In Silico Approach for Early Antimalarial Drug Discovery: De Novo Design of Virtual Multi-Strain Antiplasmodial Inhibitors
Abstract
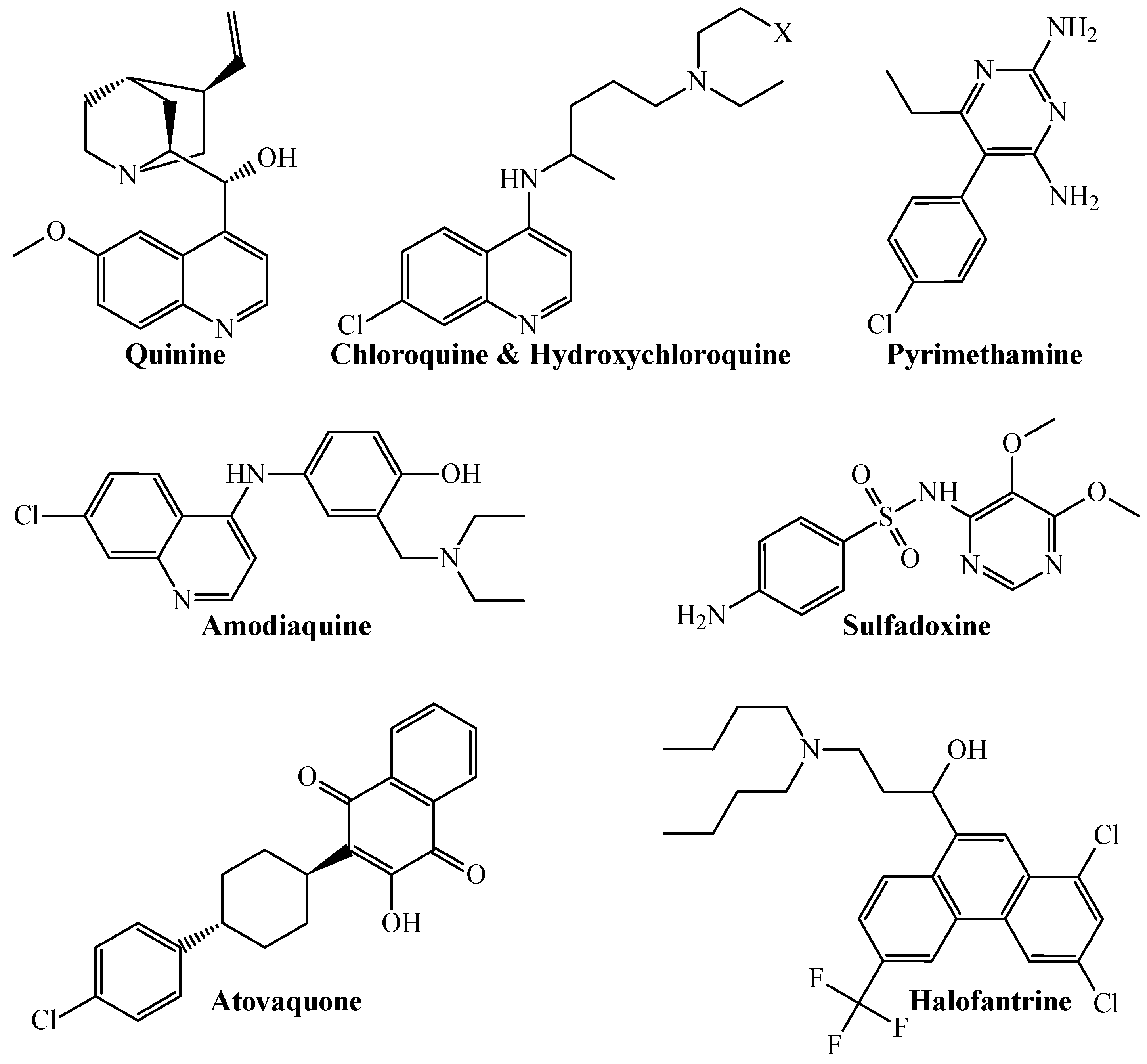
1. Introduction
2. Materials and Methods
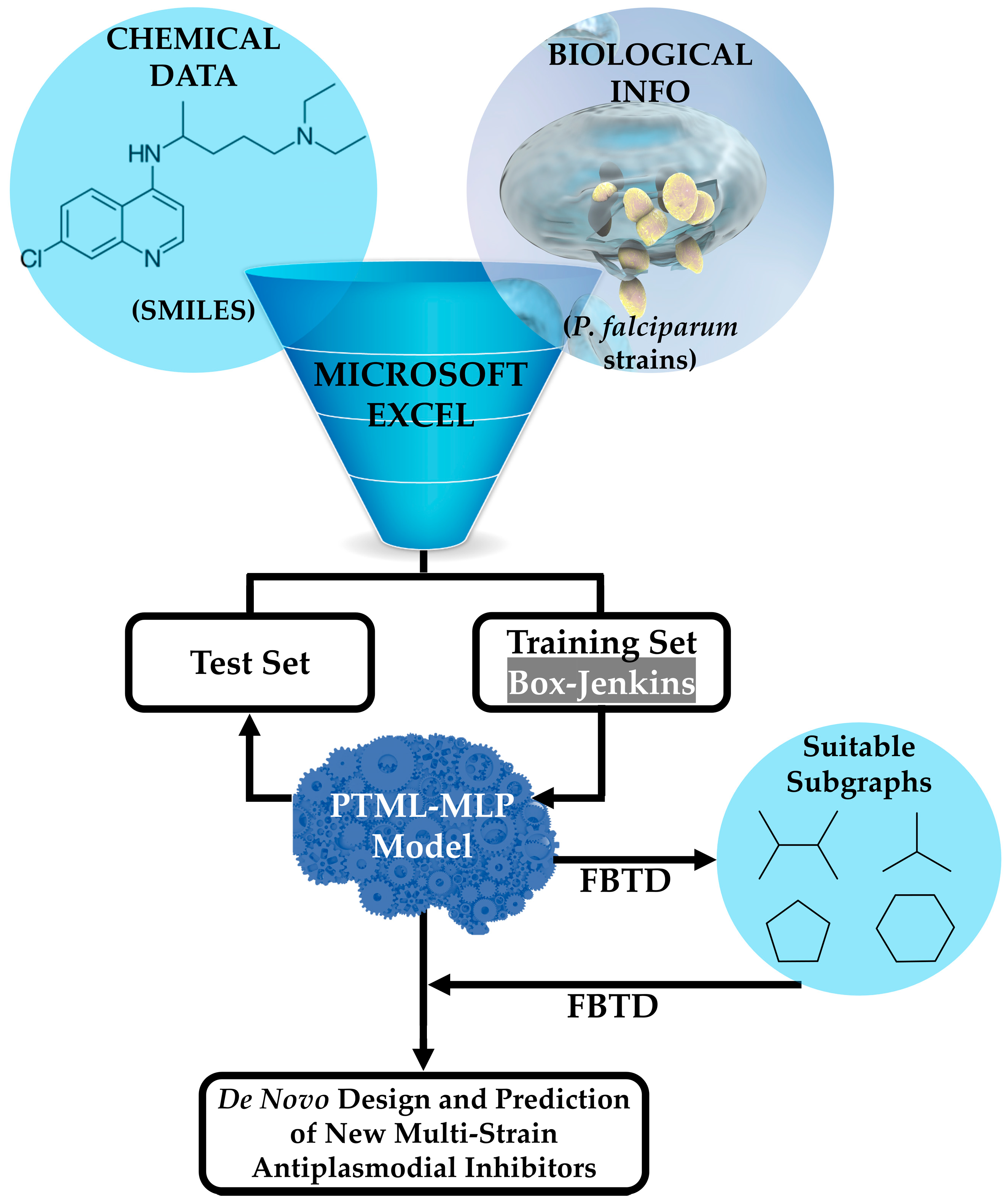
2.1. Data Curation and Topological Indices
2.2. PTML Modeling: Assessment of Performance and Applicability Domain
3. Results and Discussion
3.1. Analyzing the Performance of the PTML-MLP Model
3.2. The FBTD Approach: Interpretation of the PTML-MLP
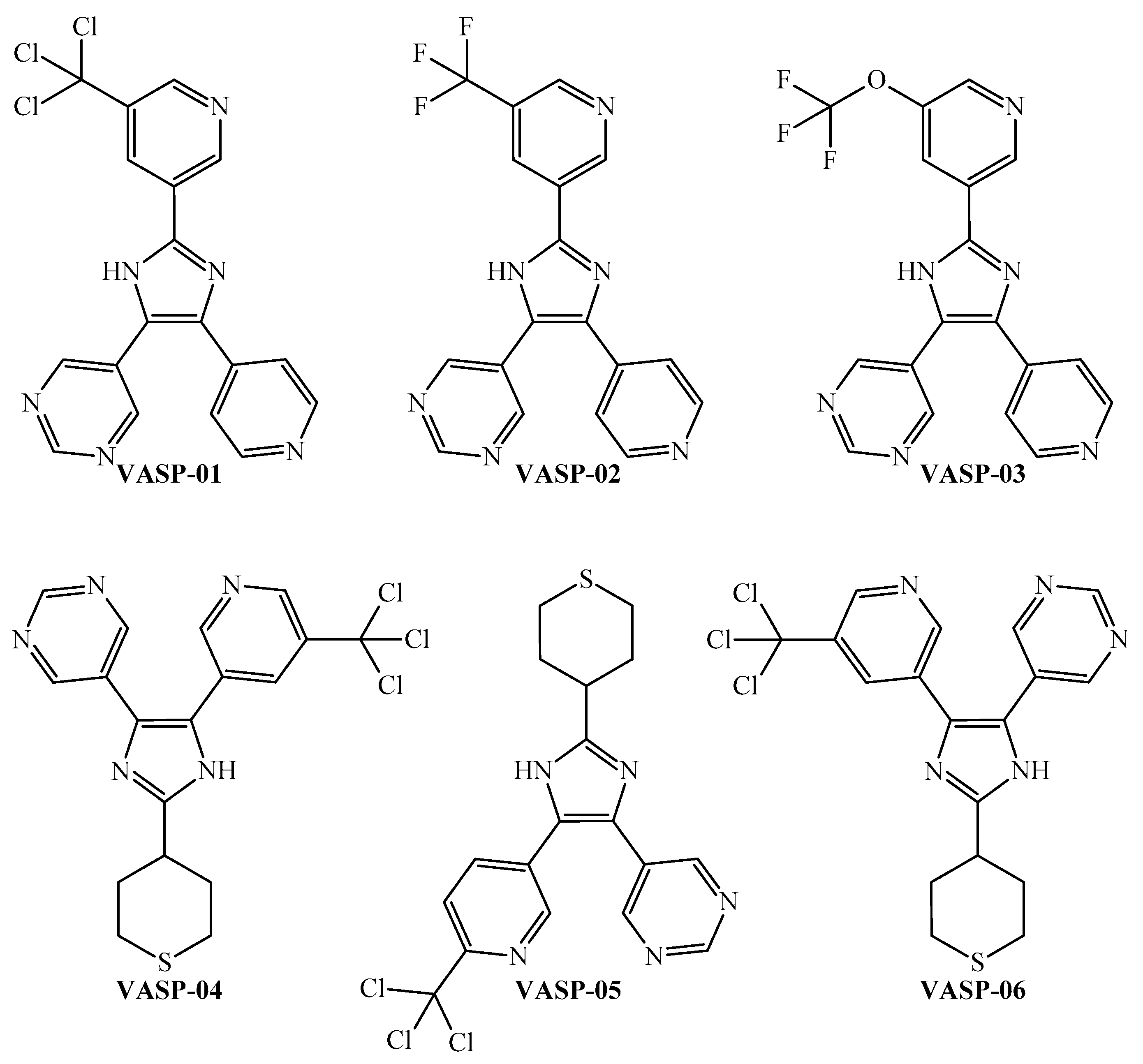
3.3. Combining the PTML-MLP and FBTD to Enable the Design of Multi-Strain Antiplasmodial Inhibitors
3.4. Druglikeness of the Designed Molecules
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siqueira-Neto, J.L.; Wicht, K.J.; Chibale, K.; Burrows, J.N.; Fidock, D.A.; Winzeler, E.A. Antimalarial drug discovery: Progress and approaches. Nat. Rev. Drug Discov. 2023, 22, 807–826. [Google Scholar] [CrossRef]
- Xia, J.; Wu, D.; Wu, K.; Zhu, H.; Sun, L.; Lin, W.; Li, K.; Zhang, J.; Wan, L.; Zhang, H.; et al. Epidemiology of Plasmodium falciparum Malaria and Risk Factors for Severe Disease in Hubei Province, China. Am. J. Trop. Med. Hyg. 2020, 103, 1534–1539. [Google Scholar] [CrossRef] [PubMed]
- Deutsch-Feldman, M.; Brazeau, N.F.; Parr, J.B.; Thwai, K.L.; Muwonga, J.; Kashamuka, M.; Tshefu Kitoto, A.; Aydemir, O.; Bailey, J.A.; Edwards, J.K.; et al. Spatial and epidemiological drivers of Plasmodium falciparum malaria among adults in the Democratic Republic of the Congo. BMJ Glob. Health 2020, 5, e002316. [Google Scholar] [CrossRef] [PubMed]
- Schafer, T.M.; Pessanha de Carvalho, L.; Inoue, J.; Kreidenweiss, A.; Held, J. The problem of antimalarial resistance and its implications for drug discovery. Expert Opin. Drug Discov. 2024, 19, 209–224. [Google Scholar] [CrossRef] [PubMed]
- Masserey, T.; Lee, T.; Golumbeanu, M.; Shattock, A.J.; Kelly, S.L.; Hastings, I.M.; Penny, M.A. The influence of biological, epidemiological, and treatment factors on the establishment and spread of drug-resistant Plasmodium falciparum. Elife 2022, 11, e77634. [Google Scholar] [CrossRef]
- Dhorda, M.; Amaratunga, C.; Dondorp, A.M. Artemisinin and multidrug-resistant Plasmodium falciparum—A threat for malaria control and elimination. Curr. Opin. Infect. Dis. 2021, 34, 432–439. [Google Scholar] [CrossRef]
- Saadeh, K.; Nantha Kumar, N.; Fazmin, I.T.; Edling, C.E.; Jeevaratnam, K. Anti-malarial drugs: Mechanisms underlying their proarrhythmic effects. Br. J. Pharmacol. 2022, 179, 5237–5258. [Google Scholar] [CrossRef]
- Lewis, J.; Gregorian, T.; Portillo, I.; Goad, J. Drug interactions with antimalarial medications in older travelers: A clinical guide. J. Travel Med. 2020, 27, taz089. [Google Scholar] [CrossRef]
- Umumararungu, T.; Nkuranga, J.B.; Habarurema, G.; Nyandwi, J.B.; Mukazayire, M.J.; Mukiza, J.; Muganga, R.; Hahirwa, I.; Mpenda, M.; Katembezi, A.N.; et al. Recent developments in antimalarial drug discovery. Bioorg. Med. Chem. 2023, 88–89, 117339. [Google Scholar] [CrossRef]
- Pandey, S.K.; Anand, U.; Siddiqui, W.A.; Tripathi, R. Drug Development Strategies for Malaria: With the Hope for New Antimalarial Drug Discovery—An Update. Adv. Med. 2023, 2023, 5060665. [Google Scholar] [CrossRef]
