FlbD: A Regulator of Hyphal Growth, Stress Resistance, Pathogenicity, and Chlamydospore Production in the Nematode-Trapping Fungus Arthrobotrys flagrans
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
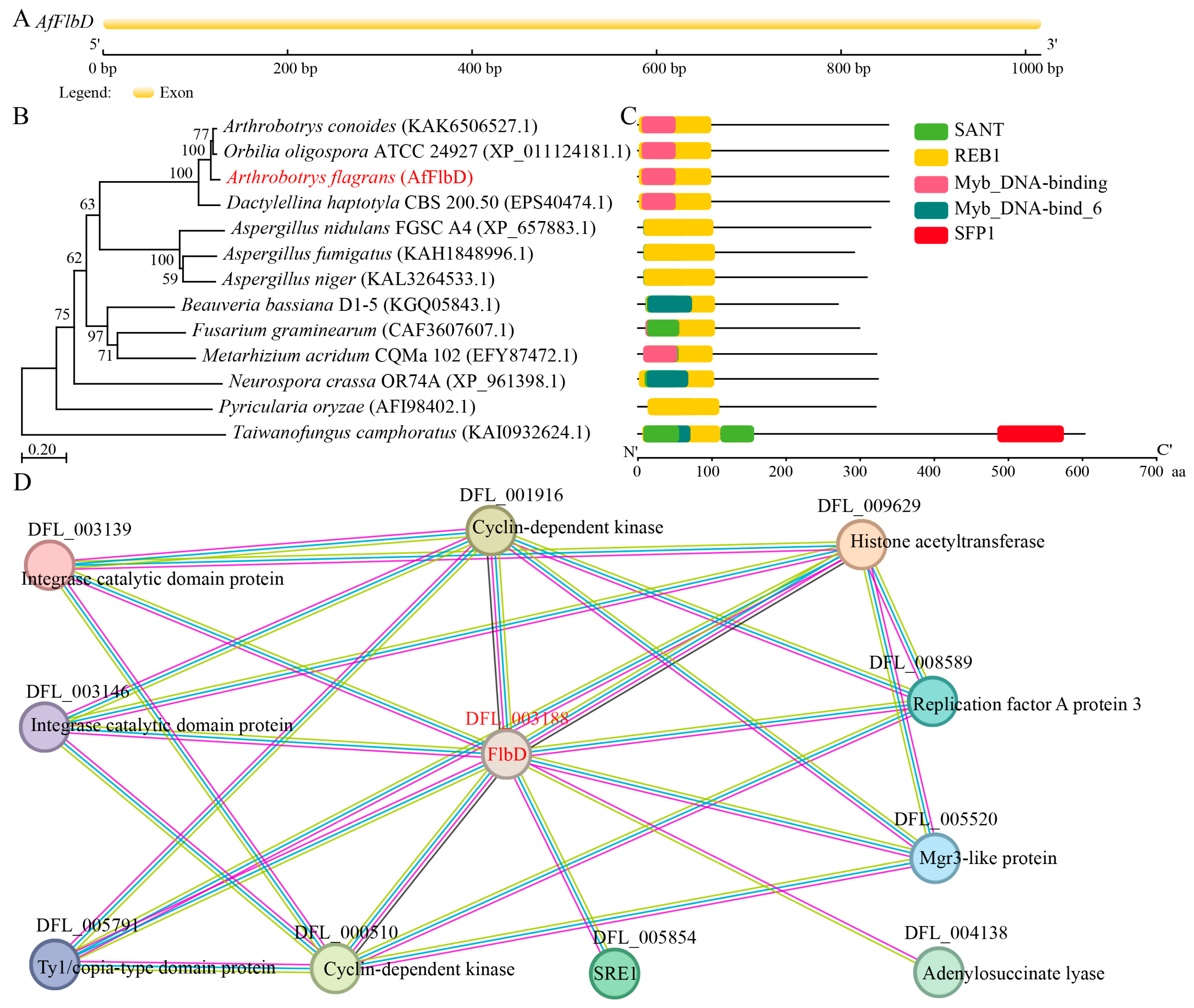
2.2. Gene Structure and Conserved Protein Domain Analysis of AfFlbD
2.3. Genomic DNA Extraction from A. flagrans
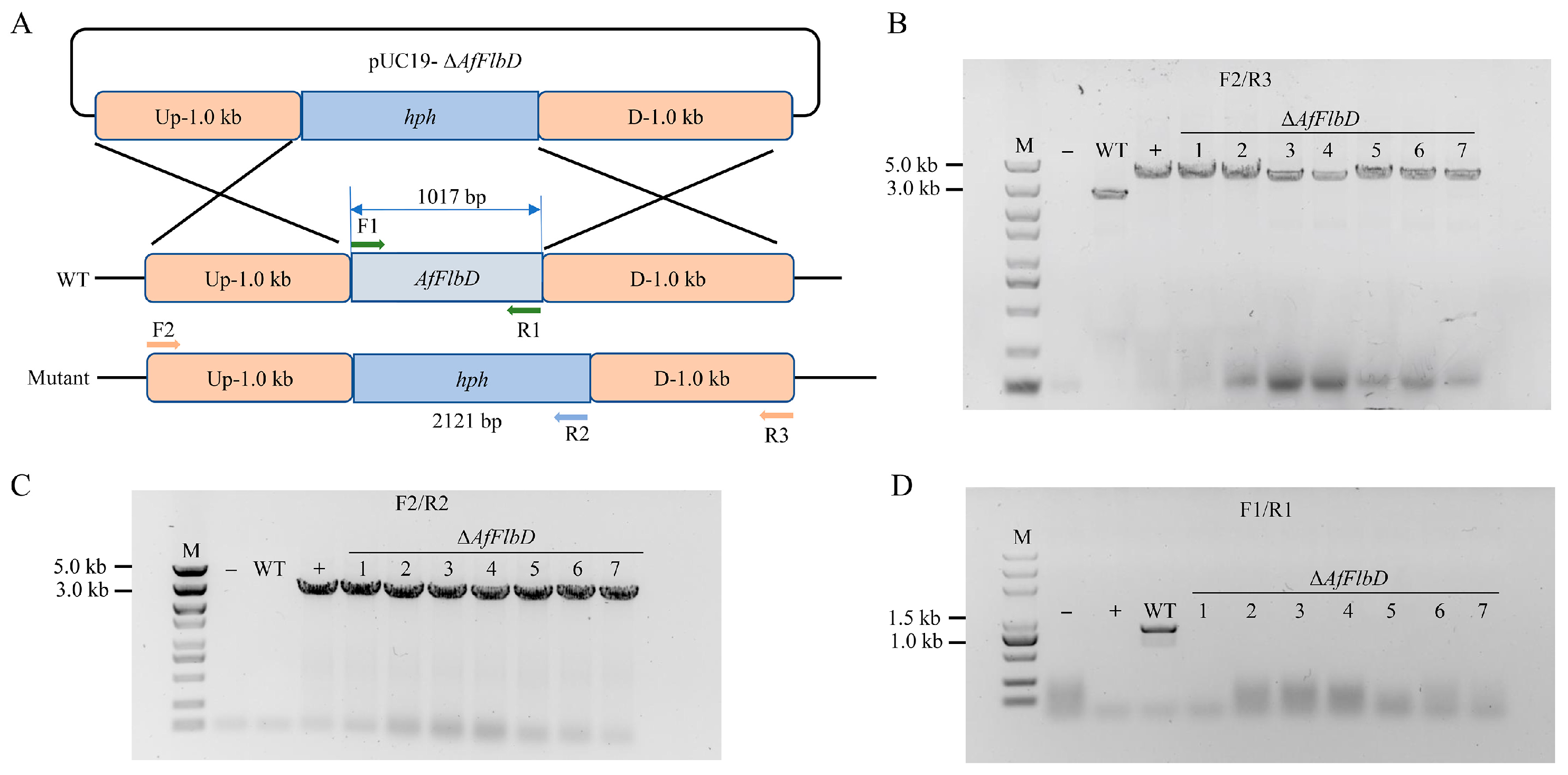
2.4. Construction of the Gene Knockout Vector
2.5. Protoplast Preparation of A. flagrans
2.6. Screening and Verification of AfFlbD Knockout Strains
2.7. Subcellular Localization of AfFlbD Protein
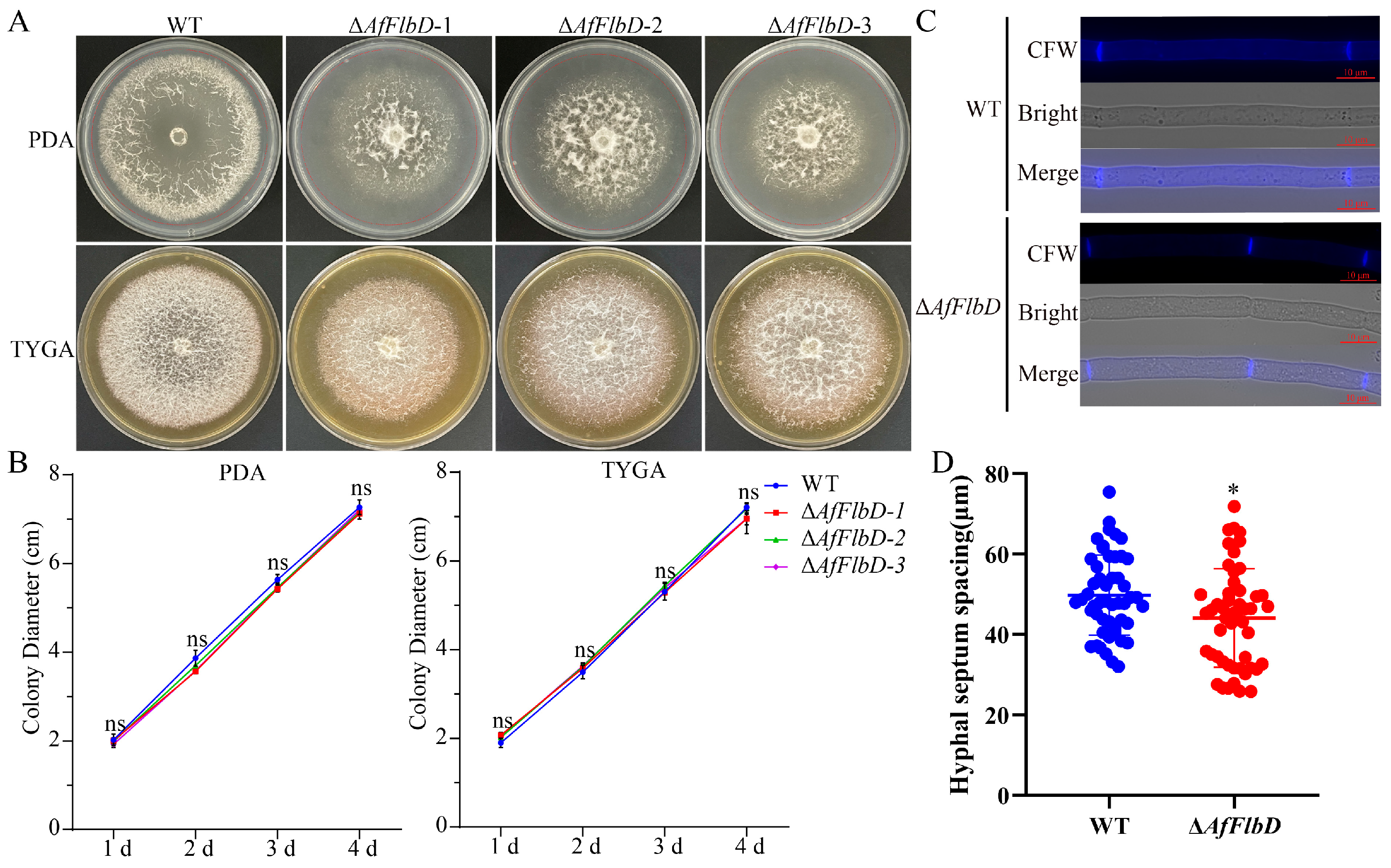
2.8. Hyphal Growth Rate Analysis
2.9. Hyphal Morphology Analysis
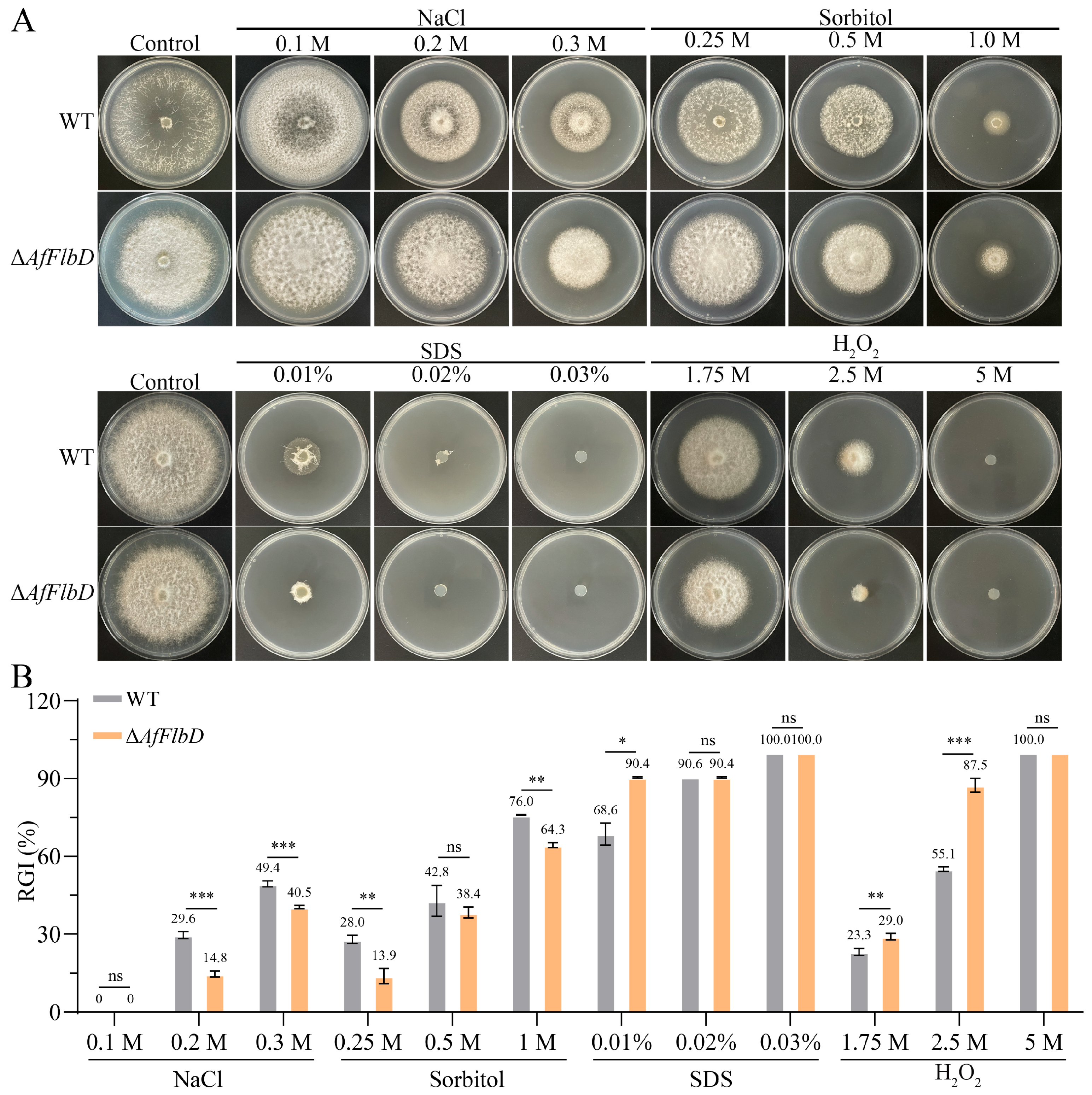
2.10. Chemical Stress Tolerance Assay
2.11. Trap Production and Pathogenicity Analysis
2.12. Chlamydospore Formation Assay
2.13. Quantitative Real-Time PCR (qRT-PCR) Assay
2.14. Data Analysis
3. Results
3.1. Gene Structure and Protein Sequence Analysis of AfFlbD
3.2. AfFlbD Regulates Growth and Stress Responses in A. flagrans
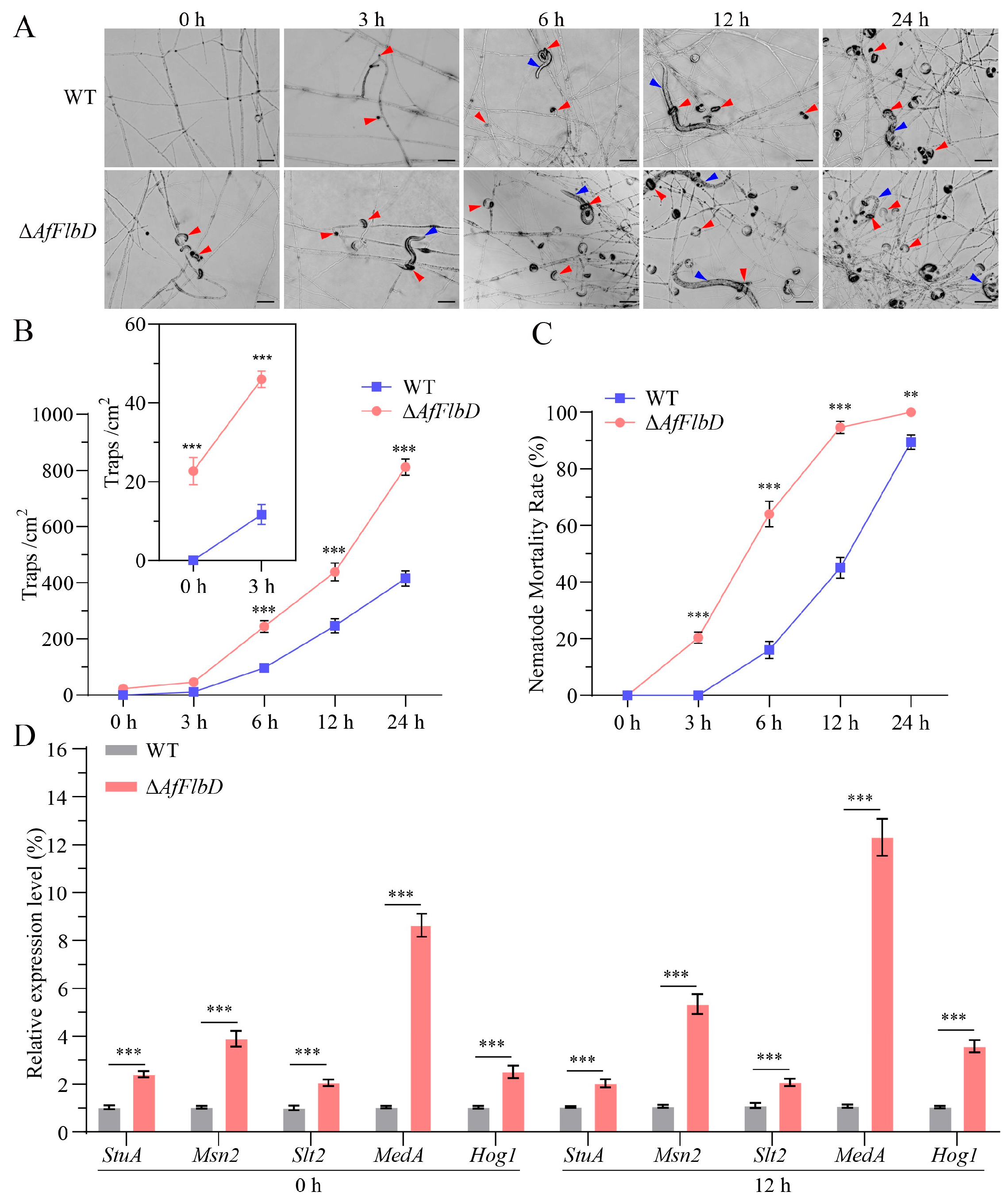
3.3. AfFlbD Negatively Regulates the Pathogenicity of A. flagrans
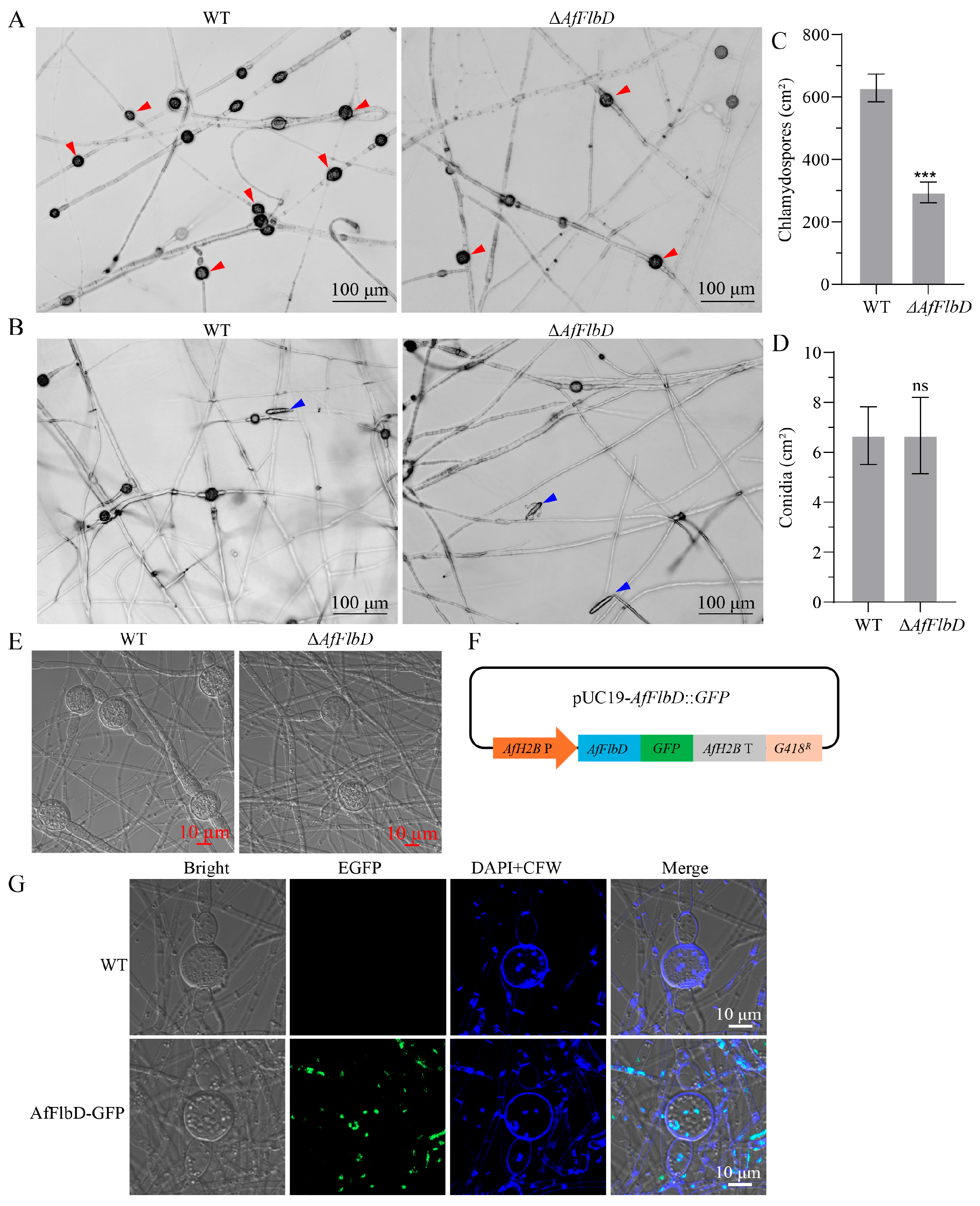
3.4. AfFlbD Regulates Chlamydospore Formation in A. flagrans
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ojeda-López, M.; Chen, W.; Eagle, C.E.; Gutiérrez, G.; Jia, W.L.; Swilaiman, S.S.; Huang, Z.; Park, H.S.; Yu, J.H.; Cánovas, D.; et al. Evolution of asexual and sexual reproduction in the aspergilli. Stud. Mycol. 2018, 91, 37–59. [Google Scholar] [CrossRef]
