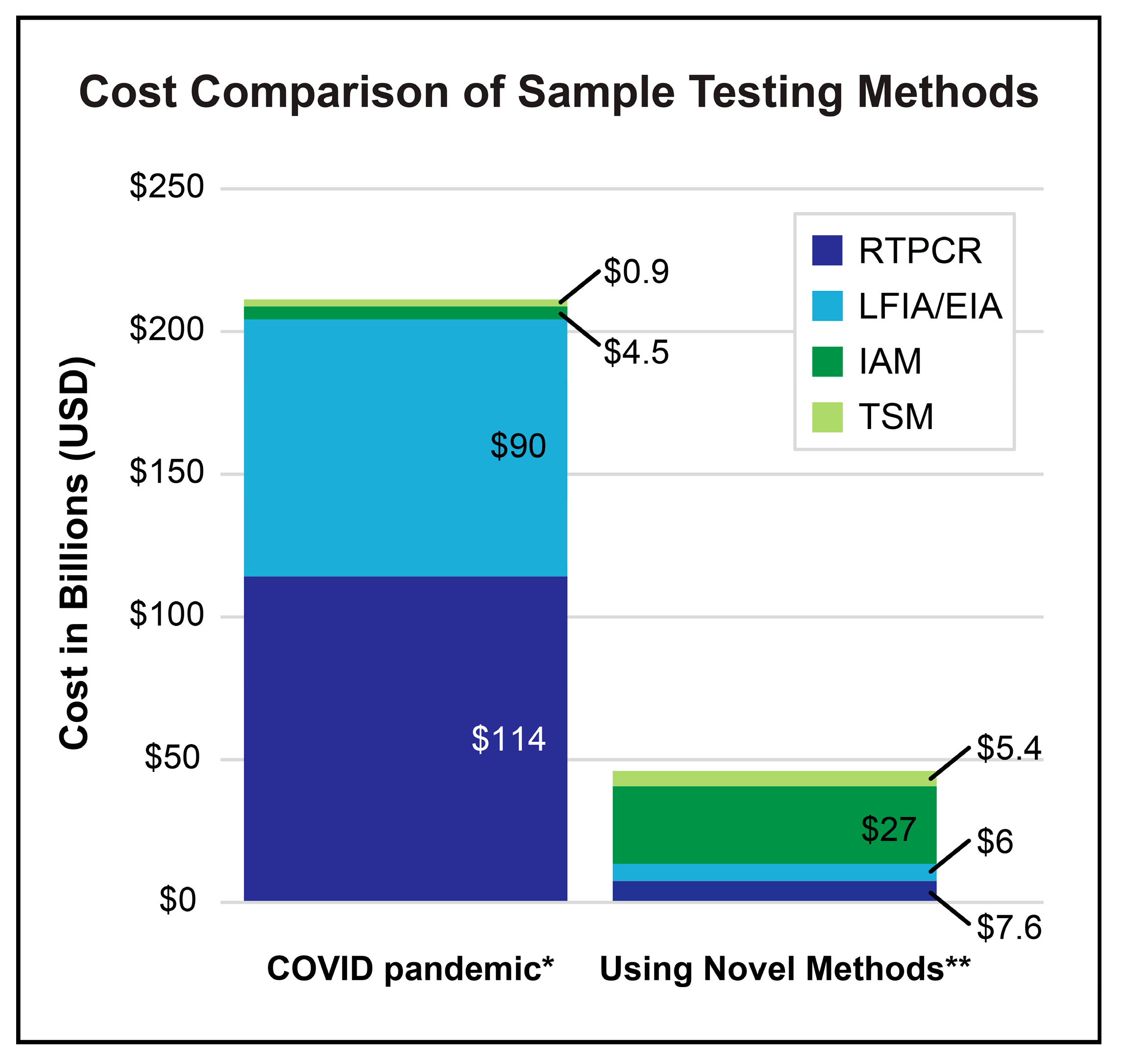
4.2. Cost of Implementation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AOID-CRISPR | All-in-One-CRISPR |
| CoV | Coronavirus |
| COVID-19 | Coronavirus Infectious Disease-2019 |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| DETTECTR | DNA Endonuclease Targeted CRISPR Trans Reporter |
| EIA | Enzymatic ImmunoAssay |
| FELUDA | FnCAS9 Editor Linked Uniform Detection Assay |
| IAM | Isothermal Amplification Methods |
| LAMP | Loop-Mediated Isothermal Amplification |
| LFIA | Lateral Flow Immunoassay |
| MERS | Middle East Respiratory Syndrome |
| MRI | Magnetic Resonance Imaging |
| NA | Nucleic Acid |
| NASBA | Nucleic Acid Sequence-Based Amplification |
| NEAR | Nicking Endonuclease Amplification Reaction |
| PET/SPECT | Positron Emission Tomography/Single Photon Emission Computed Tomography |
| POC | Point-of-Care |
| RCA | Rolling Circle Amplification |
| RNA | Ribonucleic Acid |
| RPA | Recombinase Polymerase Amplification |
| RTPCR | Real-Time reverse transcriptase Polymerase Chain Reaction |
| SARS | Severe Acute Respiratory Syndrome |
| SHERLOCK | Specific High Sensitivity Enzymatic Reporter Unlocking |
| TSM | Target Sensing Methods |
References
- Mathieu, E.; Ritchie, H.; Rodés-Guirao, L.; Appel, C.; Gavrilov, D.; Giattino, C.; Hasell, J.; Macdonald, B.; Dattani, S.; Beltekian, D.; et al. Coronavirus (COVID-19) Cases. Published online at OurWorldinData.org. 2020. Available online: https://ourworldindata.org/covid-cases (accessed on 3 June 2025).
- Respiratory Virus Hospitalization Surveillance Network (RESP-NET). RESP-NET Is a CDC System That Monitors Laboratory-Confirmed Hospitalizations Associated with COVID-19, RSV, and Flu Among Children and Adults. Available online: https://www.cdc.gov/resp-net/dashboard/ (accessed on 30 May 2025).
- Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 23 May 2025).
