The Impact of the Variability of RT-qPCR Standard Curves on Reliable Viral Detection in Wastewater Surveillance
Abstract
1. Introduction
2. Materials and Methods
2.1. RT-qPCR Standard Curve Reactions
2.2. Setting of Thresholds
2.3. Data Processing
2.4. Statistical Distributions Fitting
3. Results
3.1. RT-qPCR Standard Curves Parameters
3.2. RT-qPCR Standard Curves Efficiencies
3.3. RT-qPCR Standard Curves Variability
3.4. Distribution Fitting
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef]
- Schmidt, P.J.; Acosta, N.; Chik, A.H.S.; D’Aoust, P.M.; Delatolla, R.; Dhiyebi, H.A.; Glier, M.B.; Hubert, C.R.J.; Kopetzky, J.; Mangat, C.S.; et al. Realizing the value in “non-standard” parts of the qPCR standard curve by integrating fundamentals of quantitative microbiology. Front. Microbiol. 2023, 14, 1048661. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Niesters, H.G.M. Quantitation of Viral Load Using Real-Time Amplification Techniques. Methods 2001, 25, 419–429. [Google Scholar] [CrossRef]
- Mackay, I.M. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 2004, 10, 190–212. [Google Scholar] [CrossRef]
- Ryncarz, A.J.; Goddard, J.; Wald, A.; Huang, M.-L.; Roizman, B.; Corey, L. Development of a High-Throughput Quantitative Assay for Detecting Herpes Simplex Virus DNA in Clinical Samples. J. Clin. Microbiol. 1999, 37, 1941–1947. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union One Health 2022 Zoonoses Report. EFS2 2023, 21, e8442. [Google Scholar] [CrossRef]
- Hashemi, M.; Salayani, M.; Afshari, A.; Kafil, H.S.; Noori, S.M.A. The Global Burden of Viral Food-borne Diseases: A Systematic Review. Curr. Pharm. Biotechnol. 2023, 24, 1657–1672. [Google Scholar] [CrossRef]
- Thomas, M.K.; Murray, R.; Flockhart, L.; Pintar, K.; Pollari, F.; Fazil, A.; Nesbitt, A.; Marshall, B. Estimates of the Burden of Foodborne Illness in Canada for 30 Specified Pathogens and Unspecified Agents, Circa 2006. Foodborne Pathog. Dis. 2013, 10, 639–648. [Google Scholar] [CrossRef]
- Bosch, A.; Gkogka, E.; Le Guyader, F.S.; Loisy-Hamon, F.; Lee, A.; Van Lieshout, L.; Marthi, B.; Myrmel, M.; Sansom, A.; Schultz, A.C.; et al. Foodborne viruses: Detection, risk assessment, and control options in food processing. Int. J. Food Microbiol. 2018, 285, 110–128. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Taybeh, A.O.; Al-Nabulsi, A.; Al-Holy, M.; Hatmal, M.M.; Alzyoud, J.; Aolymat, I.; Abughoush, M.H.; Shahbaz, H.; Alzyoud, A.; et al. Common and Potential Emerging Foodborne Viruses: A Comprehensive Review. Life 2024, 14, 190. [Google Scholar] [CrossRef] [PubMed]
- Torretta, S.; Zuccotti, G.; Cristofaro, V.; Ettori, J.; Solimeno, L.; Battilocchi, L.; D’Onghia, A.; Bonsembiante, A.; Pignataro, L.; Marchisio, P.; et al. Diagnosis of SARS-CoV-2 by RT-PCR Using Different Sample Sources: Review of the Literature. Ear Nose Throat J. 2021, 100, 131S–138S. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO COVID-19 Dashboard 2024. Available online: https://data.who.int/dashboards/covid19/deaths (accessed on 20 March 2025).