- Sakura, T.; Ishii, R.; Yoshida, E.; Kita, K.; Kato, T.; Inaoka, D.K. Accelerating Antimalarial Drug Discovery with a New High-Throughput Screen for Fast-Killing Compounds. ACS Infect. Dis. 2024, 10, 4115–4126. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Rajkhowa, S.; Sinha, S.; Zaki, M.E.A. Unveiling potential repurposed drug candidates for Plasmodium falciparum through in silico evaluation: A synergy of structure-based approaches, structure prediction, and molecular dynamics simulations. Comput. Biol. Chem. 2024, 110, 108048. [Google Scholar] [CrossRef]
- Elamin, E.M.; Eshage, S.E.; Mohmmode, S.M.; Mukhtar, R.M.; Mahjoub, M.; Sadelin, E.; Shoaib, T.H.; Edris, A.; Elshamly, E.M.; Makki, A.A.; et al. Discovery of dual-target natural antimalarial agents against DHODH and PMT of Plasmodium falciparum: Pharmacophore modelling, molecular docking, quantum mechanics, and molecular dynamics simulations. SAR QSAR Environ. Res. 2023, 34, 709–728. [Google Scholar] [CrossRef] [PubMed]
- Mafethe, O.; Ntseane, T.; Dongola, T.H.; Shonhai, A.; Gumede, N.J.; Mokoena, F. Pharmacophore Model-Based Virtual Screening Workflow for Discovery of Inhibitors Targeting Plasmodium falciparum Hsp90. ACS Omega 2023, 8, 38220–38232. [Google Scholar] [CrossRef]
- Almasoudi, H.H.; Nahari, M.H. Targeting Plasmodium falciparum Schizont Egress Antigen-1 in Infected Red Blood Cells: Docking-Based Fingerprinting, Density Functional Theory, Molecular Dynamics Simulations, and Binding Free Energy Analysis. Pharmaceuticals 2025, 18, 237. [Google Scholar] [CrossRef]
- Oduselu, G.O.; Elebiju, O.F.; Ogunnupebi, T.A.; Akash, S.; Ajani, O.O.; Adebiyi, E. Employing Hexahydroquinolines as PfCDPK4 Inhibitors to Combat Malaria Transmission: An Advanced Computational Approach. Adv. Appl. Bioinform. Chem. 2024, 17, 83–105. [Google Scholar] [CrossRef]
- Withers-Martinez, C.; George, R.; Ogrodowicz, R.; Kunzelmann, S.; Purkiss, A.G.; Kjaer, S.; Walker, P.A.; Kovada, V.; Jirgensons, A.; Blackman, M.J. Structural plasticity of Plasmodium falciparum plasmepsin X to accommodate binding of potent macrocyclic hydroxyethylamine inhibitors. J. Mol. Biol. 2025, 437, 169062. [Google Scholar] [CrossRef]
- Morcoss, M.M.; Saddik, J.N.; Amin, M.E.; Mohamed, F.A.M.; El-Rashedy, A.A.; Almutairi, T.M.; Youssif, B.G.M.; Lamie, P.F. Design, synthesis, antimalarial activity, and in-silico studies of new benzimidazole/pyridine hybrids as dihydrofolate reductase inhibitors. Bioorg. Chem. 2025, 156, 108171. [Google Scholar] [CrossRef]
- Yasir, M.; Park, J.; Han, E.T.; Han, J.H.; Park, W.S.; Chun, W. Identification of Malaria-Selective Proteasome beta5 Inhibitors Through Pharmacophore Modeling, Molecular Docking, and Molecular Dynamics Simulation. Int. J. Mol. Sci. 2024, 25, 11881. [Google Scholar] [CrossRef]
- Costa, E.B.; Silva, R.C.; Espejo-Roman, J.M.; Neto, M.F.A.; Cruz, J.N.; Leite, F.H.A.; Silva, C.; Pinheiro, J.C.; Macedo, W.J.C.; Santos, C.B.R. Chemometric methods in antimalarial drug design from 1,2,4,5-tetraoxanes analogues. SAR QSAR Environ. Res. 2020, 31, 677–695. [Google Scholar] [CrossRef]
- Roche-Lima, A.; Rosado-Quinones, A.M.; Feliu-Maldonado, R.A.; Figueroa-Gispert, M.D.M.; Diaz-Rivera, J.; Diaz-Gonzalez, R.G.; Carrasquillo-Carrion, K.; Nieves, B.G.; Colon-Lorenzo, E.E.; Serrano, A.E. Antimalarial Drug Combination Predictions Using the Machine Learning Synergy Predictor (MLSyPred(c)) tool. Acta Parasitol. 2024, 69, 415–425. [Google Scholar] [CrossRef] [PubMed]