- Safder, T.; McCullough, P.; Wheelan, K.; Rahimi, G.; Zurawski, S.; Zurawski, G.; Gu, J.; Wang, X.; Balaji, U.; Berhe, M.; et al. Screening for SARS-CoV-2 via PCR and serological testing in asymptomatic healthcare workers. Bayl. Univ. Med. Cent. Proc. 2021, 34, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Binny, R.N.; Priest, P.; French, N.; Parry, M.; Lustig, A.; Hendy, S.; Maclaren, O.; Ridings, K.; Steyn, N.; Vattiato, G.; et al. Sensitivity of Reverse Transcription Polymerase Chain Reaction Tests for Severe Acute Respiratory Syndrome Coronavirus 2 Through Time. J. Infect. Dis. 2022, 227, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Dip, S.D.; Sarkar, S.L.; Setu, M.A.A.; Das, P.K.; Pramanik, M.H.A.; Rubayet Ul Alam, A.S.M.; Al-Emran, H.M.; Hossain, M.A.; Jahid, I.K. Evaluation of RT-PCR assays for detection of SARS-CoV-2 variants of concern. Sci. Rep. 2023, 13, 2342. [Google Scholar] [CrossRef]
- Eftekhari, A.; Alipour, M.; Chodari, L.; Dizaj, S.M.; Ardalan, M.; Samiei, M.; Sharifi, S.; Vahed, S.Z.; Huseynova, I.; Khalilov, R.; et al. A Comprehensive Review of Detection Methods for SARS-CoV-2. Microorganisms 2021, 9, 232. [Google Scholar] [CrossRef]
- Castellanos, M.; Somoza, A. Emerging clinically tested detection methods for COVID-19. FEBS J. 2023, 290, 3089–3104. [Google Scholar] [CrossRef]
- Vandenberg, O.; Martiny, D.; Rochas, O.; van Belkum, A.; Kozlakidis, Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021, 19, 171–183. [Google Scholar] [CrossRef]
- Cheng, M.P.; Papenburg, J.; Desjardins, M.; Kanjilal, S.; Quach, C.; Libman, M.; Dittrich, S.; Yansouni, C.P. Diagnostic Testing for Severe Acute Respiratory Syndrome-Related Coronavirus 2: A Narrative Review. Ann. Intern. Med. 2020, 172, 726–734. [Google Scholar] [CrossRef]
- Aloraij, Y.M.; Suaifan, G.A.R.Y.; Shibl, A.; Al-Kattan, K.; Zourob, M.M. Development of Rapid Aptamer-Based Screening Assay for the Detection of Covid-19 Variants. ACS Omega 2023, 8, 32877–32883. [Google Scholar] [CrossRef]
- Pascual-Iglesias, A.; Canton, J.; Ortega-Prieto, A.M.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A. An Overview of Vaccines against SARS-CoV-2 in the COVID-19 Pandemic Era. Pathogens 2021, 10, 1030. [Google Scholar] [CrossRef] [PubMed]
- Tai, J.-H.; Lee, D.-C.; Lin, H.-F.; Chao, T.-L.; Ruan, Y.; Cheng, Y.-W.; Chou, Y.-C.; Lin, Y.-Y.; Chang, S.-Y.; Chen, P.-J. Tradeoffs between proliferation and transmission in virus evolution- insights from evolutionary and functional analyses of SARS-CoV-2. Virol. J. 2025, 22, 107. [Google Scholar] [CrossRef]
- Grelewska-Nowotko, K.; Elhag, A.E.; Turowski, T.W. Transcription Kinetics in the Coronavirus Life Cycle. Wiley Interdiscip. Rev. RNA 2025, 16, e70000. [Google Scholar] [CrossRef]
- Dangi, T.; Li, S.; Penaloza-MacMaster, P. Development of a cross-protective common cold coronavirus vaccine. Biorxiv, 2025; preprint. [Google Scholar] [CrossRef]
- Haque, A.; Pant, A.B. Efforts at COVID-19 Vaccine Development: Challenges and Successes. Vaccines 2020, 8, 739. [Google Scholar] [CrossRef]
- Simancas-Racines, D.; Franco, J.V.A.; Guerra, C.V.; Felix, M.L.; Hidalgo, R.; Martinez-Zapata, M.J. Vaccines for the common cold. Cochrane Database Syst. Rev. 2017, 5, CD002190. [Google Scholar] [CrossRef] [PubMed]
- Tyrrell, D.A.; Bynoe, M.L. Cultivation of viruses from patients with colds. Lancet 1966, 1, 76–77. [Google Scholar] [CrossRef]
- Wat, D. The common cold: A review of the literature. Eur. J. Intern. Med. 2004, 15, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018, 100, 163–188. [Google Scholar]
- Payne, S. Family Coronaviridae. Viruses 2017, 17, 149–158. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef]
- Venkatesh, S.; Memish, Z.A. SARS: The new challenge to international health travel medicine. East. Mediterr. Health J. 2004, 10, 655–662. [Google Scholar] [CrossRef]
- Wang, L.; Shi, Z.; Zhang, S.; Field, H.; Daszak, P.; Eaton, B.T. Review of Bats and SARS. Emerg. Infect. Dis. 2006, 12, 1834–1840. [Google Scholar] [CrossRef] [PubMed]
- Zandi, M.; EBehboudi, S. Soltani, Role of Glycoprotein Hemagglutinin-Esterase in COVID-19 Pathophysiology? Stem Cell Rev. Rep. 2021, 17, 2359–2360. [Google Scholar] [CrossRef]
- Llanes, A.; Restrepo, C.M.; Caballero, Z.; Rajeev, S.; Kennedy, M.A.; Lleonart, R. Betacoronavirus Genomes: How Genomic Information has been Used to Deal with Past Outbreaks the COVID-19 Pandemic. Int. J. Mol. Sci. 2020, 21, 4546. [Google Scholar] [CrossRef]
- Marra, M.J.; Jones, S.J.M.; Astell, C.R.; Holt, R.A.; Brooks-Wilson, A.; Butterfield, Y.S.N.; Khattra, J.; Asano, J.K.; Barber, S.A.; Chan, S.Y.; et al. The Genome Sequence of the SARS-Associated Coronavirus. Science 2003, 300, 1399–1404. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Huang, Y.; Lau, S.K.P.; Yuen, K.-Y. Coronavirus genomics and bioinformatics analysis. Viruses 2010, 2, 1804–1820. [Google Scholar] [CrossRef]
- Niesters, H.G.M. Molecular and diagnostic clinical virology in real time. Clin. Microbiol. Infect. 2004, 10, 5–11. [Google Scholar] [CrossRef]
- Richardson, S.E.; Tellier, R.; Mahony, J. The Laboratory Diagnosis of Severe Acute Respiratory Syndrome: Emerging Laboratory Tests for an Emerging Pathogen. Clin. Biochem. Rev. 2004, 25, 133–141. [Google Scholar]
- Papaneri, A.B.; Johnson, R.F.; Wada, J.; Bollinger, L.; Jahrling, P.B.; Kuhn, J.H. Middle East respiratory syndrome: Obstacles and prospects for vaccine development. Expert Rev. Vaccines 2015, 14, 949–962. [Google Scholar] [CrossRef]