- Guo, C.T.; Luo, X.C.; Tong, S.M.; Zhou, Y.; Ying, S.H.; Feng, M.G. FluG and FluG-like FlrA coregulate manifold gene sets vital for fungal insect-pathogenic lifestyle but not involved in asexual development. mSystems 2022, 7, e0031822. [Google Scholar] [CrossRef]
- Garzia, A.; Etxebeste, O.; Herrero-García, E.; Ugalde, U.; Espeso, E.A. The concerted action of bZip and cMyb transcription factors FlbB and FlbD induces brlA expression and asexual development in Aspergillus nidulans. Mol. Microbiol. 2010, 75, 1314–1324. [Google Scholar] [CrossRef]
- Arratia-Quijada, J.; Sánchez, O.; Scazzocchio, C.; Aguirre, J. FlbD, a Myb transcription factor of Aspergillus nidulans, is uniquely involved in both asexual and sexual differentiation. Eukaryot. Cell 2012, 11, 1132–1142. [Google Scholar] [CrossRef]
- Otamendi, A.; Perez-de-Nanclares-Arregi, E.; Oiartzabal-Arano, E.; Cortese, M.S.; Espeso, E.A.; Etxebeste, O. Developmental regulators FlbE/D orchestrate the polarity site-to-nucleus dynamics of the fungal bZIP transcription factor FlbB. Cell Mol. Life Sci. 2019, 76, 4369–4390. [Google Scholar] [CrossRef] [PubMed]