- van Heerden, A.; Turon, G.; Duran-Frigola, M.; Pillay, N.; Birkholtz, L.M. Machine Learning Approaches Identify Chemical Features for Stage-Specific Antimalarial Compounds. ACS Omega 2023, 8, 43813–43826. [Google Scholar] [CrossRef] [PubMed]
- Kleandrova, V.V.; Cordeiro, M.N.D.S.; Speck-Planche, A. Perturbation-Theory Machine Learning for Multi-Objective Antibacterial Discovery: Current Status and Future Perspectives. Appl. Sci. 2025, 15, 1166. [Google Scholar] [CrossRef]
- Kleandrova, V.V.; Cordeiro, M.N.D.S.; Speck-Planche, A. Optimizing drug discovery using multitasking models for quantitative structure-biological effect relationships: An update of the literature. Expert Opin. Drug Discov. 2023, 18, 1231–1243. [Google Scholar] [CrossRef]
- Velasquez-Lopez, Y.; Ruiz-Escudero, A.; Arrasate, S.; Gonzalez-Diaz, H. Implementation of IFPTML Computational Models in Drug Discovery Against Flaviviridae Family. J. Chem. Inf. Model. 2024, 64, 1841–1852. [Google Scholar] [CrossRef]
- Santiago, C.; Ortega-Tenezaca, B.; Barbolla, I.; Fundora-Ortiz, B.; Arrasate, S.; Dea-Ayuela, M.A.; Gonzalez-Diaz, H.; Sotomayor, N.; Lete, E. Prediction of Antileishmanial Compounds: General Model, Preparation, and Evaluation of 2-Acylpyrrole Derivatives. J. Chem. Inf. Model. 2022, 62, 3928–3940. [Google Scholar] [CrossRef] [PubMed]
- Dieguez-Santana, K.; Casanola-Martin, G.M.; Torres, R.; Rasulev, B.; Green, J.R.; Gonzalez-Diaz, H. Machine Learning Study of Metabolic Networks vs ChEMBL Data of Antibacterial Compounds. Mol. Pharm. 2022, 19, 2151–2163. [Google Scholar] [CrossRef]
- Barbolla, I.; Hernandez-Suarez, L.; Quevedo-Tumailli, V.; Nocedo-Mena, D.; Arrasate, S.; Dea-Ayuela, M.A.; Gonzalez-Diaz, H.; Sotomayor, N.; Lete, E. Palladium-mediated synthesis and biological evaluation of C-10b substituted Dihydropyrrolo[1,2-b]isoquinolines as antileishmanial agents. Eur. J. Med. Chem. 2021, 220, 113458. [Google Scholar] [CrossRef]
- Vasquez-Dominguez, E.; Armijos-Jaramillo, V.D.; Tejera, E.; Gonzalez-Diaz, H. Multioutput Perturbation-Theory Machine Learning (PTML) Model of ChEMBL Data for Antiretroviral Compounds. Mol. Pharm. 2019, 16, 4200–4212. [Google Scholar] [CrossRef]
- Speck-Planche, A.; Kleandrova, V.V. Multi-Condition QSAR Model for the Virtual Design of Chemicals with Dual Pan-Antiviral and Anti-Cytokine Storm Profiles. ACS Omega 2022, 7, 32119–32130. [Google Scholar] [CrossRef]
- Baltasar-Marchueta, M.; Llona, L.; M-Alicante, S.; Barbolla, I.; Ibarluzea, M.G.; Ramis, R.; Salomon, A.M.; Fundora, B.; Araujo, A.; Muguruza-Montero, A.; et al. Identification of Riluzole derivatives as novel calmodulin inhibitors with neuroprotective activity by a joint synthesis, biosensor, and computational guided strategy. Biomed. Pharmacother. 2024, 174, 116602. [Google Scholar] [CrossRef]
- Sampaio-Dias, I.E.; Rodriguez-Borges, J.E.; Yanez-Perez, V.; Arrasate, S.; Llorente, J.; Brea, J.M.; Bediaga, H.; Vina, D.; Loza, M.I.; Caamano, O.; et al. Synthesis, Pharmacological, and Biological Evaluation of 2-Furoyl-Based MIF-1 Peptidomimetics and the Development of a General-Purpose Model for Allosteric Modulators (ALLOPTML). ACS Chem. Neurosci. 2021, 12, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Diez-Alarcia, R.; Yanez-Perez, V.; Muneta-Arrate, I.; Arrasate, S.; Lete, E.; Meana, J.J.; Gonzalez-Diaz, H. Big Data Challenges Targeting Proteins in GPCR Signaling Pathways; Combining PTML-ChEMBL Models and [(35)S]GTPgammaS Binding Assays. ACS Chem. Neurosci. 2019, 10, 4476–4491. [Google Scholar] [CrossRef]