- Ommeh, S.; Zhang, W.; Zohaib, A.; Chen, J.; Zhang, H.; Hu, B.; Ge, X.-Y.; Yang, X.-L.; Masika, M.; Obanda, V.; et al. Genetic Evidence of Middle East Respiratory Syndrome Coronavirus (MERS-Cov) and Widespread Seroprevalence among Camels in Kenya. Virol. Sin. 2018, 33, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Layqah, L.A.; Eissa, S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta 2019, 186, 224. [Google Scholar] [CrossRef] [PubMed]
- Yurdusev, E.; Trahan, P.L.; Perreault, J. Adaptation of a Model Spike Aptamer for Isothermal Amplification-Based Sensing. Sensors 2024, 24, 6875. [Google Scholar] [CrossRef]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Nascimento, E.D.; Fonseca, W.T.; de Oliveira, T.R.; de Correia, C.R.S.T.B.; Faça, V.M.; de Morais, B.P.; Silvestrini, V.C.; Pott-Junior, H.; Teixeira, F.R.; Faria, R.C. COVID-19 diagnosis by SARS-CoV-2 Spike protein detection in saliva using an ultrasensitive magneto-assay based on disposable electrochemical sensor. Sens. Actuators B Chem. 2022, 353, 131128. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhou, Y.; Chen, Y.; Xie, W.; Meng, J.; Shen, D.; He, X.; Chen, H. Rapid detection of the SARS-CoV-2 omicron variants based on high-resolution melting curve analysis. Sci. Rep. 2024, 14, 28227. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Singh, J.; Streithorst, J.; Granados, A.; Sotomayor-Gonzalez, A.; Zorn, K.; Gopez, A.; et al. Rapid Detection of 2019 Novel Coronavirus SARS-CoV-2 Using a CRISPR-based DETECTR Lateral Flow Assay. MedRxiv, 2020; preprint. [Google Scholar] [CrossRef]
- Neff, C.P.; Cikara, M.; Geiss, B.J.; Caltagirone, G.T.; Liao, A.; Atif, S.M.; Macdonald, B.; Schaden, R. Nucleocapsid protein binding DNA aptamers for detection of SARS-COV-2. Curr. Res. Biotechnol. 2023, 5, 100132. [Google Scholar] [CrossRef]
- Jamwal, S.; Gautam, A.; Elsworth, J.; Kumar, M.; Chawla, R.; Kumar, P. An updated insight into the molecular pathogenesis, secondary complications and potential therapeutics of COVID-19 pandemic. Life Sci. 2020, 257, 118105. [Google Scholar] [CrossRef]
- Haas, M.; Fürhacker, P.; Hodek, J.; Stangl, P.; Alon, I.; Kainz, K.; Fajgelj, V.; Mädel, C.; Dotzler, S.; Götzinger, F.; et al. Detection of viable SARS-CoV-2 on the hands of hospitalized children with COVID-19. Clin. Microbiol. Infect. 2023, 29, 1211–1213. [Google Scholar] [CrossRef] [PubMed]
- Leung, N.H.L. Transmissibility and transmission of respiratory viruses. Nat. Rev. Microbiol. 2021, 19, 528–545. [Google Scholar] [CrossRef] [PubMed]
- Mallakpour, S.; Azadi, E.; Hussain, C.M. Fabrication of air filters with advanced filtration performance for removal of viral aerosols and control the spread of COVID-19. Adv. Colloid Interface Sci. 2022, 303, 102653. [Google Scholar] [CrossRef] [PubMed]
- Sousan, S.; Fan, M.; Outlaw, K.; Williams, S.; Roper, R. SARS-CoV-2 Detection in air samples from inside heating ventilation air conditioning (HVAC) systems- COVIDsurveillance in student dorms. Am. J. Infect. Control 2022, 50, 330–335. [Google Scholar] [CrossRef]
- Bellarmino, N.; Cantoro, R.; Castelluzzo, M.; Correale, R.; Squillero, G.; Castelletti, F.; Ciricugno, C.; Dalla Gasperina, D.; Dentali, F.; Poggialini, G.; et al. COVID-19 detection from exhaled breath. Sci. Rep. 2024, 14, 23245. [Google Scholar] [CrossRef]
- Ding, X.; Yin, K.; Li, Z.; Sfeir, M.M.; Liu, C. Sensitive quantitative detection of SARS-CoV-2 in clinical samples using digital warm-start CRISPR assay. Biosens. Bioelectron. 2021, 184, 113218. [Google Scholar] [CrossRef]
- Roberts, A.; Chouhan, R.S.; Shahdeo, D.; Shrikrishna, N.S.; Kesarwani, V.; Horvat, M.; Gandhi, S. A Recent Update on Advanced Molecular Diagnostic Techniques for COVID-19 Pandemic: An Overview. Front. Immunol. 2021, 12, 732756. [Google Scholar] [CrossRef]
- Sun, Q.; Li, J.; Ren, H.; Pastor, L.; Loginova, Y.; Madej, R.; Taylor, K.; Wong, J.K.; Zhang, Z.; Zhang, A.; et al. Saliva as a testing specimen with or without pooling for SARS-CoV-2 detection by multiplex RT-PCR test. PLoS ONE 2021, 16, e0243183. [Google Scholar]
- Erdem, A.; Senturk, H.; Yildiz, E.; Maral, M. Optimized aptamer-based next generation biosensor for the ultra-sensitive determination of SARS-CoV-2 S1 protein in saliva samples. Int. J. Biol. Macromol. 2024, 281, 136233. [Google Scholar] [CrossRef]
- Dolgin, E. The future of at-home molecular testing. Nature 2024. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Ratnesar-Shumate, S.; Williams, G.; Green, B.; Krause, M.; Holland, B.; Wood, S.; Bohannon, J.; Boydston, J.; Freeburger, D.; Hooper, I.; et al. Simulated Sunlight Rapidly Inactivates SARS-CoV-2 on Surfaces. J. Infect. Dis. 2020, 222, 214–222. [Google Scholar] [CrossRef]
- Aslan, Y.; Atabay, M.; Chowdhury, H.K.; Göktürk, I.; Saylan, Y.; Inci, F. Aptamer-Based Point-of-Care Devices: Emerging Technologies and Integration of Computational Methods. Biosensors 2023, 13, 569. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, J.; Li, T.; Fan, T.; Meng, C.; Li, C.; Kang, J.; Chai, L.; Hao, Y.; Tang, Y.; et al. A CRISPR/Cas12a-empowered surface plasmon resonance platform for rapid and specific diagnosis of the Omicron variant of SARS-CoV-2. Natl. Sci. Rev. 2022, 9, nwac104. [Google Scholar] [CrossRef]
- Papi, M.; De Spirito, M.; Palmieri, V. Nanotechnology in the COVID-19 era: Carbon-based nanomaterials as a promising solution. Carbon 2023, 210, 118058. [Google Scholar] [CrossRef]