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Karrer, E.E.; Lincoln, J.E.; Hogenhout, S.; Bennett, A.B.; Bostock, R.M.; Martineau, B.; Lucas, W.J.; Gilchrist, D.G.; Alexander, D. In situ isolation of mRNA from individual plant cells: Creation of cell-specific cDNA libraries. Proc. Natl. Acad. Sci. USA 1995, 92, 3814–3818. [Google Scholar] [CrossRef]
- Reiter, M.; Pfaffl, M.W. Effects of Plate Position, Plate Type and Sealing Systems on Real-Time PCR Results. Biotechnol. Biotechnol. Equip. 2008, 22, 824–828. [Google Scholar] [CrossRef][Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Ståhlberg, A.; Håkansson, J.; Xian, X.; Semb, H.; Kubista, M. Properties of the Reverse Transcription Reaction in mRNA Quantification. Clin. Chem. 2004, 50, 509–515. [Google Scholar] [CrossRef]
- Gunson, R.; Collins, T.; Carman, W. Practical experience of high throughput real time PCR in the routine diagnostic virology setting. J. Clin. Virol. 2006, 35, 355–367. [Google Scholar] [CrossRef]
- Rutledge, R.G.; Stewart, D. A kinetic-based sigmoidal model for the polymerase chain reaction and its application to high-capacity absolute quantitative real-time PCR. BMC Biotechnol. 2008, 8, 47. [Google Scholar] [CrossRef]
- Shanks, O.C.; Atikovic, E.; Blackwood, A.D.; Lu, J.; Noble, R.T.; Domingo, J.S.; Seifring, S.; Sivaganesan, M.; Haugland, R.A. Quantitative PCR for Detection and Enumeration of Genetic Markers of Bovine Fecal Pollution. Appl. Environ. Microbiol. 2008, 74, 745–752. [Google Scholar] [CrossRef]
- Ahmed, W.; Bivins, A.; Bertsch, P.M.; Bibby, K.; Choi, P.M.; Farkas, K.; Gyawali, P.; Hamilton, K.A.; Haramoto, E.; Kitajima, M.; et al. Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimization and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health 2020, 17, 82–93. [Google Scholar] [CrossRef]
- Bivins, A.; Kaya, D.; Bibby, K.; Simpson, S.L.; Bustin, S.A.; Shanks, O.C.; Ahmed, W. Variability in RT-qPCR assay parameters indicates unreliable SARS-CoV-2 RNA quantification for wastewater surveillance. Water Res. 2021, 203, 117516. [Google Scholar] [CrossRef] [PubMed]
- McCluskey, K.; Jarret, R.L. (Eds.) The Biological Resources of Model Organisms; CRC Press: Boca Raton, FL, USA, 2020; ISBN 978-1-315-10099-9. [Google Scholar]
- Rogers-Broadway, K.-R.; Karteris, E. Amplification efficiency and thermal stability of qPCR instrumentation: Current landscape and future perspectives. Exp. Ther. Med. 2015, 10, 1261–1264. [Google Scholar] [CrossRef]
- Muenchhoff, M.; Mairhofer, H.; Nitschko, H.; Grzimek-Koschewa, N.; Hoffmann, D.; Berger, A.; Rabenau, H.; Widera, M.; Ackermann, N.; Konrad, R.; et al. Multicentre comparison of quantitative PCR-based assays to detect SARS-CoV-2, Germany, March 2020. Eurosurveillance 2020, 25, 2001057. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cataluña, A.; Cuevas-Ferrando, E.; Randazzo, W.; Falcó, I.; Allende, A.; Sánchez, G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021, 758, 143870. [Google Scholar] [CrossRef]
- Sahoo, S.; Mandal, S.; Das, P.; Bhattacharya, S.; Chandy, M. An analysis of the standard curve parameters of cytomegalovirus, BK virus and hepatitis B virus quantitative polymerase chain reaction from a clinical virology laboratory in eastern India. Indian J. Med. Microbiol. 2022, 40, 81–85. [Google Scholar] [CrossRef]
- Lindén, J.; Ranta, J.; Pohjanvirta, R. Bayesian modeling of reproducibility and robustness of RNA reverse transcription and quantitative real-time polymerase chain reaction. Anal. Biochem. 2012, 428, 81–91. [Google Scholar] [CrossRef]
- Schwaber, J.; Andersen, S.; Nielsen, L. Shedding light: The importance of reverse transcription efficiency standards in data interpretation. Biomol. Detect. Quantif. 2019, 17, 100077. [Google Scholar] [CrossRef]
- CDC. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. Centers for Disease Control and Prevention. 2020. Available online: https://www.fda.gov/media/134922/download (accessed on 20 March 2025).