- Wieser, J.; Adams, T.H. flbD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes. Dev. 1995, 9, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Son, H.; Kim, M.G.; Chae, S.K.; Lee, Y.W. FgFlbD regulates hyphal differentiation required for sexual and asexual reproduction in the ascomycete fungus Fusarium graminearum. J. Microbiol. 2014, 52, 930–939. [Google Scholar] [CrossRef]
- Hu, X.; Hoffmann, D.S.; Wang, M.; Schuhmacher, L.; Stroe, M.C.; Schreckenberger, B.; Elstner, M.; Fischer, R. GprC of the nematode-trapping fungus Arthrobotrys flagrans activates mitochondria and reprograms fungal cells for nematode hunting. Nat. Microbiol. 2024, 9, 1752–1763. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Ran, Y.; Zhang, K.Q.; Li, G.H. AfLaeA, a global regulator of mycelial growth, chlamydospore production, pathogenicity, secondary metabolism, and energy metabolism in the nematode-trapping fungus Arthrobotrys flagrans. Microbiol. Spectr. 2023, 11, e0018623. [Google Scholar] [CrossRef]
- Youssar, L.; Wernet, V.; Hensel, N.; Yu, X.; Hildebrand, H.G.; Schreckenberger, B.; Kriegler, M.; Hetzer, B.; Frankino, P.; Dillin, A.; et al. Intercellular communication is required for trap formation in the nematode-trapping fungus Duddingtonia flagrans. PLoS Genet. 2019, 15, e1008029. [Google Scholar] [CrossRef]
- Wernet, V.; Wäckerle, J.; Fischer, R. The STRIPAK component SipC is involved in morphology and cell-fate determination in the nematode-trapping fungus Duddingtonia flagrans. Genetics 2022, 220, iyab153. [Google Scholar] [CrossRef]