- Mackay, I.M. RTPCR detection chemistries in virology. Nucleic Acids Res. 2002, 30, 1292–1305. [Google Scholar] [CrossRef]
- Alsharksi, A.N.; Sirekbasan, S.; Gürkök-Tan, T.; Mustapha, A. From Tradition to Innovation: Diverse Molecular Techniques in the Fight Against Infectious Diseases. Diagnostics 2024, 14, 2876. [Google Scholar] [CrossRef]
- Madadelahi, M.; Agarwal, R.; Martinez-Chapa, S.O.; Madou, M.J. A roadmap to high-speed polymerase chain reaction (PCR): COVID-19 as a technology accelerator. Biosens. Bioelectron. 2024, 246, 115830. [Google Scholar] [CrossRef]
- Garzarelli, V.; Chiriacò, M.; Autuori, I.; Ferrara, F. Miniaturized Real-Time PCR systems for SARS-CoV-2 detection at the Point-of-Care. Clin. Chim. Acta 2022, 536, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, E.; Kim, B.; Cho, J.; Ryu, S.-W.; Lee, K.-A. Evaluation of diagnostic performance of SARS-CoV-2 infection using digital droplet polymerase chain reaction in individuals with or without COVID-19 symptoms. Clin. Chim. Acta 2024, 554, 117759. [Google Scholar] [CrossRef] [PubMed]
- Fujita-Rohwerder, N.; Beckmann, L.; Zens, Y.; Verma, A. Diagnostic accuracy of rapid point-of-care tests for diagnosis of current SARS-CoV-2 infections in children: A systematic review and meta-analysis. BMJ Evid. Based Med. 2022, 27, 274–287. [Google Scholar] [CrossRef] [PubMed]
- Hoeg, T.B.; Prasad, V. Rapid antigen testing for COVID-19: Decreasing diagnostic reliability, potential detrimental effects and a lack of evidence to support continued public funding of community-based testing. Public Health Pract. 2023, 6, 100451. [Google Scholar] [CrossRef]
- Medicare Part B Spending on Lab Tests Increased in 2021, Driven By Higher Volume of COVID-19 Tests, Genetic Tests, and Chemistry Tests. US Department of Health and Human Services, Office of Inspector General: Washington, DC, USA, 2022. Available online: https://oig.hhs.gov/reports/all/2022/medicare-part-b-spending-on-lab-tests-increased-in-2021-driven-by-higher-volume-of-covid-19-tests-genetic-tests-and-chemistry-tests/ (accessed on 30 May 2025).
- Quarterly Update for Clinical Laboratory Fee Schedule and Laboratory Services Subject to Reasonable Charge Payment. Centers for Medicare and Medicaid Services. Available online: https://www.cms.gov/medicare/payment/fee-schedules/clinical-laboratory-fee-schedule-clfs/files (accessed on 3 June 2025).
- Prices for COVID-19 Testing. Available online: https://www.healthsystemtracker.org/brief/prices-for-covid-19-testing (accessed on 10 January 2025).
- Medicare Administrative Contractor (MAC) COVID-19 Test Pricing, To establish amounts paid to clinical diagnostic Laboratories for high throughput detection of SARS-CoV-2. 2021. Available online: https://www.cms.gov/files/document/mac-covid-19-test-pricing.pdf (accessed on 30 May 2025).
- Islam, M.M.; Koirala, D. Toward a next-generation diagnostic tool: A review on emerging isothermal nucleic acid amplification techniques for the detection of SARS-CoV-2 and other infectious viruses. Anal. Chim. Acta 2022, 1209, 339338. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of, D.N.A. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Mautner, L.; Baillie, C.K.; Herold, H.M.; Volkwein, W.; Guertler, P.; Eberle, U.; Ackermann, N.; Sing, A.; Pavlovic, M.; Goerlich, O.; et al. Rapid point-of-care detection of SARS-CoV-2 using reverse transcription loop-mediated isothermal amplification (RT-LAMP). Virol. J. 2020, 17, 160. [Google Scholar] [CrossRef]
- Kashir, J.; Yaqinuddin, A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med. Hypotheses 2020, 141, 109786. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Tang, G.; Ismail, N.; Wang, X. Developing RT-LAMP assays for rapid diagnosis of SARS-CoV-2 in saliva. EBioMedicine 2022, 75, 103736. [Google Scholar] [CrossRef] [PubMed]
- Bokelmann, L.; Nickel, O.; Maricic, T.; Pääbo, S.; Meyer, M.; Borte, S.; Riesenberg, S. Point-of-care bulk testing for SARS-CoV-2 by combining hybridization capture with improved colorimetric LAMP. Nat. Commun. 2021, 12, 1467. [Google Scholar] [CrossRef] [PubMed]
- Spiteri, S.; Marino, F.; Girolamini, L.; Pascale, M.R.; Derelitto, C.; Caligaris, L.; Paghera, S.; Cristino, S. Loop-Mediated Isothermal Amplification (LAMP): An Innovative Approach for the Environmental Monitoring of SARS-CoV-2. Pathogens 2024, 13, 1022. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Tao, Y.; Yang, R.; Mao, K.; Zhou, H.; Xu, M.; Sun, T.; Li, X.; Shi, C.; Ge, Z.; et al. Compact highly sensitive photothermal RT-LAMP chip for simultaneous multidisease detection. Sci. Adv. 2024, 10, eadq2899. [Google Scholar] [CrossRef]
- Song, X.; Coulter, F.J.; Yang, M.; Smith, J.L.; Tafesse, F.G.; Messer, W.B.; Reif, J.H. A lyophilized colorimetric RT-LAMP test kit for rapid, low-cost, at-home molecular testing of SARS-CoV-2 and other pathogens. Sci. Rep. 2022, 12, 7043. [Google Scholar] [CrossRef]
- Amaral, C.; Antunes, W.; Moe, E.; Duarte, A.G.; Lima, L.M.P.; Santos, C.; Gomes, I.L.; Afonso, G.S.; Vieira, R.; Teles, H.S.S.; et al. A molecular test based on RT-LAMP for rapid, sensitive and inexpensive colorimetric detection of SARS-CoV-2 in clinical samples. Sci. Rep. 2021, 11, 16430. [Google Scholar] [CrossRef]
- Dao Thi, V.L.; Herbst, K.; Böttcher, M.; Kirrmaier, D.; Freistaedter, A.; Galmozzi, C.; Stanifer, M.L.; Boulant, S.; Klein, S.; Chlanda, P.; et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12, eabc7075. [Google Scholar] [CrossRef]