- Ferreira da Costa, J.; Silva, D.; Caamano, O.; Brea, J.M.; Loza, M.I.; Munteanu, C.R.; Pazos, A.; Garcia-Mera, X.; Gonzalez-Diaz, H. Perturbation Theory/Machine Learning Model of ChEMBL Data for Dopamine Targets: Docking, Synthesis, and Assay of New l-Prolyl-l-leucyl-glycinamide Peptidomimetics. ACS Chem. Neurosci. 2018, 9, 2572–2587. [Google Scholar] [CrossRef] [PubMed]
- Kleandrova, V.V.; Speck-Planche, A. PTML Modeling for Alzheimer’s Disease: Design and Prediction of Virtual Multi-Target Inhibitors of GSK3B, HDAC1, and HDAC6. Curr. Top. Med. Chem. 2020, 20, 1661–1676. [Google Scholar] [CrossRef] [PubMed]
- Tenorio-Borroto, E.; Castanedo, N.; Garcia-Mera, X.; Rivadeneira, K.; Vazquez Chagoyan, J.C.; Barbabosa Pliego, A.; Munteanu, C.R.; Gonzalez-Diaz, H. Perturbation Theory Machine Learning Modeling of Immunotoxicity for Drugs Targeting Inflammatory Cytokines and Study of the Antimicrobial G1 Using Cytometric Bead Arrays. Chem. Res. Toxicol. 2019, 32, 1811–1823. [Google Scholar] [CrossRef]
- Vazquez-Prieto, S.; Paniagua, E.; Solana, H.; Ubeira, F.M.; Gonzalez-Diaz, H. A study of the Immune Epitope Database for some fungi species using network topological indices. Mol. Divers. 2017, 21, 713–718. [Google Scholar] [CrossRef]
- Martinez-Arzate, S.G.; Tenorio-Borroto, E.; Barbabosa Pliego, A.; Diaz-Albiter, H.M.; Vazquez-Chagoyan, J.C.; Gonzalez-Diaz, H. PTML Model for Proteome Mining of B-Cell Epitopes and Theoretical-Experimental Study of Bm86 Protein Sequences from Colima, Mexico. J. Proteome Res. 2017, 16, 4093–4103. [Google Scholar] [CrossRef]
- Santana, R.; Zuluaga, R.; Ganan, P.; Arrasate, S.; Onieva, E.; Montemore, M.M.; Gonzalez-Diaz, H. PTML Model for Selection of Nanoparticles, Anticancer Drugs, and Vitamins in the Design of Drug-Vitamin Nanoparticle Release Systems for Cancer Cotherapy. Mol. Pharm. 2020, 17, 2612–2627. [Google Scholar] [CrossRef]
- Santana, R.; Zuluaga, R.; Ganan, P.; Arrasate, S.; Onieva, E.; Gonzalez-Diaz, H. Predicting coated-nanoparticle drug release systems with perturbation-theory machine learning (PTML) models. Nanoscale 2020, 12, 13471–13483. [Google Scholar] [CrossRef]
- Urista, D.V.; Carrue, D.B.; Otero, I.; Arrasate, S.; Quevedo-Tumailli, V.F.; Gestal, M.; Gonzalez-Diaz, H.; Munteanu, C.R. Prediction of Antimalarial Drug-Decorated Nanoparticle Delivery Systems with Random Forest Models. Biology 2020, 9, 198. [Google Scholar] [CrossRef] [PubMed]
- Munteanu, C.R.; Gutierrez-Asorey, P.; Blanes-Rodriguez, M.; Hidalgo-Delgado, I.; Blanco Liverio, M.J.; Castineiras Galdo, B.; Porto-Pazos, A.B.; Gestal, M.; Arrasate, S.; Gonzalez-Diaz, H. Prediction of Anti-Glioblastoma Drug-Decorated Nanoparticle Delivery Systems Using Molecular Descriptors and Machine Learning. Int. J. Mol. Sci. 2021, 22, 11519. [Google Scholar] [CrossRef]
- Cabrera-Andrade, A.; Lopez-Cortes, A.; Munteanu, C.R.; Pazos, A.; Perez-Castillo, Y.; Tejera, E.; Arrasate, S.; Gonzalez-Diaz, H. Perturbation-Theory Machine Learning (PTML) Multilabel Model of the ChEMBL Dataset of Preclinical Assays for Antisarcoma Compounds. ACS Omega 2020, 5, 27211–27220. [Google Scholar] [CrossRef]
- Cabrera-Andrade, A.; Lopez-Cortes, A.; Jaramillo-Koupermann, G.; Gonzalez-Diaz, H.; Pazos, A.; Munteanu, C.R.; Perez-Castillo, Y.; Tejera, E. A Multi-Objective Approach for Anti-Osteosarcoma Cancer Agents Discovery through Drug Repurposing. Pharmaceuticals 2020, 13, 409. [Google Scholar] [CrossRef]