- ISO 15216-1:2017; Microbiology of the Food Chain—Horizontal Method for Determination of Hepatitis A Virus and Norovirus Using Real-Time RT-PCR—Part 1: Method for Quantification. ISO: Geneva, Switzerland, 2017.
- Sano, D.; Pintó, R.M.; Omura, T.; Bosch, A. Detection of Oxidative Damages on Viral Capsid Protein for Evaluating Structural Integrity and Infectivity of Human Norovirus. Environ. Sci. Technol. 2010, 44, 808–812. [Google Scholar] [CrossRef]
- Jothikumar, N.; Cromeans, T.L.; Robertson, B.H.; Meng, X.J.; Hill, V.R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 2006, 131, 65–71. [Google Scholar] [CrossRef]
- Jothikumar, N.; Kang, G.; Hill, V.R. Broadly reactive TaqMan® assay for real-time RT-PCR detection of rotavirus in clinical and environmental samples. J. Virol. Methods 2009, 155, 126–131. [Google Scholar] [CrossRef]
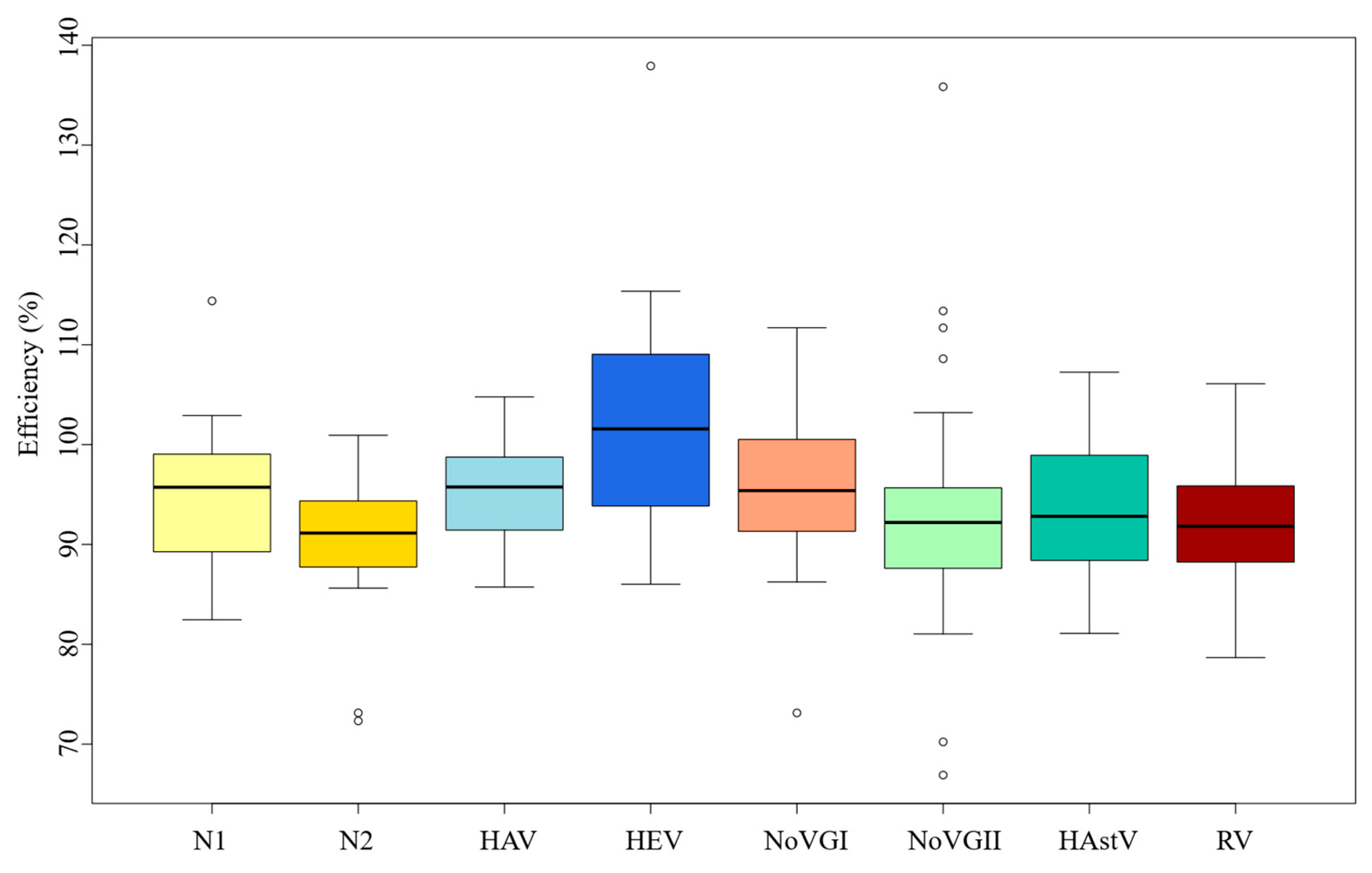
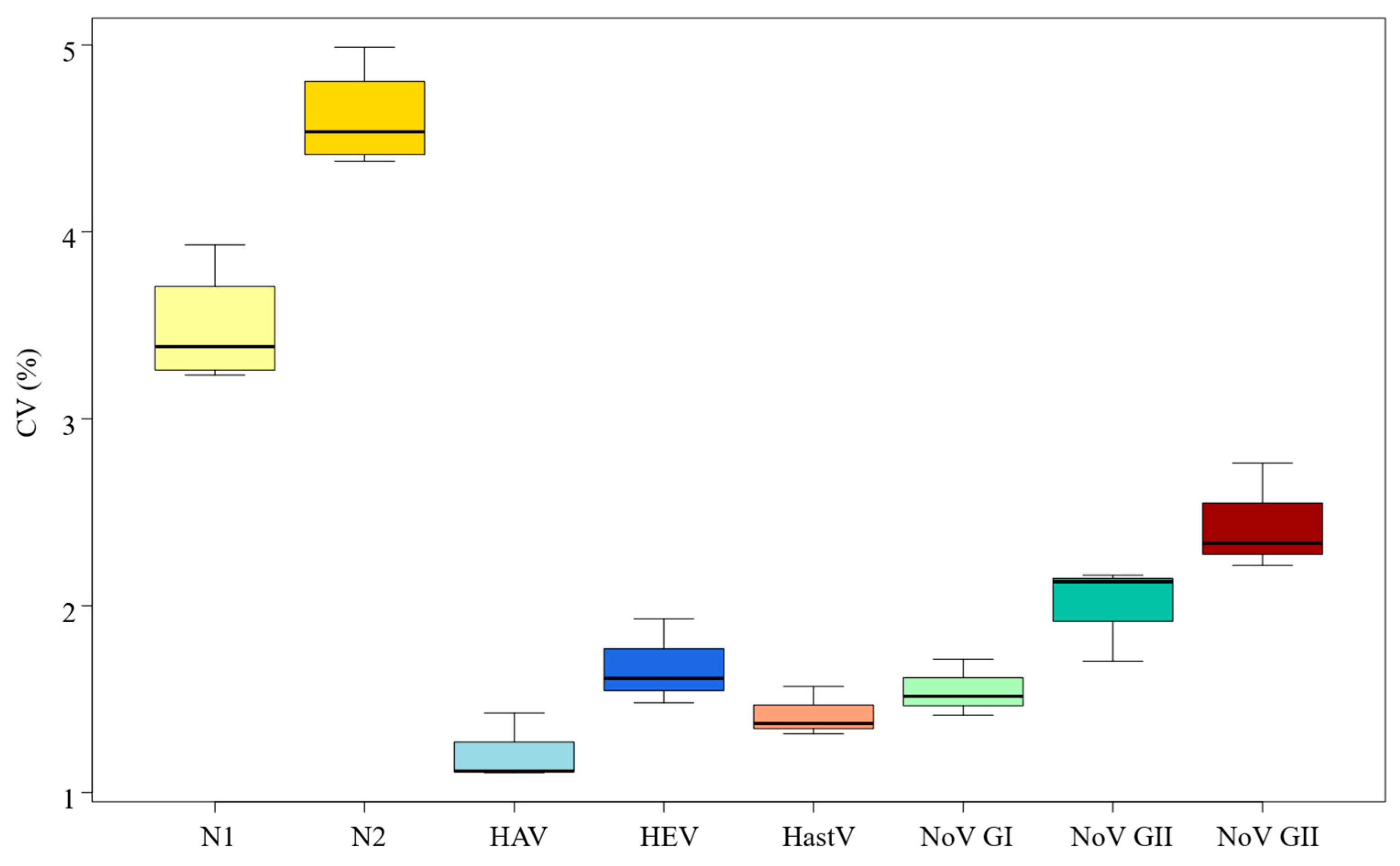
| Virus | Log gc/Reaction | CT Mean | SD | Min | Max | CV (%) |
|---|---|---|---|---|---|---|
| SARS-CoV-2 (N1 gene) | 4.40 | 21.50 | 0.84 | 19.73 | 22.91 | 3.93% |
| 3.40 | 24.97 | 0.87 | 23.44 | 26.37 | 3.49% | |
| 2.40 | 28.44 | 0.93 | 27.15 | 30.07 | 3.29% | |
| 1.40 | 31.91 | 1.03 | 30.17 | 33.76 | 3.23% | |
| SARS-CoV-2 (N2 gene) | 4.40 | 21.36 | 1.07 | 20.13 | 23.43 | 4.99% |
| 3.40 | 24.94 | 1.15 | 23.64 | 26.96 | 4.62% | |