- Jang, M.; Kim, S. Inhibition of Non-specific Amplification in Loop-Mediated Isothermal Amplification via Tetramethylammonium Chloride. Biochip J. 2022, 16, 326–333. [Google Scholar] [CrossRef]
- Soroka, M.; Wasowicz, B.; Rymaszewska, A. Loop-Mediated Isothermal Amplification (LAMP): The Better Sibling of PCR? Cells 2021, 10, 1931. [Google Scholar] [CrossRef]
- Yang, N.; Zhang, H.; Han, X.; Liu, Z.; Lu, Y. Advancements and applications of loop-mediated isothermal amplification technology: A comprehensive overview. Front. Microbiol. 2024, 15, 1406632. [Google Scholar] [CrossRef]
- Quyen, T.L.; Vinayaka, A.C.; Golabi, M.; Ngoc, H.V.; Bang, D.D.; Wolff, A. Elimination of Carryover Contamination in Real-Time Reverse Transcriptase Loop-Mediated Isothermal Amplification for Rapid Detection of the SARS-CoV-2 Virus in Point-of-Care Testing. Front. Cell. Infect. Microbiol. 2022, 12, 856553. [Google Scholar] [CrossRef]
- Lau, Y.L.; Ismail, I.B.; Mustapa, N.I.B.; Lai, M.Y.; Soh, T.S.T.; Hassan, A.H.; Peariasamy, K.M.; Lee, Y.L.; Goh, P.P. A Sensitive Reverse Transcription Loop-Mediated Isothermal Amplification Assay for Direct Visual Detection of SARS-CoV-2. Am. J. Trop. Med. Hyg. 2020, 103, 2350–2352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, Y.; Zeng, Y.; Zhang, C. Evolution of the Probe-Based Loop-Mediated Isothermal Amplification (LAMP) Assays in Pathogen Detection. Diagnostics 2023, 13, 1530. [Google Scholar] [CrossRef]
- Tamanaha, E.; Zhang, Y.; Tanner, N.A. Profiling RT-LAMP tolerance of sequence variation for SARS-CoV-2 RNA detection. PLoS ONE 2022, 17, e0259610. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wei, Y.; Ye, C.; Cao, J.; Zhou, X.; Xie, M.; Qing, J.; Chen, Z. Application of recombinase polymerase amplification with lateral flow assay to pathogen point-of-care diagnosis. Front. Cell. Infect. Microbiol. 2024, 14, 1475922. [Google Scholar] [CrossRef]
- Lau, Y.L.; Ismail, I.B.; Mustapa, N.I.B.; Lai, M.Y.; Soh, T.S.T.; Hassan, A.H.; Peariasamy, K.M.; Lee, Y.L.; Abdul Kahar, M.K.B.; Chong, J.; et al. Development of a reverse transcription recombinase polymerase amplification assay for rapid and direct visual detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). PLoS ONE 2021, 16, e0245164. [Google Scholar]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. Trac Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef]
- Qian, J.; Boswell, S.A.; Chidley, C.; Lu, Z.-X.; Pettit, M.E.; Gaudio, B.L.; Fajnzylber, J.M.; Ingram, R.T.; Ward, R.H.; Li, J.Z.; et al. An enhanced isothermal amplification assay for viral detection. Nat. Commun. 2020, 11, 5920. [Google Scholar] [CrossRef]
- Higgins, M.; Ravenhall, M.; Ward, D.; Phelan, J.; Ibrahim, A.; Forrest, M.S.; Clark, T.G.; Campino, S. PrimedRPA: Primer design for recombinase polymerase amplification assays. Bioinformatics 2019, 35, 682–684. [Google Scholar] [CrossRef]
- Tan, M.; Liao, C.; Liang, L.; Yi, X.; Zhou, Z.; Wei, G. Recent advances in recombinase polymerase amplification: Principle, advantages, disadvantages and applications. Front. Cell. Infect. Microbiol. 2022, 12, 1019071. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Tang, X.; Chen, T.; Chen, G. Types and Applications of Nicking Enzyme-Combined Isothermal Amplification. Int. J. Mol. Sci. 2022, 23, 4620. [Google Scholar] [CrossRef]
- Maples, B.K.; Holmberg, R.C.; Miller, A.P.; Provins, J.W.; Roth, R.B.; Mandell, J.G. Nicking and Extension Amplification Reaction for the Exponential Amplification of Nucleic Acids. U.S. Patent US20090081670A1, 11 April 2017. [Google Scholar]
- James, A.S.; Alawneh, J.I. COVID-19 Infection Diagnosis: Potential Impact of Isothermal Amplification Technology to Reduce Community Transmission of SARS-CoV-2. Diagnostics 2020, 10, 399. [Google Scholar] [CrossRef]
- Menova, P.; Raindlova, V.; Hocek, M. Scope and limitations of the nicking enzyme amplification reaction for the synthesis of base-modified oligonucleotides and primers for PCR. Bioconjug. Chem. 2013, 24, 1081–1093. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.Q. Rolling replication of short DNA circles. Proc. Natl. Acad. Sci. USA 1995, 92, 4641–4645. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Zhu, Z.; Wang, L.; Gong, S.; Li, H.; Xu, W.; Zhang, C.; Zhang, M.; Zhou, J. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 2020, 165, 112356. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Yadav, K.; Kumar, A.; Tyagi, C.; Varshney, U.; Singh, L. A Novel Rolling Circle Amplification-Based Detection of SARS-CoV-2 with Multi-Region Padlock Hybridization. Diagnostics 2022, 12, 2252. [Google Scholar] [CrossRef]
- Kyung, K.; Kim, J.; Park, J.; Lee, D.; Shin, J.; Park, S. Fluorometric Detection of SARS-CoV-2 Single-Nucleotide Variant L452R Using Ligation-Based Isothermal Gene Amplification. Bioengineering 2023, 10, 1116. [Google Scholar] [CrossRef]
- Deiman, B.; Pvan Aarle, P. Sillekens, Characteristics and applications of nucleic acid sequence-based amplification (NASBA). Mol. Biotechnol. 2002, 20, 163–179. [Google Scholar] [CrossRef]
- Ju, Y.; Hou, Y.; Zhang, Y.; Hu, Y.; Guo, S.; Zhao, Q.; Wu, X.; Wang, X.; Cui, X. Ultrasensitive version of nucleic acid sequence-based amplification (NASBA) utilizing a nicking and extension chain reaction system. Nanoscale 2021, 13, 10785–10791. [Google Scholar] [CrossRef] [PubMed]
