Molecular and Drug Resistance Characteristics of Haemophilus influenzae Carried by Pediatric Patients with Adenoid Hypertrophy
Abstract
1. Introduction
2. Material and Methods
2.1. Data Collection
2.2. H. influenzae Isolation
2.3. Typing of the H. influenzae Strains by q-PCR Method
2.4. Identification of H. influenzae
2.5. MLST Analysis and Minimum Spanning Tree Analysis
2.6. Detection of β-Lactam Resistance-Related Genes
2.7. Antibiotic Susceptibility Test (AST)
2.8. Analysis of the Amino Acid Sequences of the acrRAB Gene Cluster Coding for the Multi-Drug Efflux Pump and the Gene ompP2 of Porin
2.9. Statistical Analysis
3. Results
3.1. Relevant Data from the Enrolled Patients
3.2. Capsular Genotyping Results
3.3. MLST Results, ftsI Allele Grouping Data, and Presence of the β-lactamases blaTEM-1 and blaROB-1
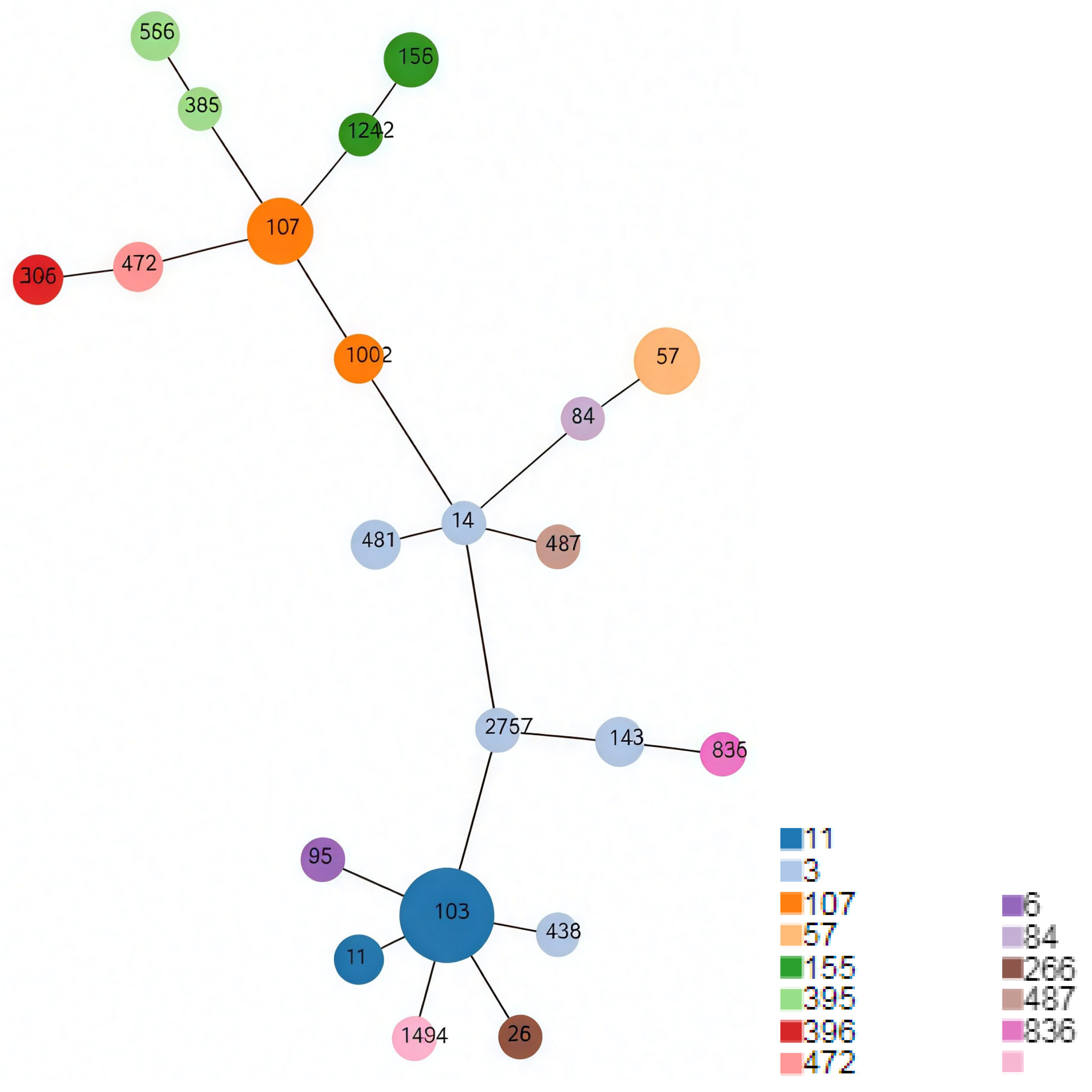
3.4. Minimum Spanning Tree
3.5. Mutation Patterns of the ftsI Alleles
3.6. Summary of the AST Results
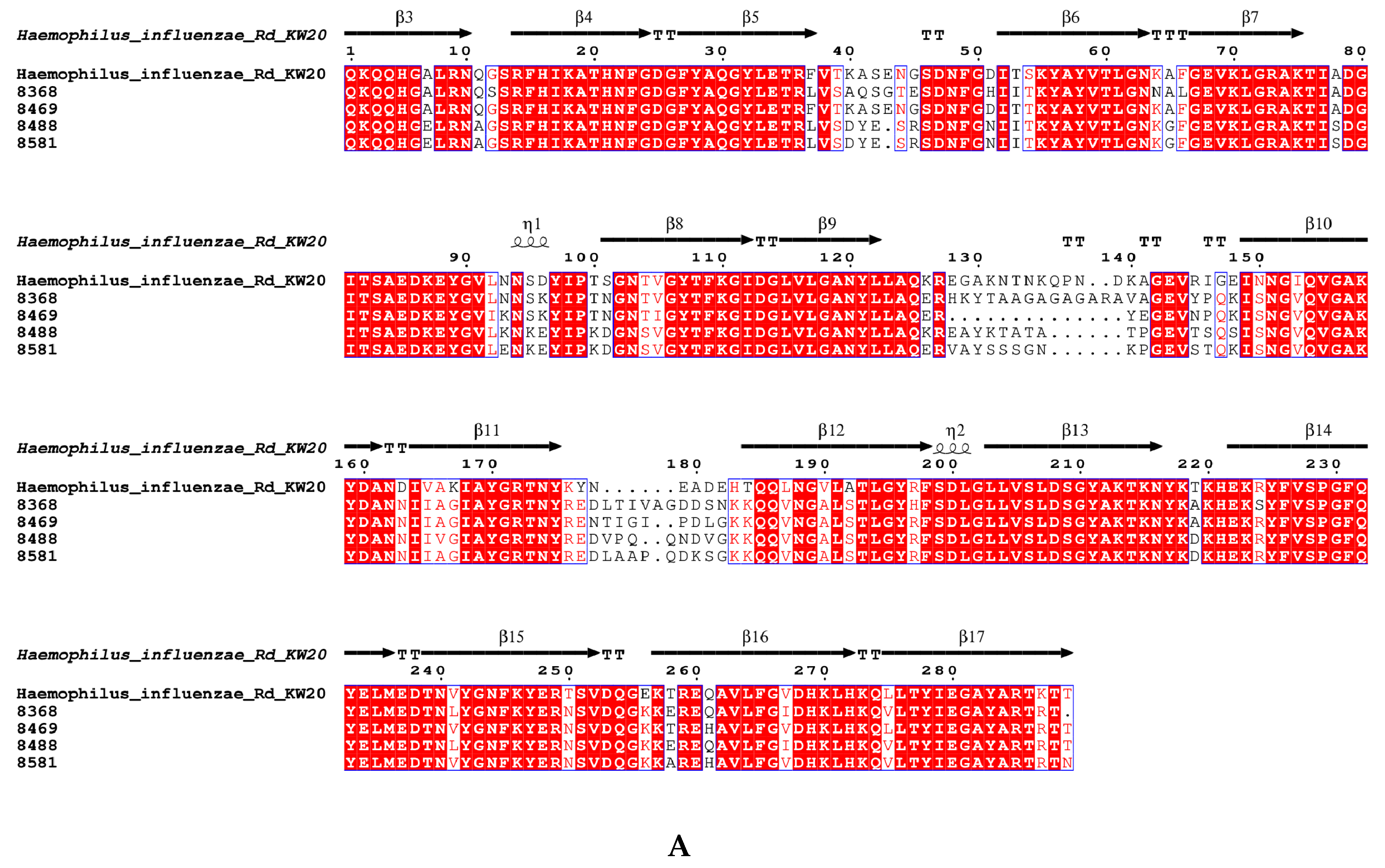
3.7. The Amino Acid Sequence Pattern of acrRAB Gene Clusters and the Gene ompP2 Coding for Porin
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Niedzielski, A.; Chmielik, L.; Mielnik-Niedzielska, G.; Kasprzyk, A.; Bogusławska, J. Adenoid hypertrophy in children. A narrative review of pathogenesis and clinical relevance. BMJ Paediatr. Open 2023, 7, e001710. [Google Scholar] [CrossRef]
- Johnston, J.J.; Douglas, R. Adenotonsillar microbiome: An update. Postgrad. Med. J. 2018, 94, 398–403. [Google Scholar] [CrossRef]
- Johnston, J.; Hoggard, M.; Biswas, K.; Astudillo-Garcia, C.; Radcliff, F.J.; Mahadevan, M.; Douglas, R.G. Pathogen reservoir hypothesis investigated by analyses of the adenotonsillar and middle ear microbiota. Int. J. Pediatr. Otorhinolaryngol. 2019, 118, 103–109. [Google Scholar] [CrossRef]
- Ren, T.; Glatt, D.U.; Nguyen, T.N.; Allen, E.K.; Early, S.V.; Sale, M.; Winther, B.; Wu, M. 16S rRNA survey revealed complex bacterial communities and evidence of bacterial interference on human adenoids. Environ. Microbiol. 2013, 15, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Goktas, O.; Bessler, C.; Loening-Baucke, V.; Hale, L.P.; Andree, H.; Weizenegger, M.; Holzl, M.; Scherer, H.; Lochs, H. Spatial organisation of microbiota in quiescent adenoiditis and tonsillitis. J. Clin. Pathol. 2007, 60, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Cobos, S.; Campos, J.; Lazaro, E.; Roman, F.; Cercenado, E.; Garcia-Rey, C.; Perez-Vazquez, M.; Oteo, J.; de Abajo, F. Ampicillin-resistant non-beta-lactamase-producing Haemophilus influenzae in Spain. recent emergence of clonal isolates with increased resistance to cefotaxime and cefixime. Antimicrob. Agents Chemother. 