- Bediaga, H.; Arrasate, S.; Gonzalez-Diaz, H. PTML Combinatorial Model of ChEMBL Compounds Assays for Multiple Types of Cancer. ACS Comb. Sci. 2018, 20, 621–632. [Google Scholar] [CrossRef]
- Kleandrova, V.V.; Cordeiro, M.N.D.S.; Speck-Planche, A. Perturbation Theory Machine Learning Model for Phenotypic Early Antineoplastic Drug Discovery: Design of Virtual Anti-Lung-Cancer Agents. Appl. Sci. 2024, 14, 9344. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Felix, E.; Magarinos, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [PubMed]
- Mok, N.Y.; Brenk, R. Mining the ChEMBL database: An efficient chemoinformatics workflow for assembling an ion channel-focused screening library. J. Chem. Inf. Model. 2011, 51, 2449–2454. [Google Scholar] [CrossRef]
- Quadros, H.C.; Herrmann, L.; Manaranche, J.; Paloque, L.; Borges-Silva, M.C.; Dziwornu, G.A.; D’Alessandro, S.; Chibale, K.; Basilico, N.; Benoit-Vical, F.; et al. Characterization of antimalarial activity of artemisinin-based hybrid drugs. Antimicrob. Agents Chemother. 2024, 68, e0014324. [Google Scholar] [CrossRef]
- Herrmann, L.; Leidenberger, M.; Sacramento de Morais, A.; Mai, C.; Capci, A.; da Cruz Borges Silva, M.; Plass, F.; Kahnt, A.; Moreira, D.R.M.; Kappes, B.; et al. Autofluorescent antimalarials by hybridization of artemisinin and coumarin: In vitro/in vivo studies and live-cell imaging. Chem. Sci. 2023, 14, 12941–12952. [Google Scholar] [CrossRef] [PubMed]
- Kore, M.; Acharya, D.; Sharma, L.; Vembar, S.S.; Sundriyal, S. Development and experimental validation of a machine learning model for the prediction of new antimalarials. BMC Chem. 2025, 19, 28. [Google Scholar] [CrossRef]
- Estrada, E.; Gutiérrez, Y. MODESLAB, v1.5; Santiago de Compostela, Spain. 2004. Available online: https://insilicomoleculardesign.com/software-databases/ (accessed on 10 May 2025).
- Todeschini, R.; Consonni, V. Handbook of Molecular Descriptors; WILEY-VCH Verlag GmbH: Hoboken, NJ, USA, 2000. [Google Scholar]
- Todeschini, R.; Consonni, V. (Eds.) Molecular Descriptors for Chemoinformatics; WILEY-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2009; Volumes I and II. [Google Scholar]
- Kleandrova, V.V.; Cordeiro, M.N.D.S.; Speck-Planche, A. In Silico Approach for Antibacterial Discovery: PTML Modeling of Virtual Multi-Strain Inhibitors Against Staphylococcus aureus. Pharmaceuticals 2025, 18, 196. [Google Scholar] [CrossRef] [PubMed]
- Urias, R.W.; Barigye, S.J.; Marrero-Ponce, Y.; Garcia-Jacas, C.R.; Valdes-Martini, J.R.; Perez-Gimenez, F. IMMAN: Free software for information theory-based chemometric analysis. Mol. Divers. 2015, 19, 305–319. [Google Scholar] [CrossRef]
- Stahura, F.L.; Godden, J.W.; Bajorath, J. Differential Shannon entropy analysis identifies molecular property descriptors that predict aqueous solubility of synthetic compounds with high accuracy in binary QSAR calculations. J. Chem. Inf. Comput. Sci. 2002, 42, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef]
- Press, W.H.; Flannery, B.P.; Teukolsky, S.A.; Vetterling, W.T. Numerical Recipes in C: The art of Scientific Computing, 1st ed.; Cambridge University Press: New York, NY, USA, 1988. [Google Scholar]
- STATISTICA (Data Analysis Software System); v13.5.0.17; TIBCO-Software-Inc.: Palo Alto, CA, USA, 2018.