- Xing, W.; Su, J.; Huang, L.; Xu, J.; Song, H.; Xu, J.; Wu, X.; Huang, J.; Xu, Z.; Zhang, C.; et al. A High-Throughput, Multi-Index Isothermal Amplification Platform for Rapid Detection of 19 Types of Common Respiratory Viruses Including SARS-CoV-2. Engineering 2020, 6, 1130–1140. [Google Scholar] [CrossRef]
- Honsvall, B.K.; Robertson, L.J. From research lab to standard environmental analysis tool: Will NASBA make the leap? Water Res. 2017, 109, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, P.; Prasad, D. Isothermal nucleic acid amplification and its uses in modern diagnostic technologies. 3 Biotech 2023, 13, 200. [Google Scholar] [CrossRef]
- Aufdembrink, L.M.; Tran, N.; Berensmeier, S.; Tan, W.; Fitzgerald, D.J.; Grate, J.W. Highly specific, multiplexed isothermal pathogen detection with fluorescent aptamer readout. RNA 2020, 26, 1283–1290. [Google Scholar] [CrossRef]
- Rahman, M.R.; Hosen, M.J.; Islam, M.R.; Uzzaman, M.S.; Ahmad, S.; Hossain, M.S.; Nessa, A.; Ahmed, K.U.; Islam, S.; Nabi, A.H.M.N.; et al. CRISPR is a useful biological tool for detecting nucleic acid of SARS-CoV-2 in human clinical samples. Biomed. Pharmacother. 2021, 140, 111772. [Google Scholar] [CrossRef]
- Balaban Hanoglu, S.; Ozkan-Ariksoysal, D.; Buyukserin, F.; Cetinkaya, M.; Sayin, A.; Akgun, O.M. Detection strategies of infectious diseases via peptide-based electrochemical biosensors. Bioelectrochemistry 2024, 160, 108784. [Google Scholar] [CrossRef]
- Menon, S.; Suresh, S.; Lalitha, K.V.; Ramalingam, R.; Balasubramanian, K. Recent advances challenges in electrochemical biosensors for emerging re-emerging infectious diseases. J. Electroanal. Chem. 2020, 878, 114596. [Google Scholar] [CrossRef]
- Fozouni, P.; Son, S.; de León Derby, M.D.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333 e9. [Google Scholar] [CrossRef] [PubMed]
- Khan, W.A.; Barney, R.E.; Tsongalis, G.J. CRISPR-cas13 enzymology rapidly detects SARS-CoV-2 fragments in a clinical setting. J. Clin. Virol. 2021, 145, 105019. [Google Scholar] [CrossRef] [PubMed]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Surarit, R.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 15, 1311, Erratum in Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qi, X.; Ma, W.; Liu, G.; Li, S.; Zhang, H.; Chen, L.; Cao, C.; Gao, X.; Zhu, Z.; et al. Strategies to Improve Multi-enzyme Compatibility Coordination in One-Pot SHERLOCK. Anal. Chem. 2023, 95, 10522–10531. [Google Scholar] [CrossRef]
- Ding, X.; Yin, K.; Li, Z.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef]
- Ghouneimy, A.; Rahim, F.; Liu, S.; Li, Y. CRISPR-Based Diagnostics: Challenges and Potential Solutions toward Point-of-Care Applications. ACS Synth. Biol. 2023, 12, 1–16. [Google Scholar] [CrossRef]
- Phutela, R.; Rao, M.; Garg, S.; Giri, S.; Kumari, A.; Bhardwaj, S.; Yadav, P.; Malhotra, B.D. FnCas9 Editor Linked Uniform Detection Assay for COVID-19. Methods Mol. Biol. 2022, 2511, 149–159. [Google Scholar]
- Azhar, M.; Singh, P.; Kaur, M.; Rani, R.; Tiwari, S. Rapid and accurate nucleobase detection using FnCas9 and its application in COVID-19 diagnosis. Biosens. Bioelectron. 2021, 183, 113207. [Google Scholar] [CrossRef]
- Kumar, M.; Garg, S.; Hegde, N.R.; Chakraborty, S.; Malhotra, B.D. FnCas9-based CRISPR diagnostic for rapid and accurate detection of major SARS-CoV-2 variants on a paper strip. Elife 2021, 10, e67130. [Google Scholar] [CrossRef]
- Roueinfar, M.; Khosravi, M.; Shabani, A.M.H.; Namaki, S.; Alavi, M.S.; Hosseinkhani, S. An Update of Nucleic Acids Aptamers Theranostic Integration with CRISPR/Cas Technology. Molecules 2022, 27, 1114. [Google Scholar] [CrossRef]
- Konstantakos, V.; Stelzer, E.H.K.; Plattner, H. CRISPR-Cas9 gRNA efficiency prediction: An overview of predictive tools and the role of deep learning. Nucleic Acids Res. 2022, 50, 3616–3637. [Google Scholar] [CrossRef]
- Cummins, B.M.; Ligler, F.S.; Walker, G.M. Point-of-care diagnostics for niche applications. Biotechnol. Adv. 2016, 34, 161–176. [Google Scholar] [CrossRef]
- Chen, A.; Yang, S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 2015, 71, 230–242. [Google Scholar] [CrossRef]
- Kolovskaya, O.S.; Obraztsova, A.S.; Zavyalova, M.V.; Smolina, N.P.; Kurbatov, L.K.; Terskikh, V.V. Monitoring of breast cancer progression via aptamer-based detection of circulating tumor cells in clinical blood samples. Front. Mol. Biosci. 2023, 10, 1184285. [Google Scholar] [CrossRef]
- Diaz-Fernandez, A.; Arenas-Arrocena, M.C.; Shukla, S.; Susa, M.; Iyer, S.; Wentz, T.; Dronina, J.; Henson, B.; Patel, S.; Galipeau, J. Impedimetric aptamer-based glycan PSA score for discrimination of prostate cancer from other prostate diseases. Biosens. Bioelectron. 2021, 175, 112872. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; He, Q.; Zhang, Y.; Song, Y.; Ma, J.; Liu, H.; Zhang, T. Aptamer Technologies in Neuroscience, Neuro-Diagnostics and Neuro-Medicine Development. Molecules 2024, 29, 1124. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Huang, H.; Zhang, L.; Sun, S.; Zhang, Y.; Zhang, J.; Ma, J. Wearable aptamer-field-effect transistor sensing system for noninvasive cortisol monitoring. Sci. Adv. 2022, 8, eabk0967. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Liu, Y.; Chen, Y.; Zhang, W.; Xu, W.; Zhang, X.; Chen, H. A wearable aptamer nanobiosensor for non-invasive female hormone monitoring. Nat. Nanotechnol. 2024, 19, 330–337. [Google Scholar] [CrossRef]