2007, 51, 2564–2573. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, G.; Kim, J.H.; Kim, M.N.; Lee, J. Characterization of Ceftriaxone-Resistant Haemophilus influenzae Among Korean Children. J. Korean Med. Sci. 2024, 39, e136. [Google Scholar] [CrossRef]
- Skaare, D.; Allum, A.; Anthonisen, I.; Jenkins, A.; Lia, A.; Strand, L.; Tveten, Y.; Kristiansen, B. Mutant ftsI genes in the emergence of penicillin-binding protein-mediated beta-lactam resistance in Haemophilus influenzae in Norway. Clin. Microbiol. Infect. 2010, 16, 1117–1124. [Google Scholar] [CrossRef]
- Dabernat, H.; Delmas, C.; Seguy, M.; Pelissier, R.; Faucon, G.; Bennamani, S.; Pasquier, C. Diversity of beta-lactam resistance-conferring amino acid substitutions in penicillin-binding protein 3 of Haemophilus influenzae. Antimicrob. Agents Chemother. 2002, 46, 2208–2218. [Google Scholar] [CrossRef]
- Yuan, M.; Ma, M.; Jiang, H.; Fan, M.; Sun, Y.; Zhou, B.; Feng, X.; Yang, J.; Su, M.; He, X. Characterization of Serotypes and Molecular Drug Resistance Patterns of Haemophilus influenzae in Kunming Children. Pol. J. Microbiol. 2023, 72, 125–131. [Google Scholar] [CrossRef]
- Su, P.; Cheng, W.; Ho, C. Molecular characterization of multidrug-resistant non-typeable Haemophilus influenzae with high-level resistance to cefuroxime, levofloxacin, and trimethoprim-sulfamethoxazole. BMC Microbiol. 2023, 23, 178. [Google Scholar] [CrossRef] [PubMed]
- Søndergaard, A.; Nørskov-Lauritsen, N. Contribution of PBP3 Substitutions and TEM-1, TEM-15, and ROB-1 Beta-Lactamases to Cefotaxime Resistance in Haemophilus influenzae and Haemophilus parainfluenzae. Microb. Drug Resist. 2016, 22, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Søndergaard, A.; Lund, M.; Nørskov-Lauritsen, N. TEM-1-encoding small plasmids impose dissimilar fitness costs on Haemophilus influenzae and Haemophilus parainfluenzae. Microbiology 2015, 161, 2310–2315. [Google Scholar] [CrossRef] [PubMed]
- Skaare, D.; Lia, A.; Hannisdal, A.; Tveten, Y.; Matuschek, E.; Kahlmeter, G.; Kristiansen, B.E. Haemophilus influenzae with Non-Beta-Lactamase-Mediated Beta-Lactam Resistance. Easy To Find but Hard to Categorize. J. Clin. Microbiol. 2015, 53, 3589. [Google Scholar] [CrossRef]