- Yu, X.; Hu, X.; Pop, M.; Wernet, N.; Kirschhöfer, F.; Brenner-Weiß, G.; Keller, J.; Bunzel, M.; Fischer, R. Fatal attraction of Caenorhabditis elegans to predatory fungi through 6-methyl-salicylic acid. Nat. Commun. 2021, 12, 5462. [Google Scholar] [CrossRef] [PubMed]
- Wernet, N.; Wernet, V.; Fischer, R. The small-secreted cysteine-rich protein CyrA is a virulence factor participating in the attack of Caenorhabditis elegans by Duddingtonia flagrans. PLoS Pathog. 2021, 17, e1010028. [Google Scholar] [CrossRef] [PubMed]
- Emser, J.; Wernet, N.; Hetzer, B.; Wohlmann, E.; Fischer, R. The cysteine-rich virulence factor NipA of Arthrobotrys flagrans interferes with cuticle integrity of Caenorhabditis elegans. Nat. Commun. 2024, 15, 5795. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.Q.; Ferraz, C.M.; Ribeiro, N.R.C.; Ulfeldt, K.B.; Ribeiro, J.C.C.; Merizio, M.F.; Rossi, G.A.M.; Aguiar, A.; Araújo, J.V.; Soares, F.E.F.; et al. Efficacy of Duddingtonia flagrans (Bioverm®) on the biological control of buffalo gastrointestinal nematodes. Exp. Parasitol. 2023, 253, 108592. [Google Scholar] [CrossRef]
- Pandit, R.; Patel, R.; Patel, N.; Bhatt, V.; Joshi, C.; Singh, P.K.; Kunjadia, A. RNA-Seq reveals the molecular mechanism of trapping and killing of root-knot nematodes by nematode-trapping fungi. World J. Microbiol. Biotechnol. 2017, 33, 65. [Google Scholar] [CrossRef]
- Wernet, V.; Fischer, R. Establishment of Arthrobotrys flagrans as biocontrol agent against the root pathogenic nematode Xiphinema index. Environ. Microbiol. 2023, 25, 283–293. [Google Scholar] [CrossRef]