- Hu, C.; Wang, J.; Sun, W.; Wang, S.; Wang, L.; Wang, X. Viral aptamer screening and aptamer-based biosensors for virus detection: A review. Int. J. Biol. Macromol. 2024, 276, 133935. [Google Scholar] [CrossRef]
- Lou, B.; Zhang, D.; Liu, Y.; Chen, J.; Wu, Y. Aptamer-based biosensors for virus protein detection. Trends Anal. Chem. 2022, 157, 116738. [Google Scholar] [CrossRef]
- Ni, S.; Zhao, J.; Dong, C.; Wang, J.; Li, S.; Wu, X.; Zhang, Y.; Liu, Y.; Wang, X.; Zhao, Y. Recent Progress in Aptamer Discoveries and Modifications for Therapeutic Applications. ACS Appl. Mater. Interfaces 2021, 13, 9500–9519. [Google Scholar] [CrossRef]
- Yang, L.F.; Zhao, J.; Chen, H.; Wang, X.; Wu, X.; Li, S.; Ni, S.; Zhang, Y. Aptamers 101: Aptamer discovery and in vitro applications in biosensors and separations. Chem. Sci. 2023, 14, 4961–4978. [Google Scholar] [CrossRef]
- Sheraz, M.; Khan, I.U.; Khan, S.; Iqbal, M.; Ul Haq, I.; Amin, M.U.; Hussain, M.; Mehmood, A.; Akhtar, M.W. Recent Developments in Aptamer-Based Sensors for Diagnostics. Sensors 2024, 24, 7432. [Google Scholar] [CrossRef]
- Chaibun, T.; Piyawattanametha, W.; Vivatvisakul, V.; Kanatharana, P.; Dungchai, W.; Chailapakul, O. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021, 12, 802. [Google Scholar] [CrossRef]
- Heo, W.; Lee, K.; Lee, S.; Kim, H.; Kang, D.; Han, S. Electrochemical biosensor for nucleic acid amplification-free and sensitive detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA via CRISPR/Cas13a trans-cleavage reaction. Biosens. Bioelectron. 2022, 201, 113960. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, Y.; Zhou, Y.; Cai, L.; Liu, C.; Liu, Z.; Zheng, L. Recent Advances in Signal Amplification to Improve Electrochemical Biosensing for Infectious Diseases. Front. Chem. 2022, 10, 911678. [Google Scholar] [CrossRef] [PubMed]
- Torres-Salvador, F.; Ojeda, J.; Castro, C.; Gerasimova, Y.; Chumbimuni-Torres, K. A Single Electrochemical Biosensor Designed to Detect Any Virus. Anal. Chem. 2024, 96, 5752–5756. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Lin, X.; He, C.; Zeng, W.; Luo, Y.; Liu, C.; Liu, Z.; Yang, M.; Kuang, Y.; Huang, Q. Recent Progresses in Electrochemical DNA Biosensors for SARS-CoV-2 Detection. Front. Bioeng. Biotechnol. 2022, 10, 952510. [Google Scholar] [CrossRef] [PubMed]
- Adachi, T.; Nakamura, S.; Michishita, A.; Kawahara, D.; Yamamoto, M.; Hamada, M.; Nakamura, Y. RaptGen-Assisted Generation of an RNA/DNA Hybrid Aptamer against SARS-CoV-2 Spike Protein. Biochemistry 2024, 63, 906–912. [Google Scholar] [CrossRef]
- Dong, Y.; Wang, J.; Chen, L.; Chen, H.; Dang, S.; Li, F. Aptamer-based assembly systems for SARS-CoV-2 detection and therapeutics. Chem. Soc. Rev. 2024, 53, 6830–6859. [Google Scholar] [CrossRef]
- Service, R. In ‘milestone,’ FDA OKs simple, accurate coronavirus test that could cost just $5. 2020. Available online: https://www.science.org/content/article/milestone-fda-oks-simple-accurate-coronavirus-test-could-cost-just-5 (accessed on 30 May 2025).
- Renuse, S.; Vanderboom, P.M.; Maus, A.D.; Kemp, J.V.; Gurtner, K.M.; Madugundu, A.K.; Chavan, S.; Peterson, J.A.; Madden, B.J.; Mangalaparthi, K.K.; et al. A mass spectrometry-based targeted assay for detection of SARS-CoV-2 antigen from clinical specimens. EBioMedicine 2021, 69, 103465. [Google Scholar] [CrossRef]
- Bojórquez-Velázquez, E.; Llamas-García, M.L.; Elizalde-Contreras, J.M.; Zamora-Briseño, J.A.; Ruiz-May, E. Mass Spectrometry Approaches for SARS-CoV-2 Detection: Harnessing for Application in Food and Environmental Samples. Viruses 2022, 14, 872. [Google Scholar] [CrossRef]
- Fistera, D.; Kikull, K.; Risse, J.; Herrmann, A.; Brachmann, M.; Kill, C. Point-of-care PCR testing of SARS-CoV-2 in the emergency department: Influence on workflow and efficiency. PLoS ONE 2023, 18, e0288906. [Google Scholar] [CrossRef]
- Sheridan, C. Fast, portable tests come online to curb coronavirus pandemic. Nat. Biotechnol. 2020, 38, 509–522. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, K.; Zhang, H.; Zhao, Y.; Wen, J.; Zhao, M.; Li, X.; Li, Z. A tube-based biosensor for DNA and RNA detection. Sci. Adv. 2025, 11, eadu2271. [Google Scholar] [CrossRef]
- Liu, K.-Z.; Tian, G.; Ko, A.C.-T.; Geissler, M.; Malic, L.; Moon, B.-U.; Clime, L.; Veres, T. Microfluidic methods for the diagnosis of acute respiratory tract infections. Analyst 2025, 150, 9–33. [Google Scholar] [CrossRef]
- Christaki, E. New technologies in predicting, preventing and controlling emerging infectious diseases. Virulence 2015, 6, 558–565. [Google Scholar] [CrossRef]
- Ogden, N.H.; AbdelMalik, P.; Pulliam, J.R.C. Emerging infectious diseases: Prediction and detection. Can. Commun. Dis. Rep. 2017, 43, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.Z.; Senthil, R.; Ramalingam, V.; Gopal, R. Predicting Infectious Disease Outbreaks with Machine Learning Epidemiological Data. J. Adv. Zool. India 2023, 44, 110–121. [Google Scholar] [CrossRef]
- Zhao, A.P.; Li, S.; Cao, Z.; Hu, P.J.-H.; Wang, J.; Xiang, Y.; Xie, D.; Lu, X. AI for science: Predicting infectious diseases. J. Saf. Sci. Resil. 2024, 5, 130–146. [Google Scholar] [CrossRef]
- Balish, A.; Warnes, C.; Wu, K.; Barnes, N.; Emery, S.; Berman, L.; Shu, B.; Lindstrom, S.; Xu, X.; Uyeki, T.; et al. Evaluation of Rapid Influenza Diagnostic Tests for Detection of Novel Influenza A (H1N1) in MMWR; CDC: Atlanta, GA, USA, 2009; pp. 826–829.