- Ubukata, K.; Shibasaki, Y.; Yamamoto, K.; Chiba, N.; Hasegawa, K.; Takeuchi, Y.; Sunakawa, K.; Inoue, M.; Konno, M. Association of amino acid substitutions in penicillin-binding protein 3 with beta-lactam resistance in beta-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob. Agents Chemother. 2001, 45, 1693–1699. [Google Scholar] [CrossRef]
- Skaare, D.; Anthonisen, I.; Kahlmeter, G.; Matuschek, E.; Natås, O.; Steinbakk, M.; Sundsfjord, A.; Kristiansen, B. Emergence of clonally related multidrug resistant Haemophilus influenzae with penicillin-binding protein 3-mediated resistance to extended-spectrum cephalosporins, Norway, 2006 to 2013. Euro Surveill. 2014, 19, 20986. [Google Scholar] [CrossRef]
- Zanella, R.; Bokermann, S.; Galhardo, M.; Gava, C.; Almeida, S.; Pereira, G.; de Lemos, A. Trends in serotype distribution and antimicrobial susceptibility pattern of invasive Haemophilus influenzae isolates from Brazil, 2009–2021. Int. Microbiol. 2024, 28, 157–163. [Google Scholar] [CrossRef]
- Häuser, S.; Wegele, C.; Stump-Guthier, C.; Borkowski, J.; Weiss, C.; Rohde, M.; Ishikawa, H.; Schroten, H.; Schwerk, C.; Adam, R. Capsule and fimbriae modulate the invasion of Haemophilus influenzae in a human blood-cerebrospinal fluid barrier model. Int. J. Med. Microbiol. 2018, 308, 829–839. [Google Scholar] [CrossRef]
- Ulanova, M.; Tsang, R. Haemophilus influenzae serotype a as a cause of serious invasive infections. Lancet Infect. Dis. 2014, 14, 70–82. [Google Scholar] [CrossRef]
- Slack, M.P.E.; Cripps, A.W.; Grimwood, K.; Mackenzie, G.A.; Ulanova, M. Invasive Haemophilus influenzae Infections after 3 Decades of Hib Protein Conjugate Vaccine Use. Clin. Microbiol. Rev. 2021, 34, e00028-21. [Google Scholar] [CrossRef]
- Van Eldere, J.; Slack, M.P.; Ladhani, S.; Cripps, A.W. Non-typeable Haemophilus influenzae, an under-recognised pathogen. Lancet Infect. Dis. 2014, 14, 1281–1292. [Google Scholar] [CrossRef]
- Ortiz-Romero, M.; Espejo-García, M.; Alfayate-Miguelez, S.; Ruiz-López, F.; Zapata-Hernandez, D.; Gonzalez-Pacanowska, A. Epidemiology of Nasopharyngeal Carriage by Haemophilus influenzae in Healthy Children. A Study in the Mediterranean Coast Region. Pediatr. Infect. Dis. J. 2017, 36, 919–923. [Google Scholar] [CrossRef] [PubMed]
- Langereis, J.; de Jonge, M. Unraveling Haemophilus influenzae virulence mechanisms enable discovery of new targets for antimicrobials and vaccines. Curr. Opin. Infect. Dis. 2020, 33, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Davis, G.S.; Sandstedt, S.A.; Patel, M.; Marrs, C.F.; Gilsdorf, J.R. Use of bexB to detect the capsule locus in Haemophilus influenzae. J. Clin. Microbiol. 2011, 49, 2594–2601. [Google Scholar] [CrossRef]