- Fischer, R.; Requena, N. Small-secreted proteins as virulence factors in nematode-trapping fungi. Trends Microbiol. 2022, 30, 615–617. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Y.; Yang, X.; Yang, E.; Xu, H.; Chen, Y.; Chagan, I.; Yan, J. Alternative splicing of heat shock transcription factor 2 regulates the expression of laccase gene family in response to copper in Trametes trogii. Appl. Environ. Microbiol. 2021, 87, e00055-21. [Google Scholar] [CrossRef]
- Linghu, S.X.; Zhang, Y.; Zuo, J.F.; Mo, M.H.; Li, G.H. AfSwi6 regulates the stress response, chlamydospore production, and pathogenicity in the nematode-trapping fungus Arthrobotrys flagrans. Microorganisms 2024, 12, 1765. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Liu, Q.; Zhu, M.; Zhu, L.; Yang, J. The Hog1-Nmd5 signaling pathway regulates asexual development, lipid metabolism, stress response, trap morphogenesis, and secondary metabolism of Arthrobotrys oligospora. Virulence 2025, 16, 2468294. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, Y.; Duan, S.; Bai, N.; Zhu, M.; Yang, J. Cellular communication and fusion regulate cell fusion, trap morphogenesis, conidiation, and secondary metabolism in Arthrobotrys oligospora. Microbiol. Res. 2024, 278, 127516. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, Z.; Luo, Y.; Yang, E.; Xu, H.; Chagan, I.; Yan, J. The manganese peroxidase gene family of Trametes trogii: Gene identification and expression patterns using various metal ions under different culture conditions. Microorganisms 2021, 9, 2595. [Google Scholar] [CrossRef]