| 2.40 | 28.51 | 1.27 | 27.07 | 30.76 | 4.45% | |
| 1.40 | 32.08 | 1.40 | 30.45 | 34.99 | 4.38% | |
| Hepatitis A | 4.14 | 22.86 | 0.33 | 21.91 | 23.55 | 1.43% |
| 3.14 | 26.31 | 0.29 | 25.34 | 26.87 | 1.11% | |
| 2.14 | 29.76 | 0.33 | 28.77 | 30.43 | 1.11% | |
| Hepatitis E | 3.13 | 27.45 | 0.41 | 26.79 | 28.38 | 1.48% |
| 2.13 | 30.74 | 0.50 | 29.64 | 32.01 | 1.61% | |
| 1.13 | 34.03 | 0.66 | 32.30 | 35.63 | 1.93% | |
| Norovirus Genogroup I | 4.14 | 25.54 | 0.44 | 24.50 | 26.59 | 1.71% |
| 3.14 | 28.99 | 0.41 | 28.12 | 29.85 | 1.41% | |
| 2.14 | 32.44 | 0.49 | 31.61 | 33.28 | 1.51% | |
| Norovirus Genogroup II | 4.14 | 21.81 | 0.46 | 21.02 | 22.92 | 2.13% |
| 3.14 | 25.35 | 0.43 | 24.48 | 26.24 | 1.70% | |
| 2.14 | 28.89 | 0.62 | 27.79 | 30.41 | 2.16% | |
| Human Astrovirus | 4.14 | 23.12 | 0.32 | 22.49 | 23.99 | 1.37% |
| 3.14 | 26.62 | 0.35 | 25.87 | 27.55 | 1.31% | |
| 2.14 | 30.13 | 0.47 | 29.09 | 31.11 | 1.57% | |
| Rotavirus | 3.40 | 20.99 | 0.58 | 19.56 | 21.79 | 2.76% |
| 2.40 | 24.53 | 0.57 | 23.10 | 25.35 | 2.33% | |
| 1.40 | 28.07 | 0.62 | 26.29 | 29.14 | 2.22% |
| Distribution | Virus | Log gc/Reaction | Min | Max | Perc 2.5th | Perc 97.5th |
|---|---|---|---|---|---|---|
| Uniform | N1 SARS-CoV-2 | 1.40 | 30.17 | 33.76 | 30.35 | 33.58 |
| 2.40 | 27.15 | 30.07 | 27.30 | 29.92 | ||
| 3.40 | 23.44 | 26.37 | 23.59 | 26.22 | ||
| 4.40 | 19.73 | 22.91 | 19.89 | 22.75 | ||