- WHO. Laboratory Methods for Diagnosis of Meningitis Caused by Neisseria Meningitidis, Streptococcus Pneumoniae, and Haemophlius Influenzae, 2nd ed.; WHO Press: Geneva, Switzerland, 2011. [Google Scholar]
- Weisburg, W.; Barns, S.; Pelletier, D.; Lane, D. 16S ribosomal DNA amplification for phylogenetic study.pdf. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Meats, E.; Feil, E.J.; Stringer, S.; Cody, A.J.; Goldstein, R.; Kroll, J.S.; Popovic, T.; Spratt, B.G. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 2003, 41, 1623–1636. [Google Scholar] [CrossRef]
- Nascimento, M.; Sousa, A.; Ramirez, M.; Francisco, A.; Carriço, J.; Vaz, C. PHYLOViZ 2.0. providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 2017, 33, 128–129. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 34th ed.; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2024. [Google Scholar]
- EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters (Version 14.0); European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2024. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2016. [Google Scholar]
- Hayashi, K.; Nakashima, R.; Sakurai, K.; Kitagawa, K.; Yamasaki, S.; Nishino, K.; Yamaguchi, A. AcrB-AcrA Fusion Proteins That Act as Multidrug Efflux Transporters. J. Bacteriol. 2015, 198, 332–342. [Google Scholar] [CrossRef]
- Zwama, M.; Yamaguchi, A.; Nishino, K. Phylogenetic functional characterisation of the Haemophilus influenzae multidrug efflux pump AcrB. Commun. Biol. 2019, 2, 340. [Google Scholar] [CrossRef]
- Cherkaoui, A.; Diene, S.M.; Renzoni, A.; Emonet, S.; Renzi, G.; Francois, P.; Schrenzel, J. Imipenem heteroresistance in nontypeable Haemophilus influenzae is linked to a combination of altered PBP3, slow drug influx and direct efflux regulation. Clin. Microbiol. Infect. 2017, 23, 118.e9–118.e119. [Google Scholar] [CrossRef]
- Gouet, P.; Courcelle, E.; Stuart, D.; Métoz, F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 1999, 15, 305–308. [Google Scholar] [CrossRef]
- Chen, W.; Yin, G.; Chen, Y.; Wang, L.; Wang, Y.; Zhao, C.; Wang, W.; Ye, J. Analysis of factors that influence the occurrence of otitis media with effusion in pediatric patients with adenoid hypertrophy. Front. Pediatr. 2023, 11, 1098067. [Google Scholar] [CrossRef]