- Schneider, G.; Wrede, P. Artificial neural networks for computer-based molecular design. Prog. Biophys. Mol. Biol. 1998, 70, 175–222. [Google Scholar] [CrossRef]
- Manallack, D.T.; Livingstone, D.J.; A-Razzak, M.; Glen, R.C. Neural Networks and Expert Systems in Molecular Design. In Advanced Computer––Assisted Techniques in Drug Discovery; van de Waterbeemd, H., Ed.; Methods and Principles in Medicinal Chemistry; VCH Verlagsgesellschaft mbH: Weinheim, Germany, 1994; pp. 293–331. [Google Scholar]
- Chicco, D.; Jurman, G. The Matthews correlation coefficient (MCC) should replace the ROC AUC as the standard metric for assessing binary classification. BioData Min. 2023, 16, 4. [Google Scholar] [CrossRef]
- Rende, M.; Pistilli, A.; Stabile, A.M.; Terenzi, A.; Cattaneo, A.; Ugolini, G.; Sanna, P. Role of nerve growth factor and its receptors in non-nervous cancer growth: Efficacy of a tyrosine kinase inhibitor (AG879) and neutralizing antibodies antityrosine kinase receptor A and antinerve growth factor: An in-vitro and in-vivo study. Anti-Cancer Drugs 2006, 17, 929–941. [Google Scholar] [CrossRef]
- He, H.; Yumiko, H.; Aviv, G.; Yoshihiro, Y.; Hiroyuki, M.; Yuko, K.; Toshiaki, K.; Ching-Yi, H.; Hsing-Jien, K.; Guillaume, L.; et al. The Tyr-Kinase Inhibitor AG879, that Blocks the ETK-PAK1 Interaction, Suppresses the RAS-induced PAK1 Activation and Malignant Transformation. Cancer Biol. Ther. 2004, 3, 96–101. [Google Scholar] [CrossRef]
- Zhou, X.; Lin, H.; Lin, H. Global Sensitivity Analysis. In Encyclopedia of GIS; Shekhar, S., Xiong, H., Eds.; Springer: Boston, MA, USA, 2008; pp. 408–409. [Google Scholar]
- Estrada, E. Spectral moments of the edge adjacency matrix in molecular graphs. 1. Definition and applications for the prediction of physical properties of alkanes. J. Chem. Inf. Comput. Sci. 1996, 36, 844–849. [Google Scholar] [CrossRef]
- Estrada, E. Spectral moments of the edge adjacency matrix in molecular graphs. 2. Molecules containing heteroatoms and QSAR applications. J. Chem. Inf. Comput. Sci. 1997, 37, 320–328. [Google Scholar] [CrossRef]
- Estrada, E. Spectral moments of the edge adjacency matrix in molecular graphs. 3. Molecules containing cycles. J. Chem. Inf. Comput. Sci. 1998, 38, 23–27. [Google Scholar] [CrossRef]
- Estrada, E. How the parts organize in the whole? A top-down view of molecular descriptors and properties for QSAR and drug design. Mini Rev. Med. Chem. 2008, 8, 213–221. [Google Scholar] [CrossRef]
- Estrada, E.; Patlewicz, G.; Gutierrez, Y. From knowledge generation to knowledge archive. A general strategy using TOPS-MODE with DEREK to formulate new alerts for skin sensitization. J. Chem. Inf. Comput. Sci. 2004, 44, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Estrada, E.; Molina, E. Automatic extraction of structural alerts for predicting chromosome aberrations of organic compounds. J. Mol. Graph. Model. 2006, 25, 275–288. [Google Scholar] [CrossRef]
- Estrada, E. Physicochemical Interpretation of Molecular Connectivity Indices. J. Phys. Chem. A 2002, 106, 9085–9091. [Google Scholar] [CrossRef]
- Kier, L.B.; Murray, W.J.; Hall, L.H. Molecular connectivity. 4. Relationships to biological activities. J. Med. Chem. 1975, 18, 1272–1274. [Google Scholar] [CrossRef]
- Kier, L.B.; Hall, L.H. Molecular connectivity VII: Specific treatment of heteroatoms. J. Pharm. Sci. 1976, 65, 1806–1809. [Google Scholar] [CrossRef]
- Hall, L.H.; Kier, L.B. Structure-activity studies using valence molecular connectivity. J. Pharm. Sci. 1977, 66, 642–644. [Google Scholar] [CrossRef]
- Kier, L.B.; Hall, L.H. Derivation and significance of valence molecular connectivity. J. Pharm. Sci. 1981, 70, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Kier, L.B.; Hall, L.H. Intermolecular accessibility: The meaning of molecular connectivity. J. Chem. Inf. Comput. Sci. 2000, 40, 792–795. [Google Scholar] [CrossRef]
- Kier, L.B.; Hall, L.H. Molecular connectivity: Intermolecular accessibility and encounter simulation. J. Mol. Graph. Model. 2001, 20, 76–83. [Google Scholar] [CrossRef]
- Estrada, E. Edge adjacency relationship and a novel topological index related to molecular volume. J. Chem. Inf. Comput. Sci. 1995, 35, 31–33. [Google Scholar] [CrossRef]
- Estrada, E. Edge adjacency relationships in molecular graphs containing heteroatoms: A new topological index related to molar volume. J. Chem. Inf. Comput. Sci. 1995, 35, 701–707. [Google Scholar] [CrossRef]
- Estrada, E.; Rodríguez, L. Edge-Connectivity Indices in QSPR/QSAR Studies. 1. Comparison to Other Topological Indices in QSPR Studies. J. Chem. Inf. Comput. Sci. 1999, 39, 1037–1041. [Google Scholar] [CrossRef]
- Estrada, E. Edge-Connectivity Indices in QSPR/QSAR Studies. 2. Accounting for Long-Range Bond Contributions. J. Chem. Inf. Comput. Sci. 1999, 39, 1042–1048. [Google Scholar] [CrossRef]
- Estrada, E.; Guevara, N.; Gutman, I. Extension of Edge Connectivity Index. Relationships to Line Graph Indices and QSPR Applications. J. Chem. Inf. Comput. Sci. 1998, 38, 428–431. [Google Scholar] [CrossRef]
- Amrane, D.; Gellis, A.; Hutter, S.; Prieri, M.; Verhaeghe, P.; Azas, N.; Vanelle, P.; Primas, N. Synthesis and Antiplasmodial Evaluation of 4-Carboxamido- and 4-Alkoxy-2-Trichloromethyl Quinazolines. Molecules 2020, 25, 3929. [Google Scholar] [CrossRef]