- Xie, M.; Wang, Y.; Tang, L.; Yang, L.; Zhou, D.; Li, Q.; Niu, X.; Zhang, K.Q.; Yang, J. AoStuA, an APSES transcription factor, regulates the conidiation, trap formation, stress resistance and pathogenicity of the nematode-trapping fungus Arthrobotrys oligospora. Environ. Microbiol. 2019, 21, 4648–4661. [Google Scholar] [CrossRef]
- Zhen, Z.; Xing, X.; Xie, M.; Yang, L.; Yang, X.; Zheng, Y.; Chen, Y.; Ma, N.; Li, Q.; Zhang, K.Q.; et al. MAP kinase Slt2 orthologs play similar roles in conidiation, trap formation, and pathogenicity in two nematode-trapping fungi. Fungal Genet. Biol. 2018, 116, 42–50. [Google Scholar] [CrossRef]
- Bai, N.; Xie, M.; Liu, Q.; Zhu, Y.; Yang, X.; Zhang, K.Q.; Yang, J. AoMedA has a complex regulatory relationship with AoBrlA, AoAbaA, and AoWetA in conidiation, trap formation, and secondary metabolism in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Environ. Microbiol. 2023, 89, e0098323. [Google Scholar] [CrossRef]
- Liu, Q.; Jiang, K.; Duan, S.; Zhao, N.; Shen, Y.; Zhu, L.; Zhang, K.Q.; Yang, J. Identification of a transcription factor AoMsn2 of the Hog1 signaling pathway contributes to fungal growth, development and pathogenicity in Arthrobotrys oligospora. J. Adv. Res. 2025, 68, 1–15. [Google Scholar] [CrossRef]