| N2 SARS-CoV-2 | 1.40 | 30.45 | 34.99 | 30.68 | 34.76 | |
| 2.40 | 27.07 | 30.76 | 27.25 | 30.57 | ||
| 3.40 | 23.64 | 26.96 | 26.79 | 23.81 | ||
| 4.40 | 20.13 | 23.43 | 20.29 | 23.27 | ||
| NoVGI | 2.40 | 31.61 | 33.28 | 31.70 | 33.20 | |
| Weibull | Virus | Log gc/reaction | Shape (S.E.) | Scale (S.E.) | Perc 2.5th | Perc 97.5th |
| HAV | 2.40 | 105.76 (14.24) | 29.91 (0.05) | 29.08 | 30.22 | |
| 3.40 | 109.98 (14.82) | 26.44 (0.05) | 25.74 | 26.71 | ||
| 4.40 | 80.63 (10.79) | 23.01 (0.06) | 22.18 | 23.32 | ||
| RV | 1.40 | 59.86 (8.33) | 28.33 (0.09) | 26.96 | 28.86 | |
| 2.40 | 61.10 (8.87) | 24.77 (0.08) | 23.59 | 25.22 | ||
| 3.40 | 51.74 (7.74) | 21.24 (0.08) | 20.05 | 21.69 | ||
| Normal | Virus | Log gc/reaction | Mean (S.E) | S.D (S.E) | Perc 2.5th | Perc 97.5th |
| HEV | 1.13 | 34.03 (0.12) | 0.65 (0.08) | 32.97 | 35.09 | |
| 2.13 | 30.74 (0.09) | 0.49 (0.06) | 29.94 | 31.54 | ||
| 3.13 | 27.45 (0.07) | 0.40 (0.05) | 26.79 | 28.11 | ||
| NoV GII | 2.40 | 28.89 (0.11) | 0.61 (0.08) | 27.88 | 29.90 | |
| 3.40 | 25.34 (0.08) | 0.42 (0.06) | 24.65 | 26.04 | ||
| 4.40 | 21.81 (0.08) | 0.46 (0.06) | 21.06 | 22.56 | ||
| HAstV | 2.40 | 30.13 (0.09) | 0.46 (0.06) | 29.36 | 30.89 | |
| 3.40 | 26.63 (0.06) | 0.34 (0.04) | 26.06 | 27.19 | ||
| 4.40 | 23.12 (0.06) | 0.31 (0.04) | 22.61 | 23.63 | ||
| NoV GI | 3.40 | 28.99 (0.07) | 0.40 (0.05) | 28.33 | 29.65 | |