- Desroches, J.; Kieffer, C.; Primas, N.; Hutter, S.; Gellis, A.; El-Kashef, H.; Rathelot, P.; Verhaeghe, P.; Azas, N.; Vanelle, P. Discovery of new hit-molecules targeting Plasmodium falciparum through a global SAR study of the 4-substituted-2-trichloromethylquinazoline antiplasmodial scaffold. Eur. J. Med. Chem. 2017, 125, 68–86. [Google Scholar] [CrossRef]
- Amrane, D.; Primas, N.; Arnold, C.S.; Hutter, S.; Louis, B.; Sanz-Serrano, J.; Azqueta, A.; Amanzougaghene, N.; Tajeri, S.; Mazier, D.; et al. Antiplasmodial 2-thiophenoxy-3-trichloromethyl quinoxalines target the apicoplast of Plasmodium falciparum. Eur. J. Med. Chem. 2021, 224, 113722. [Google Scholar] [CrossRef] [PubMed]
- Overington, J. ChEMBL. An interview with John Overington, team leader, chemogenomics at the European Bioinformatics Institute Outstation of the European Molecular Biology Laboratory (EMBL-EBI). Interview by Wendy A. Warr. J. Comput. Aided Mol. Des. 2009, 23, 195–198. [Google Scholar] [CrossRef]
- Irwin, J.J.; Shoichet, B.K. ZINC—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Hersey, A.; Chambers, J.; Bellis, L.; Patricia Bento, A.; Gaulton, A.; Overington, J.P. Chemical databases: Curation or integration by user-defined equivalence? Drug Discov. Today Technol. 2015, 14, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Maggiora, G.; Vogt, M.; Stumpfe, D.; Bajorath, J. Molecular similarity in medicinal chemistry. J. Med. Chem. 2014, 57, 3186–3204. [Google Scholar] [CrossRef]
- Martin, Y.C.; Kofron, J.L.; Traphagen, L.M. Do structurally similar molecules have similar biological activity? J. Med. Chem. 2002, 45, 4350–4358. [Google Scholar] [CrossRef]
- Mauri, A. alvaDesc: A Tool to Calculate and Analyze Molecular Descriptors and Fingerprints. In Ecotoxicological QSARs; Roy, K., Ed.; Springer: New York, NY, USA, 2020; pp. 801–820. [Google Scholar]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Dong, J.; Wang, N.N.; Yao, Z.J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.P.; Cao, D.S. ADMETlab: A platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminformatics 2018, 10, 29. [Google Scholar] [CrossRef]



| Codes a,b,c | Symbols | Concepts |
|---|---|---|
| DGB01 | D[SM(Mol)4]tg | Multi-label graph index derived from the bond-based spectral moment of the 4th order, weighted by atom-based molar refractivities. |
| DGB02 | D[NSM(Hyd)3]tg | Multi-label graph index derived from the normalized bond-based spectral moment of the 3rd order, weighted by atom-based hydrophobicities. |
| DGB03 | D[NSM(Mol)1]tg | Multi-label graph index derived from the normalized bond-based spectral moment of the 1st order, weighted by atom-based molar refractivities. |
| DGB04 | D[NSM(Gas)1]tg | Multi-label graph index derived from the normalized bond-based spectral moment of the 1st order, weighted by Gasteiger-Marsili atomic charges. |
| DGB05 | D[Ne(P)1]tg | Multi-label graph index derived from the normalized bond-based connectivity of the 1st order, containing only path subgraphs. |
| DGB06 | D[Ne(P)2]tg | Multi-label graph index derived from the normalized bond-based connectivity of the 2nd order, containing only path subgraphs. |
| DGB07 | D[Ne(P)6]tg | Multi-label graph index derived from the normalized bond-based connectivity of the 6th order, containing only path subgraphs. |
| DGB08 | D[Ne(Ch)6]tg | Multi-label graph index derived from the normalized bond-based connectivity of the 6th order, containing only cycle (ring) subgraphs. |
| DGB09 | D[SM(Psa)4]ds | Multi-label graph index derived from the bond-based spectral moment of the 4th order, weighted by atom-based polar surface areas. |
| DGB10 | D[SM(Ato)7]ds | Multi-label graph index derived from the bond-based spectral moment of the 7th order, weighted by atomic weights. |
| DGB11 | D[Xv(Ch)5]ds | Multi-label graph index derived from the atom-based valence connectivity of the 5th order, containing only cycle (ring) subgraphs. |
| DGB12 | D[Xv(PC)6]ds | Multi-label graph index derived from the atom-based valence connectivity of the 6th order, containing only path-cluster subgraphs. |
| DGB13 | D[e(C)4]ds | Multi-label graph index derived from the bond-based connectivity of the 4th order, containing only cluster subgraphs. |
| DGB14 | D[NSM(Std)1]ds | Multi-label graph index derived from the normalized bond-based spectral moment of the 1st order, weighted by the standard bond distances. |
| DGB15 | D[NSM(Dip)1]ds | Multi-label graph index derived from the normalized bond-based spectral moment of the 1st order, weighted by the bond dipole moments. |
| DGB16 | D[NSM(Dip)7]ds | Multi-label graph index derived from the normalized bond-based spectral moment of the 7th order, weighted by the bond dipole moments. |
| DGB17 | D[NSM(Hyd)1]ds | Multi-label graph index derived from the normalized bond-based spectral moment of the 1st order, weighted by atom-based hydrophobicities. |
| DGB18 | D[NSM(Psa)1]ds | Multi-label graph index derived from the normalized bond-based spectral moment of the 1st order, weighted by atom-based polar surface areas. |
| DGB19 | D[NSM(Ato)1]ds | Multi-label graph index derived from the normalized bond-based spectral moment of the 1st order, weighted by atomic weights. |