- Gullett, J.C.; Nolte, F.S. Quantitative nucleic acid amplification methods for viral infections. Clin. Chem. 2015, 61, 72–78. [Google Scholar] [CrossRef]
- Dinnes, J.; Sharma, P.; Berhane, S.; van Wyk, S.S.; Nyaaba, N.; Domen, J.; Taylor, M.; Cunningham, J.; Davenport, C.; Dittrich, S.; et al. Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2022, 7, CD013705. [Google Scholar]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Underlying Cause of Death, 2018–2023. Available online: https://wonder.cdc.gov/ucd-icd10-expanded.html (accessed on 30 May 2025).
- Maxwell, A. Labs With Questionably High Billing for Additional Tests Alongside COVID-19 Tests Warrant Further Scrutiny; US Department of Health and Human Services, Office of Inspector General: Washington, DC, USA, 2022.
- Torreele, E. The rush to create a COVID-19 vaccine may do more harm than good. BMJ 2020, 370, m3209. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, G.L. The Common Cold. Prim. Care 1996, 23, 657–675. [Google Scholar] [CrossRef]
- McCullough, P.A.; Arunthamakun, J. Disconnect between community testing and hospitalization for SARS-CoV-2 (COVID-19) infection. Bayl. Univ. Med. Cent. Proc. 2020, 33, 481. [Google Scholar] [CrossRef] [PubMed]
- Giles, J.R.; zu Erbach-Schoenberg, E.; Tatem, A.J.; Gardner, L.; Bjørnstad, O.N.; Metcalf, C.J.E.; Wesolowski, A. The duration of travel impacts the spatial dynamics of infectious diseases. Proc. Natl. Acad. Sci. USA 2020, 117, 22572–22579. [Google Scholar] [CrossRef]
- Achonu, C.; Laporte, A.; Gardam, M.A. The financial impact of controlling a respiratory virus outbreak in a teaching hospital. Can. J. Public Health 2005, 96, 52–54. [Google Scholar] [CrossRef] [PubMed]
- American Medical Association. Current Procedural Terminology Category I and Proprietary Laboratory Analyses Codes for Severe Acute Respiratory Syndrome Coronavirus 2; American Medical Association: Chicago, IL, USA, 2022. [Google Scholar]
- Jiang, J.X.; Makary, M.; Bai, G.; Gupta, S.; Anderson, G. Commercial COVID-19 PCR Test Price in US Hospitals. J. Gen. Intern. Med. 2023, 38, 1341–1343. [Google Scholar] [CrossRef]
- Hermosilla, J.; Alonso-García, A.; Salmerón-García, A.; Cabeza-Barrera, J.; Medina-Castillo, A.L.; Pérez-Robles, R.; Navas, N. Analysing the In-Use Stability of mRNA-LNP COVID-19 Vaccines Comirnaty (Pfizer) and Spikevax (Moderna): A Comparative Study of the Particulate. Vaccines 2023, 11, 1635. [Google Scholar] [CrossRef]
- Parry, P.I.; Lefringhausen, A.; Turni, C.; Neil, C.J.; Cosford, R.; Hudson, N.J.; Gillespie, J. ‘Spikeopathy’: COVID-19 Spike Protein Is Pathogenic from Both Virus and Vaccine mRNA. Biomedicines 2023, 11, 2287. [Google Scholar] [CrossRef] [PubMed]
- Seneff, S.; Nigh, G.; Creighton, A.; Maganga, G.; Kennett, G.; Stevens, G.; Bhattacharyya, S.; Ingraham, N.E.; Bhattacharjee, C.; Chatterjee, D. A Potential Role of the Spike Protein in Neurodegenerative Diseases: A Narrative Review. Cureus 2023, 15, e34872. [Google Scholar] [CrossRef]
- Trougakos, I.P.; Terpos, E.; Alexopoulos, H.; Politou, M.; Paraskevis, D.; Scorilas, A.; Kastritis, E.; Andreakos, E.; Dimopoulos, M.A. Adverse effects of COVID-19 mRNA vaccines: The spike hypothesis. Trends Mol. Med. 2022, 28, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Napit, R.; Olson, A.; Shi, Y.; Sanjay, S.T.; Liu, Y.; Ao, Z.; Mu, Q.; Dong, Z.; Wu, Y.; Wen, W. Aptasensors and Advancement in Molecular Recognition Technology. Adv. Mater. Technol. 2025, 10, 2400504. [Google Scholar] [CrossRef]

| Method. | Template | Key Enzymes | Primer(s) | Temp. (°C) | Main Features |
|---|---|---|---|---|---|
| LAMP | DNA or RNA | Bst DNA polymerase (± RT) | 4–6 | 60–65 | High specificity, loop structures, turbidimetric/colorimetric detection |
| RPA | DNA | Recombinase, SSB, strand-displacing DNA polymerase | 2 | 37–42 | Rapid (20–40min), low temperature, portable |
| NEAR | DNA | strand-displacing DNA polymerase, nicking endonuclease | 2 | 55–59 | Ultra-rapid, uses endonuclease for nicking, suitable for point-of-care |
| RCA | Circular DNA | Ligase, Phi29 DNA polymerase | 1 | 30–42 | Long concatemer products, simple design, high yield |
| NASBA | RNA | Reverse transcriptase, T7 RNA polymerase, RNase H | 2 | 40–41 | RNA as the main target, transcription-based, isothermal |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papaneri, A.; Cui, G.; Chen, S.-H. Next-Generation Nucleic Acid-Based Diagnostics for Viral Pathogens: Lessons Learned from the SARS-CoV-2 Pandemic. Microorganisms 2025, 13, 1905. https://doi.org/10.3390/microorganisms13081905
Papaneri A, Cui G, Chen S-H. Next-Generation Nucleic Acid-Based Diagnostics for Viral Pathogens: Lessons Learned from the SARS-CoV-2 Pandemic. Microorganisms. 2025; 13(8):1905. https://doi.org/10.3390/microorganisms13081905
Chicago/Turabian StylePapaneri, Amy, Guohong Cui, and Shih-Heng Chen. 2025. "Next-Generation Nucleic Acid-Based Diagnostics for Viral Pathogens: Lessons Learned from the SARS-CoV-2 Pandemic" Microorganisms 13, no. 8: 1905. https://doi.org/10.3390/microorganisms13081905
APA StylePapaneri, A., Cui, G., & Chen, S.-H. (2025). Next-Generation Nucleic Acid-Based Diagnostics for Viral Pathogens: Lessons Learned from the SARS-CoV-2 Pandemic. Microorganisms, 13(8), 1905. https://doi.org/10.3390/microorganisms13081905