- Kuo, C.Y.; Chen, S.A.; Hsueh, Y.P. The high osmolarity glycerol (HOG) pathway functions in osmosensing, trap morphogenesis and conidiation of the nematode-trapping fungus Arthrobotrys oligospora. J. Fungi 2020, 6, 191. [Google Scholar] [CrossRef]
- Chen, X.; Moran Torres, J.P.; Wösten, H.A.B. The role of the Flb protein family in the life cycle of Aspergillus niger. Antonie Van. Leeuwenhoek 2024, 117, 58. [Google Scholar] [CrossRef]
- Matheis, S.; Yemelin, A.; Scheps, D.; Andresen, K.; Jacob, S.; Thines, E.; Foster, A.J. Functions of the Magnaporthe oryzae Flb3p and Flb4p transcription factors in the regulation of conidiation. Microbiol. Res. 2017, 196, 106–117. [Google Scholar] [CrossRef]
- Guo, C.T.; Luo, X.C.; Ying, S.H.; Feng, M.G. Differential roles of five fluffy genes (flbA-flbE) in the lifecycle in vitro and in vivo of the insect-pathogenic fungus Beauveria bassiana. J. Fungi 2022, 8, 334. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.C.; Li, X.M.; Zhao, N.; Yang, L.; Zhang, K.Q.; Yang, J.K. Regulatory mechanism of trap formation in the nematode-trapping fungi. J. Fungi 2022, 8, 406. [Google Scholar] [CrossRef]
- Seo, J.A.; Guan, Y.; Yu, J.H. FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics 2006, 172, 1535–1544. [Google Scholar] [CrossRef]
- Mei, X.; Wang, X.; Li, G. Pathogenicity and volatile nematicidal metabolites from Duddingtonia flagrans against Meloidogyne incognita. Microorganisms 2021, 9, 2268. [Google Scholar] [CrossRef]