- Duell, B.; Su, Y.; Riesbeck, K. Host-pathogen interactions of nontypeable Haemophilus influenzae. from commensal to pathogen. FEBS Lett. 2016, 590, 3840–3853. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, H.; Zhang, Y.; Zhao, P.; Li, Y. Aerosolization inhalation of non-typeable Haemophilus influenzae outer membrane vesicles contributing to neutrophilic asthma. Front. Microbiol. 2023, 26, 1226633. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, B.; Goff, S.; Boguniewicz, J.; Gitomer, S. Postmeningitic pediatric hearing loss from non-type b Haemophilus influenzae. Am. J. Otolaryng 2024, 45, 104104. [Google Scholar] [CrossRef] [PubMed]
- Frankel, C.; Robinson, J.; Khan, S.; Alghounaim, M.; McDonald, J.; Lopez, A.; Fanella, S.; Gunawan, J.; Wong, J.; Comeau, J.; et al. A Pediatric Investigators Collaborative Network on Infections in Children (PICNIC) multi-centre Canadian descriptive analysis of Haemophilus influenzae bacteremia in children. Emerging serotypes. Can. Commun. Dis. Rep. 2023, 49, 368–374. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Y.; Cheng, J.; Zhao, X.; Liang, Y.; Wu, J. Molecular epidemiology and antimicrobial resistance of Haemophilus influenzae in Guiyang, Guizhou, China. Front. Public Health 2022, 10, 947051. [Google Scholar] [CrossRef]
- Dong, Q.; Shi, W.; Cheng, X.; Chen, C.; Meng, Q.; Yao, K.; Qian, S. Widespread of non-typeable Haemophilus influenzae with high genetic diversity after two decades use of Hib vaccine in China. J. Clin. Lab. Anal. 2020, 34, e23145. [Google Scholar] [CrossRef]
- Li, X.; Xiao, S.; Gu, F.; He, W.; Ni, Y.; Han, L. Molecular Epidemiology and Antimicrobial Resistance of Haemophilus influenzae in Adult Patients in Shanghai, China. Front. Public Health 2020, 8, 95. [Google Scholar] [CrossRef]
- Zuo, L.; He, L.; Huang, A.; Liu, Y.; Zhang, A.; Wang, L.; Song, Y.; Geng, J. Risk factors and antibiotic sensitivity of aerobic bacteria in Chinese children with adenoid hypertrophy. BMC Pediatr. 2022, 22, 553. [Google Scholar] [CrossRef]
- Cerquetti, M.; Giufrè, M.; Cardines, R.; Mastrantonio, P. First characterization of heterogeneous resistance to imipenem in invasive nontypeable Haemophilus influenzae isolates. Antimicrob. Agents Chemother. 2007, 51, 3155–3161. [Google Scholar] [CrossRef]

| Age Group (Number of AH Patients) | Number of Strains | Ratio (95% Confidence Interval) | Chi-Square Test (Trend) | ||
|---|---|---|---|---|---|
| 2–5 years old (59) | 18 | 30.51% (20.33–43.06%) |  |  | p = 0.0003 |
| 6–9 years old (201) | 27 | 13.43% (9.46–18.78%) | |||
| 10–15 years old (126) | 11 | 8.73% (4.99–14.85%) | |||
| CC | ST | Number |
|---|---|---|
| 3 | 14 | 1 |
| 143 | 2 | |