| DGB20 | D[NXv(P)3]ds | Multi-label graph index derived from the normalized atom-based valence connectivity of the 3rd order, containing only path subgraphs. |
| DGB21 | D[NXv(C)3]ds | Multi-label graph index derived from the normalized atom-based valence connectivity of the 3rd order, containing only cluster subgraphs. |
| DGB22 | D[NXv(Ch)6]ds | Multi-label graph index derived from the normalized atom-based valence connectivity of the 6th order, containing only cycle (ring) subgraphs. |
| DGB23 | D[Ne(C)6]ds | Multi-label graph index derived from the normalized bond-based connectivity of the 6th order, containing only cluster subgraphs. |
| DGB24 | D[Ne(PC)6]ds | Multi-label graph index derived from the normalized bond-based connectivity of the 6th order, containing only path-cluster subgraphs. |
| DGB25 | D[NK(Alpha)3]ds | Multi-label graph index derived from the normalized alpha-modified shape descriptor of the 3rd order, containing only path subgraphs. |
| SYMBOLS a | Training Set | Test Set |
|---|---|---|
| NActive | 3613 | 1204 |
| TP | 3393 | 1074 |
| Sn | 93.91% | 89.20% |
| NInactive | 3584 | 1194 |
| TN | 3260 | 1029 |
| Sp | 90.96% | 86.18% |
| nMCC | 0.925 | 0.877 |
| Codes a | Average Values | Tendency b | |
|---|---|---|---|
| Active | Inactive | ||
| DGB01 | 8.220 × 10−3 | −1.054 × 10−1 | Increase |
| DGB02 | 1.195 × 10−2 | −5.387 × 10−2 | Increase |
| DGB03 | −2.959 × 10−4 | −1.455 × 10−2 | Increase |
| DGB04 | 5.718 × 10−3 | −6.431 × 10−2 | Increase |
| DGB05 | −9.300 × 10−3 | 1.488 × 10−1 | Decrease |
| DGB06 | 3.953 × 10−3 | −2.272 × 10−1 | Increase |
| DGB07 | 6.967 × 10−3 | −2.938 × 10−1 | Increase |
| DGB08 | −7.765 × 10−3 | −8.953 × 10−2 | Increase |
| DGB09 | 3.773 × 10−3 | 1.589 × 10−1 | Decrease |
| DGB10 | 1.687 × 10−4 | 1.049 × 10−1 | Decrease |
| DGB11 | −4.437 × 10−4 | 2.400 × 10−2 | Decrease |
| DGB12 | 4.613 × 10−3 | 1.039 × 10−2 | Decrease |
| DGB13 | 1.253 × 10−2 | −4.819 × 10−2 | Increase |
| DGB14 | 2.450 × 10−4 | −1.427 × 10−1 | Increase |
| DGB15 | −1.374 × 10−3 | 2.750 × 10−1 | Decrease |
| DGB16 | 2.155 × 10−3 | 1.896 × 10−1 | Decrease |
| DGB17 | 4.187 × 10−3 | −1.435 × 10−1 | Increase |
| DGB18 | 5.651 × 10−5 | 2.241 × 10−1 | Decrease |
| DGB19 | 8.620 × 10−3 | 1.171 × 10−1 | Decrease |
| DGB20 | 2.873 × 10−3 | 4.588 × 10−2 | Decrease |
| DGB21 | 4.693 × 10−3 | 1.153 × 10−1 | Decrease |
| DGB22 | 4.674 × 10−3 | −1.728 × 10−1 | Increase |
| DGB23 | 2.511 × 10−3 | 1.447 × 10−1 | Decrease |
| DGB24 | 1.987 × 10−3 | 2.057 × 10−3 | Decrease |
| DGB25 | 1.196 × 10−3 | 8.020 × 10−2 | Decrease |
| tga | dsb | ProbAct (%) c,d | |||||
|---|---|---|---|---|---|---|---|
| VASP-01 | VASP-02 | VASP-03 | VASP-04 | VASP-05 | VASP-06 | ||
| P. falciparum (7G8) | Drug-resistant | 56.83 | 44.11 | 46.14 | 74.24 | 73.70 | 74.24 |
| P. falciparum (Dd2) | Drug-resistant | 56.91 | 45.83 | 49.81 | 82.26 | 81.00 | 82.26 |
| P. falciparum (D6) | Drug-sensitive | 64.45 | 59.43 | 65.36 | 83.91 | 82.58 | 83.91 |
| P. falciparum (3D7) | Drug-sensitive | 64.38 | 49.00 | 57.75 | 82.21 | 81.12 | 82.21 |
| P. falciparum (W2) | Drug-resistant | 66.42 | 38.16 | 45.59 | 80.33 | 79.08 | 80.33 |
| P. falciparum (D10) | Drug-sensitive | 69.34 | 44.91 | 51.04 | 74.30 | 72.86 | 74.30 |
| P. falciparum (FCB) | Drug-resistant | 61.36 | 87.06 | 87.14 | 64.44 | 66.67 | 64.44 |
| P. falciparum (K1) | Drug-resistant | 69.60 | 41.70 | 47.34 | 78.69 | 77.99 | 78.69 |
| P. falciparum (NF54) | Drug-sensitive | 66.98 | 51.01 | 55.71 | 79.98 | 79.18 | 79.98 |
| ID | Physicochemical Properties a | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| MW | TNA | NRB | HBD | HBA | MR | PSA | MLOGP | ALOGP | |
| VASP-01 | 417.70 | 38 | 3 | 1 | 5 | 106.26 | 80.24 | 2.9041 | 2.8515 |
| VASP-02 | 368.35 | 38 | 3 | 1 | 8 | 91.760 | 80.24 | 2.568 | 2.2614 |
| VASP-03 | 384.35 | 39 | 4 | 1 | 9 | 93.361 | 89.47 | 1.8088 | 3.439 |
| VASP-04 | 440.81 | 43 | 3 | 1 | 4 | 113.17 | 92.65 | 3.6301 | 3.4394 |
| VASP-05 | 440.81 | 43 | 3 | 1 | 4 | 112.80 | 92.65 | 3.6301 | 3.8679 |
| VASP-06 | 440.81 | 43 | 3 | 1 | 4 | 113.17 | 92.65 | 3.6301 | 3.4394 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleandrova, V.V.; Cordeiro, M.N.D.S.; Speck-Planche, A. In Silico Approach for Early Antimalarial Drug Discovery: De Novo Design of Virtual Multi-Strain Antiplasmodial Inhibitors. Microorganisms 2025, 13, 1620. https://doi.org/10.3390/microorganisms13071620
Kleandrova VV, Cordeiro MNDS, Speck-Planche A. In Silico Approach for Early Antimalarial Drug Discovery: De Novo Design of Virtual Multi-Strain Antiplasmodial Inhibitors. Microorganisms. 2025; 13(7):1620. https://doi.org/10.3390/microorganisms13071620
Chicago/Turabian StyleKleandrova, Valeria V., M. Natália D. S. Cordeiro, and Alejandro Speck-Planche. 2025. "In Silico Approach for Early Antimalarial Drug Discovery: De Novo Design of Virtual Multi-Strain Antiplasmodial Inhibitors" Microorganisms 13, no. 7: 1620. https://doi.org/10.3390/microorganisms13071620
APA StyleKleandrova, V. V., Cordeiro, M. N. D. S., & Speck-Planche, A. (2025). In Silico Approach for Early Antimalarial Drug Discovery: De Novo Design of Virtual Multi-Strain Antiplasmodial Inhibitors. Microorganisms, 13(7), 1620. https://doi.org/10.3390/microorganisms13071620