- Elling, A.A. Major emerging problems with minor meloidogyne species. Phytopathology 2013, 103, 1092–1102. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Peng, S.-Q.; He, W.-T.; Gao, F.-F.; Shi, Q.-F.; Li, G.-H. FlbD: A Regulator of Hyphal Growth, Stress Resistance, Pathogenicity, and Chlamydospore Production in the Nematode-Trapping Fungus Arthrobotrys flagrans. Microorganisms 2025, 13, 1847. https://doi.org/10.3390/microorganisms13081847
Zhang Y, Peng S-Q, He W-T, Gao F-F, Shi Q-F, Li G-H. FlbD: A Regulator of Hyphal Growth, Stress Resistance, Pathogenicity, and Chlamydospore Production in the Nematode-Trapping Fungus Arthrobotrys flagrans. Microorganisms. 2025; 13(8):1847. https://doi.org/10.3390/microorganisms13081847
Chicago/Turabian StyleZhang, Yu, Shun-Qiao Peng, Wang-Ting He, Fei-Fei Gao, Qian-Fei Shi, and Guo-Hong Li. 2025. "FlbD: A Regulator of Hyphal Growth, Stress Resistance, Pathogenicity, and Chlamydospore Production in the Nematode-Trapping Fungus Arthrobotrys flagrans" Microorganisms 13, no. 8: 1847. https://doi.org/10.3390/microorganisms13081847
APA StyleZhang, Y., Peng, S.-Q., He, W.-T., Gao, F.-F., Shi, Q.-F., & Li, G.-H. (2025). FlbD: A Regulator of Hyphal Growth, Stress Resistance, Pathogenicity, and Chlamydospore Production in the Nematode-Trapping Fungus Arthrobotrys flagrans. Microorganisms, 13(8), 1847. https://doi.org/10.3390/microorganisms13081847