| 436 | 1 | |
| 481 | 2 | |
| 2757 | 1 | |
| 6 | 95 | 1 |
| 11 | 103 | 11 |
| 11 | 2 | |
| 57 | 57 | 5 |
| 84 | 84 | 1 |
| 107 | 107 | 5 |
| 1002 | 2 | |
| 155 | 155 | 3 |
| 1242 | 1 | |
| 266 | 266 | 1 |
| 395 | 395 | 1 |
| 556 | 2 | |
| 396 | 396 | 2 |
| 472 | 472 | 2 |
| 487 | 487 | 1 |
| 836 | 836 | 1 |
| - | 1494 | 1 |
| ftsI Allele | ftsI Group | Number of Strains | Position 350 | Position 357 | Position 377 | Position 385 | Position 389 | Position 490 | Position 502 | Position 517 | Position 526 | blaTEM-1 | blaROB-1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 | III+ | 2 | Asp350Asn | Ser357Asn | Met377Ile | Ser385Thr | Leu389Phe | Gly490Glu | Asn526Lys | + | - | ||
| 26 | III-like+ | 4 | Asp350Asn | Met377Ile | Ser385Thr | Leu389Phe | Arg517His | + | - | ||||
| 2 | + | + | |||||||||||
| 33 | III-like | 2 | Asp350Asn | Ser357Asn | Met377Ile | Ser385Thr | Arg517His | + | + | ||||
| 1 | + | - | |||||||||||
| 1 | - | - | |||||||||||
| 40 | III+ | 2 | Asp350Asn | Ser357Asn | Met377Ile | Ser385Thr | Leu389Phe | Asn526Lys | + | - | |||
| 1 | - | + | |||||||||||
| 88 | III-like+ | 13 | Asp350Asn | Ser357Asn | Met377Ile | Ser385Thr | Leu389Phe | Arg517His | + | - | |||
| 1 | + | + | |||||||||||
| 98 | IIa | 1 | Asn526Lys | + | - | ||||||||
| 107 | III-like+ | 2 | Asp350Asn | Ser357Asn | Met377Ile | Ser385Thr | Leu389Phe | Arg517His | + | + | |||
| 1 | + | - | |||||||||||
| 142 | IIa | 1 | Asn526Lys | + | - | ||||||||
| 185 | III+ | 2 | Asp350Asn | Ser357Asn | Met377Ile | Ser385Thr | Leu389Phe | Gly490Glu | Asn526Lys | + | - | ||
| 194 | III-like+ | 3 | Asp350Asn | Ser357Asn | Met377Ile | Ser385Thr | Leu389Phe | Arg517His | + | - | |||
| 197 | Miscellaneous | 1 | Asp350Asn | Met377Ile | Ser385Thr | Leu389Phe | Asn526His | + | - | ||||
| 200 | III+ | 1 | Asp350Asn | Ser357Asn | Met377Ile | Ser385Thr | Leu389Phe | Asn526Lys | + | - | |||
| 202 | III-like+ | 1 | Asp350Asn | Ser357Asn | Met377Ile | Ser385Thr | Leu389Phe | Arg517His | + | - | |||
| 275 | WT | 1 | Asp350Asn | + | - | ||||||||
| 331 | III-like | 1 | Asp350Asn | Ser357Asn | Ser385Thr | Arg517His | + | + | |||||
| 1 | + | - | |||||||||||
| 336 | WT | 2 | Asp350Asn | + | - | ||||||||
| 370 | III-like+ | 1 | Met377Ile | Ser385Thr | Leu389Phe | Arg517His | + | - | |||||
| 374 | III+IIb | 1 | Asp350Asn | Ser357Asn | Met377Ile | Ser385Thr | Leu389Phe | Ala502Val | Asn526Lys | + | - |
| Antibiotic | Susceptibility by CLSI BPs | Susceptibility by EUCAST BPs | Mode of MIC (µg/mL) | Median and Range of MIC (µg/mL) |