| 4.40 | 25.54 (0.08) | 0.43 (0.06) | 26.25 | 28.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casado-Martín, L.; Hernández, M.; Yeramian, N.; Pérez, D.; Eiros, J.M.; Valero, A.; Rodríguez-Lázaro, D. The Impact of the Variability of RT-qPCR Standard Curves on Reliable Viral Detection in Wastewater Surveillance. Microorganisms 2025, 13, 776. https://doi.org/10.3390/microorganisms13040776
Casado-Martín L, Hernández M, Yeramian N, Pérez D, Eiros JM, Valero A, Rodríguez-Lázaro D. The Impact of the Variability of RT-qPCR Standard Curves on Reliable Viral Detection in Wastewater Surveillance. Microorganisms. 2025; 13(4):776. https://doi.org/10.3390/microorganisms13040776
Chicago/Turabian StyleCasado-Martín, Lorena, Marta Hernández, Nadine Yeramian, Daniel Pérez, José M. Eiros, Antonio Valero, and David Rodríguez-Lázaro. 2025. "The Impact of the Variability of RT-qPCR Standard Curves on Reliable Viral Detection in Wastewater Surveillance" Microorganisms 13, no. 4: 776. https://doi.org/10.3390/microorganisms13040776
APA StyleCasado-Martín, L., Hernández, M., Yeramian, N., Pérez, D., Eiros, J. M., Valero, A., & Rodríguez-Lázaro, D. (2025). The Impact of the Variability of RT-qPCR Standard Curves on Reliable Viral Detection in Wastewater Surveillance. Microorganisms, 13(4), 776. https://doi.org/10.3390/microorganisms13040776