|---|---|---|---|---|
| MEM | 95.9% | 100% | 0.38 | 0.25 (from 0.032 to 1.5) |
| CRO | 100% | 59.2% | 0.048 | 0.094 (from 0.008 to 1) |
| IMP | 42.9% | 91.8% | 0.5 | 0.75 (from 0.048 to ≥32) |
| AMP | 10.2% | 10.2% | ≥256 | ≥256 (from 0.5 to ≥256) |
| SAM | 28.6% | 14.3% | 4 | 4 (from 0.25 to ≥32) |
| LEV | 100% | 98.0% | 0.032 | 0.032 (from 0.008 to 1) |
| TET | 100% | 100% | 0.25 | 0.25 (from 0.064 to 0.25) |
| SXT | 32.7% | 32.7% | ≥32 | 1 (from 0.016 to ≥32) |
| AZM | 34.7% | * | ≥256 | 64 (from 0.5 to ≥256) |
| Bank Number | MEM | CRO | IMP | AMP | SAM | LEV | TET | SXT | AZM | ftsI Group | blaTEM-1 | blaROB-1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8488 | 0.38 | 0.024 | 4 | 256 | 4 | 0.032 | 0.25 | 32 | 256 | IIa | + | − |
| 8469 | 1 | 0.125 | 8 | 256 | 4 | 0.032 | 0.25 | 32 | 64 | III+ | + | − |
| 8581 | 0.38 | 0.19 | ≥32 | 256 | 16 | 0.032 | 0.25 | 16 | 256 | III+ | + | − |
| 8368 | 0.25 | 1 | 4 | 256 | 4 | 1 | 0.25 | 32 | 256 | Miscellaneous | + | − |
| Bank Number | acrRAB Gene Cluster and AcrAB Multi-Drug Efflux Pump | ompP2 and Porin | ||
|---|---|---|---|---|
| Average Identity (%) for Nucleotides | Average Identity (%) for Amino Acids | Average Identity (%) for Nucleotides | Average Identity (%) for Amino Acids | |
| 8368 | 97.41 | 93.77 | 83.33 | 77.10 |
| 8469 | 96.72 | 92.10 | 86.85 | 82.94 |
| 8488 | 95.20 | 88.12 | 84.47 | 77.82 |
| 8581 | 97.85 | 94.94 | 85.20 | 77.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, N.; Qin, J.-H.; Zhao, X.-Y.; Liu, L. Molecular and Drug Resistance Characteristics of Haemophilus influenzae Carried by Pediatric Patients with Adenoid Hypertrophy. Microorganisms 2025, 13, 1764. https://doi.org/10.3390/microorganisms13081764
Xiao N, Qin J-H, Zhao X-Y, Liu L. Molecular and Drug Resistance Characteristics of Haemophilus influenzae Carried by Pediatric Patients with Adenoid Hypertrophy. Microorganisms. 2025; 13(8):1764. https://doi.org/10.3390/microorganisms13081764
Chicago/Turabian StyleXiao, Nan, Jia-Hao Qin, Xiu-Ying Zhao, and Lin Liu. 2025. "Molecular and Drug Resistance Characteristics of Haemophilus influenzae Carried by Pediatric Patients with Adenoid Hypertrophy" Microorganisms 13, no. 8: 1764. https://doi.org/10.3390/microorganisms13081764
APA StyleXiao, N., Qin, J.-H., Zhao, X.-Y., & Liu, L. (2025). Molecular and Drug Resistance Characteristics of Haemophilus influenzae Carried by Pediatric Patients with Adenoid Hypertrophy. Microorganisms, 13(8), 1764. https://doi.org/10.3390/microorganisms13081764






